Computational Analysis of Nanocarriers in the Tumor Microenvironment for the Treatment of Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. Results on Selected Samples
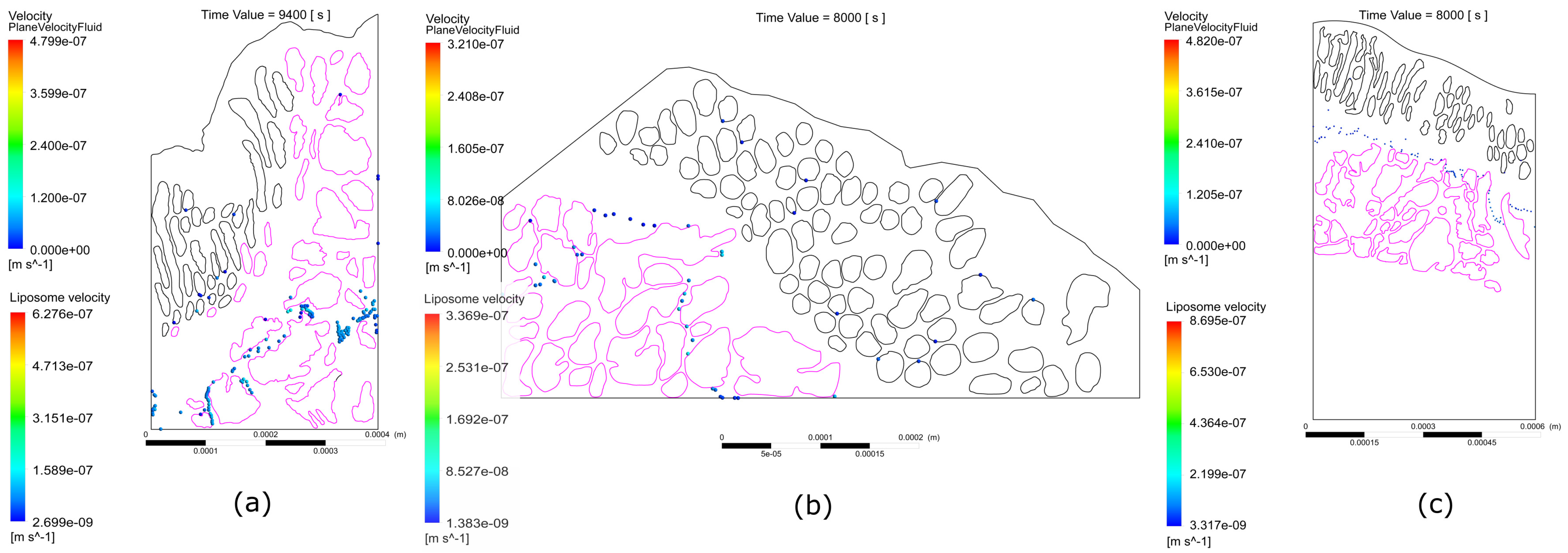
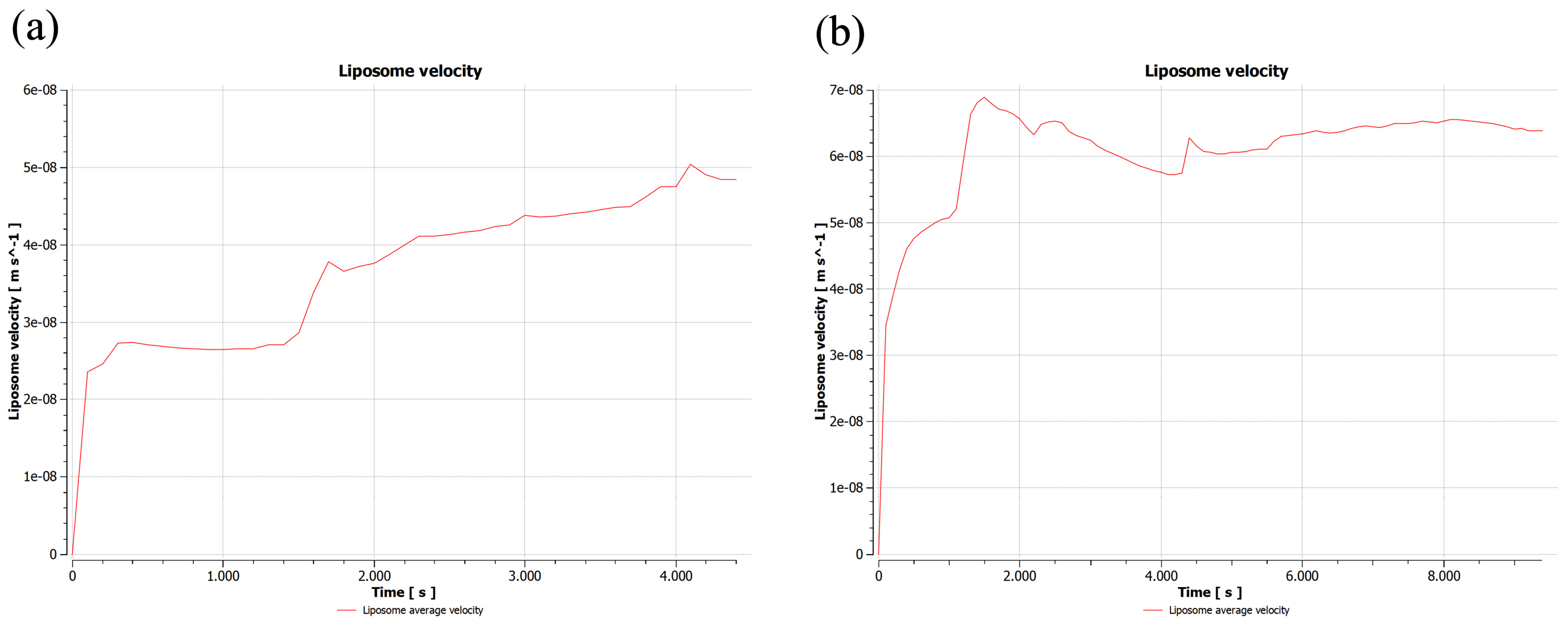
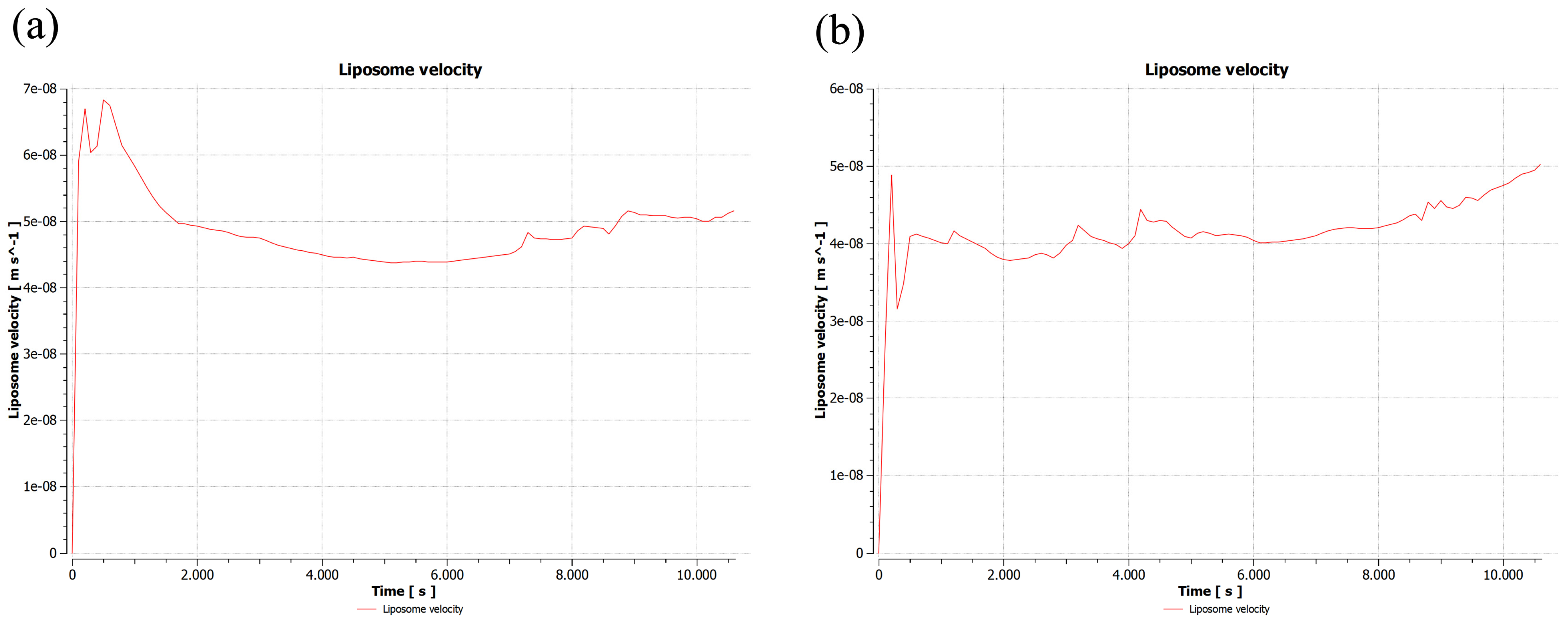
2.1.1. First Sample
2.1.2. Second Sample
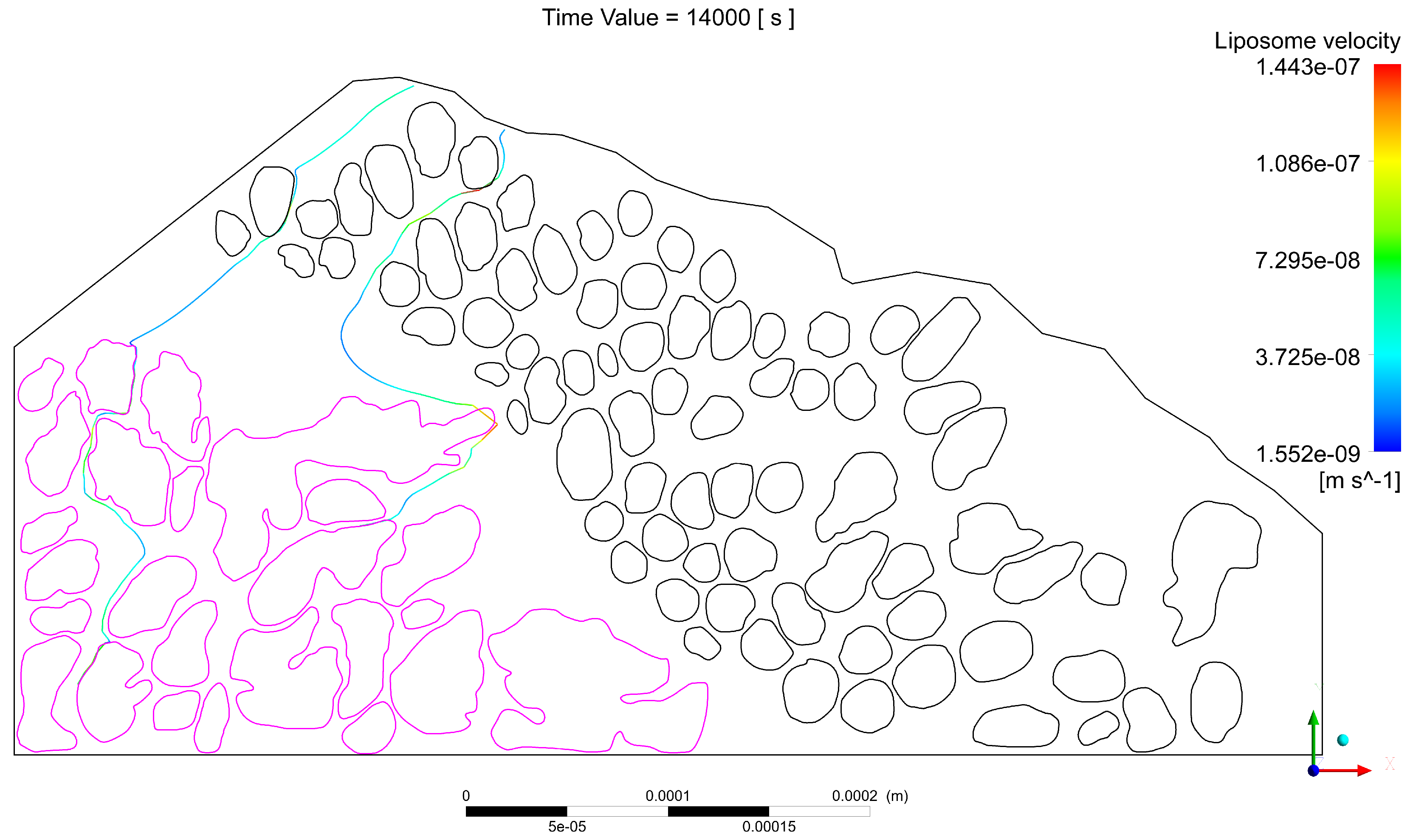
2.1.3. Third Sample
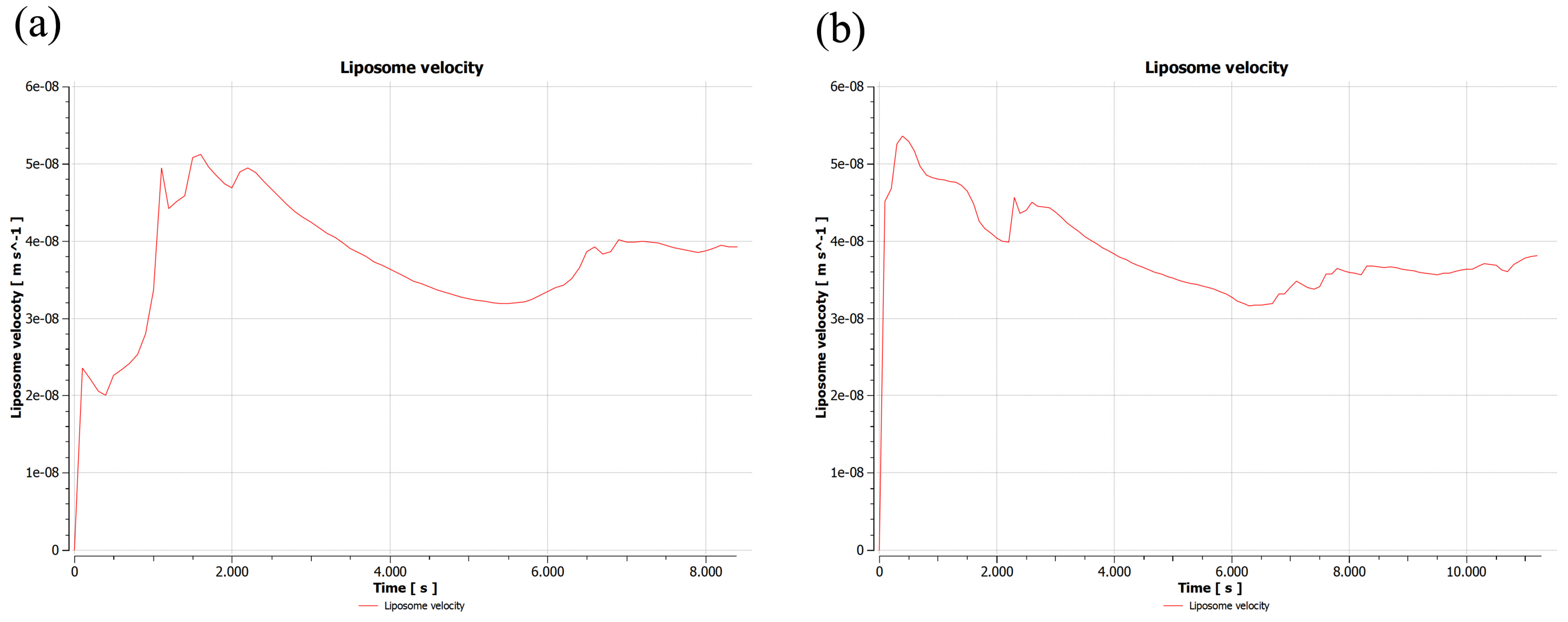
2.2. Validation of 2D Sample Results
3. Discussion
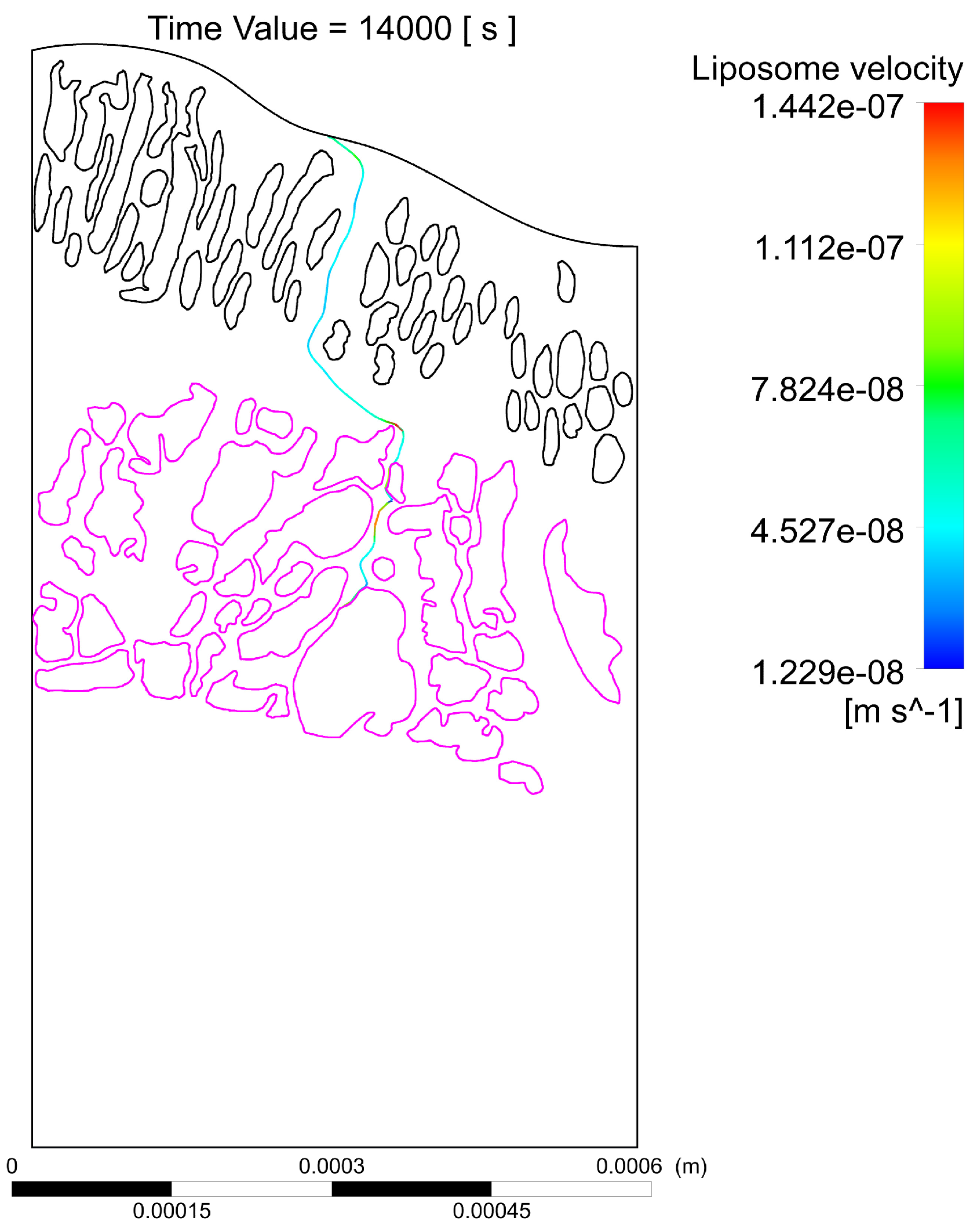
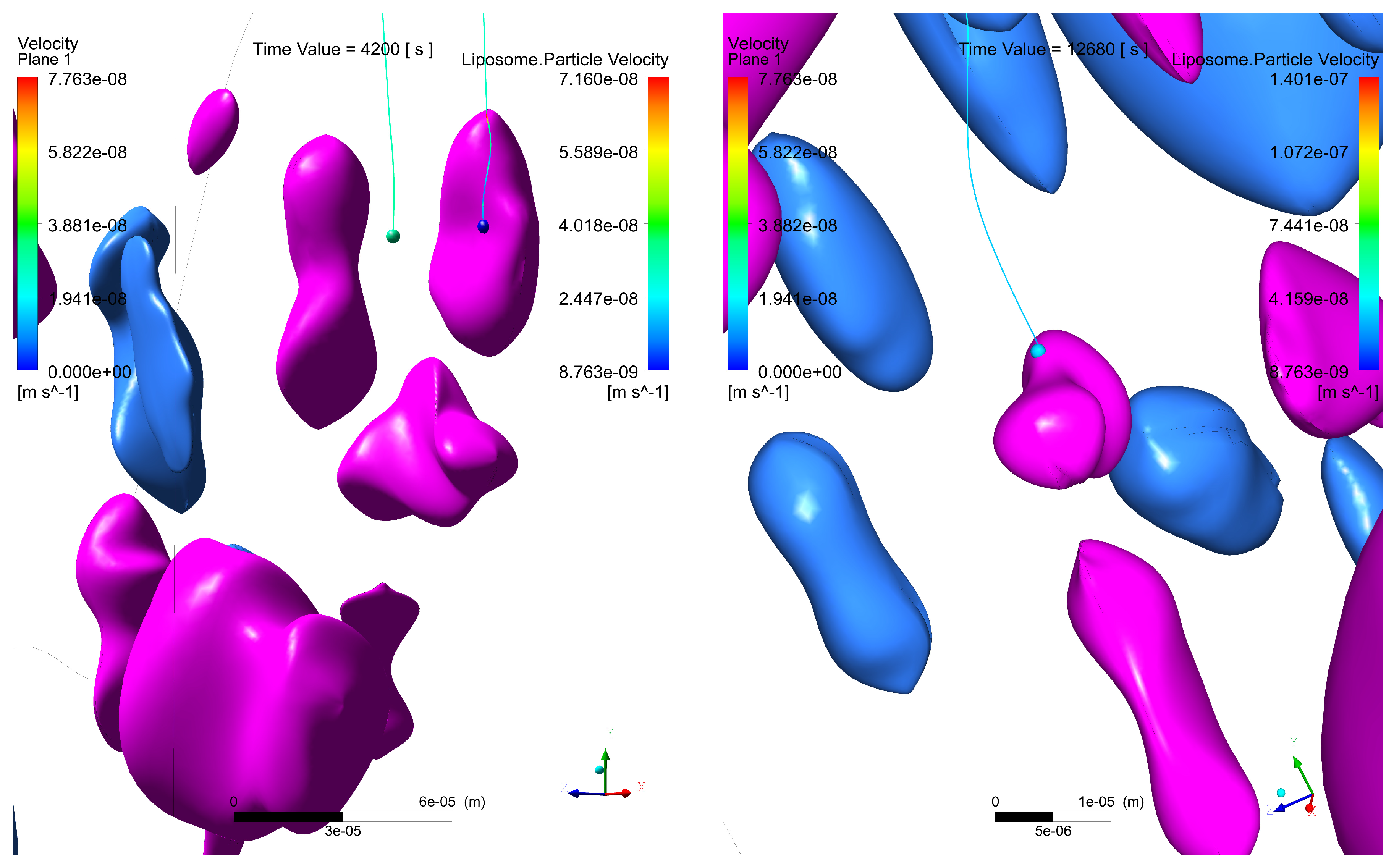
- The interstitial space between at least one pair of glands is narrow, which increases the velocity of the interstitial fluid, driving the liposomes through the streamlines at speeds sufficient to escape the electrostatic surface attraction forces of the cancerous glands;
- The interstitial space between at least one pair of glands is narrow enough so that the liposomes transiting in between, despite increasing their speed, have a trajectory close enough to the boundary layer of the gland surface, which slows down their speed and allows attractive forces to act on the liposome, increasing the probability of engagement with the cancerous region;
- A pair of glands is sufficiently separated so that the interstitial fluid does not show significant increases in velocity, which can cause liposomes to transit in regions distant from the cancerous glands, making it difficult for the two bodies to attract, and as a result, decreasing the effectiveness of the coupling;
- Even though a pair of glands is sufficiently separated, changes in velocities and trajectories produced by morphological conditions in more superficial layers can give rise to the transit of liposomes in regions with low velocities that are close to the surface of the gland. Malignant glands benefit from the attraction and subsequent coupling between positively charged liposomes and negatively charged cancerous regions, especially when the glands are found in deeper regions of the tumor and with a more acidic pH, thus increasing the negative charge.
4. Materials and Methods
- Mobility of the fluid: allows for the identification of the behavior of the fluid through the sample and its corresponding velocities;
- Maximum depth reached by the particles: used to predict the time required to achieve a complete diffusion of liposomes in a determined tumor;
- Time: The time variable is essential to estimate how long it takes for the dose that achieves contact with the surface of the cancerous tissue to break through to the maximum depth of the sample. It is also especially important to determine the time required to register the first and last coupling between a liposome and a cancerous region.
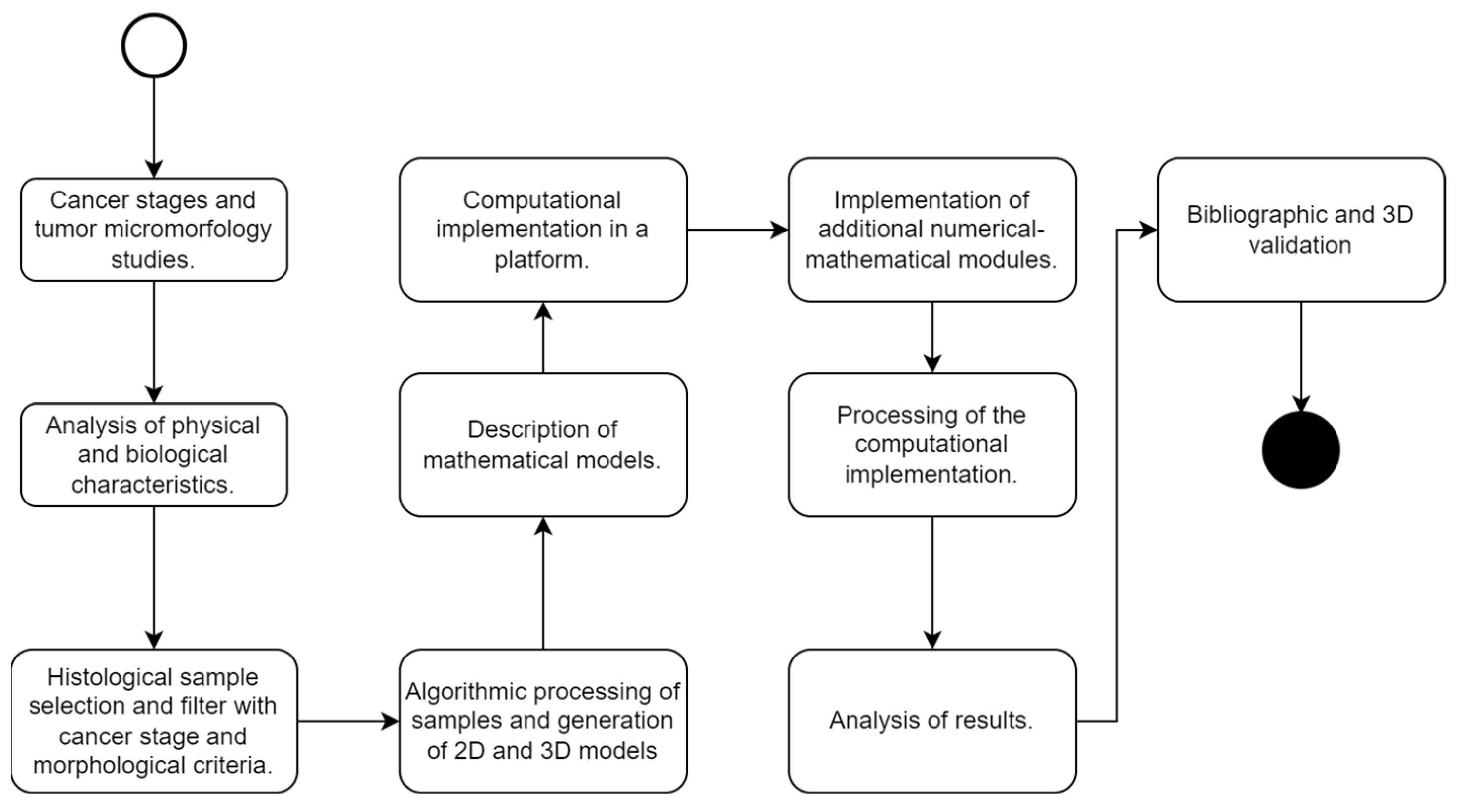
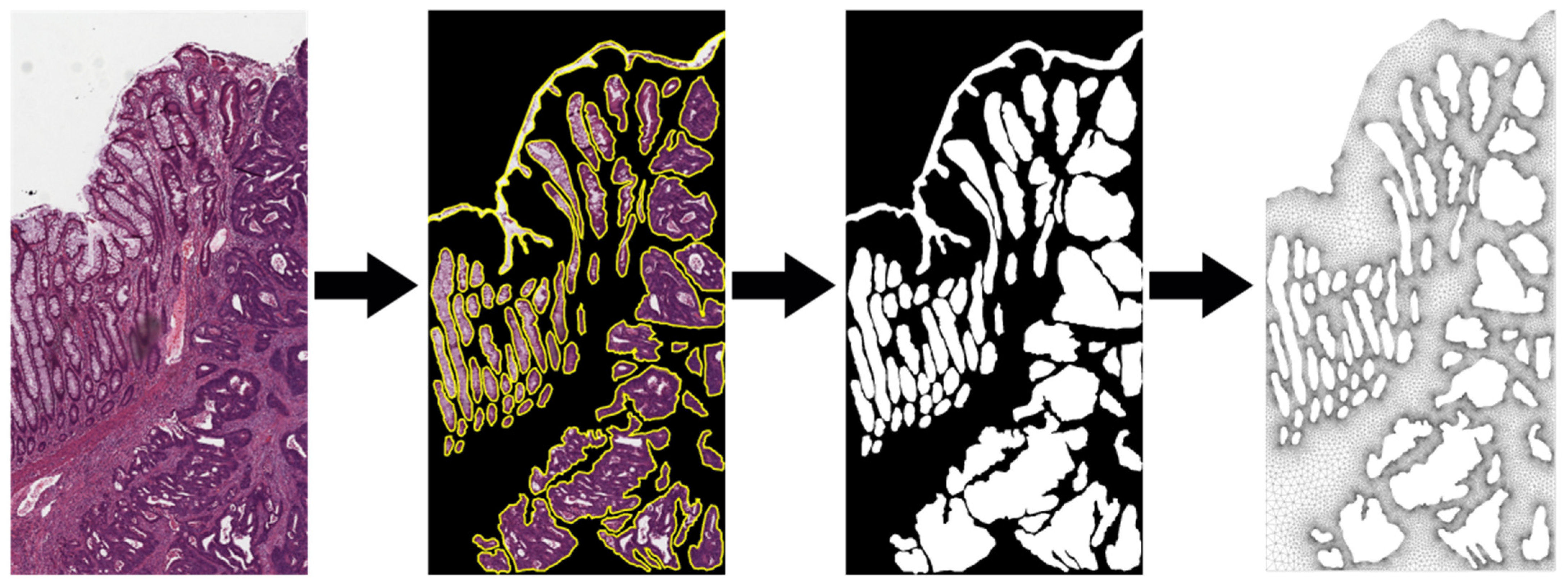
4.1. Selection and Processing of the Samples to Be Studied
- Level of glandular deformation: the identification of the regions of malignancy using the geometric shape of the cellular glands, through the characteristics of glandular aberration; glands with more asymmetric characteristics and a higher Best Alignment Metric (BAM) value tended to be classified as malignant [44];
- Stage of the disease in TNM: T (local extension of the primary tumor at the time of diagnosis); N (regional lymph node status); and M (distant metastatic disease, including non-regional lymph nodes). The selected samples must belong to a stage of the disease between stages II and III;
- Presence of benign and malignant glands in the same sample: to demonstrate the selective behavior of the nanoparticles, the histological sample must have both benign and natural cell groups, as well as malignant cell regions.
4.2. Calculation of Interstitial Fluid Mass Flow
4.3. Calculation of the Mass Flow of Liposomes
4.4. Models for Interstitial Fluid
4.5. Model for the Movement of Particles
- : drag forces acting on the particle;
- : buoyancy forces;
- : forces due to rotation;
- : Force associated with the pressure gradient. This is the force applied to the particle due to the pressure gradient in the fluid surrounding the particle caused by the acceleration of the fluid. This is only significant when the density of the fluid is comparable to or greater than the density of the particle;
- : force associated with the electrostatic interaction of the particle immersed in the electric field produced by the negative charge on the surface of the malignant cell.
- = fluid viscosity;
- = particle density;
- = particle diameter;
- Re = Reynolds number;
- = particle velocity;
- = fluid velocity;
- = drag coefficient;
- = particle mass flow;
- = timestep or step of time.
- = additional forces of interaction, such as the electrostatic force that induces selective coupling [51].
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johari, B.; Rezaeejam, H.; Moradi, M.; Taghipour, Z.; Saltanatpour, Z.; Mortazavi, Y.; Nasehi, L. Increasing the colon cancer cells sensitivity toward radiation therapy via application of Oct4–Sox2 complex decoy oligodeoxynucleotides. Mol. Biol. Rep. 2020, 47, 6793–6805. [Google Scholar] [CrossRef] [PubMed]
- Asadi, Z.; Fathi, M.; Rismani, E.; Bigdelou, Z.; Johari, B. Application of decoy oligodeoxynucleotides strategy for inhibition of cell growth and reduction of metastatic properties in nonresistant and erlotinib-resistant SW480 cell line. Cell Biol. Int. 2021, 45, 1001–1014. [Google Scholar] [CrossRef] [PubMed]
- Basave, H.N.L.; Sarmiento, I.V.; Takahashi, A.M.L.; Rosciano, A.E.P.; Molina, J.M.R.; Ruiz, G.C.; Romero, C.D.; Ruan, J.M.; Crocifoglio, V.A. Cáncer de colon y recto. In Manual de Oncología, 6th ed.; Herrera-Gómez, Á., Ñamendys-Silva, S.A., Meneses-García, A., Eds.; McGraw-Hill Education: New York, NY, USA, 2018. [Google Scholar]
- Kwaan, M.R.; Stewart Sr, D.B.; Dunn, K.B. Colon, recto y ano. In Schwartz. Principios de Cirugía, 11th ed.; Brunicardi, F.C., Andersen, D.K., Billiar, T.R., Dunn, D.L., Kao, L.S., Hunter, J.G., Matthews, J.B., Pollock, R.E., Eds.; McGraw-Hill Education: New York, NY, USA, 2020. [Google Scholar]
- Hernández, H.R.; Jacobo, S.R.; Reyes Romero, M.A. Cáncer colorrectal: Factores epidemiológicos, epigenéticos, métodos de diagnóstico y tratamiento. In Gastroenterología, 3rd ed.; Méndez-Sánchez, N., Ed.; McGraw-Hill Education: New York, NY, USA, 2018. [Google Scholar]
- Wellstein, A.; Giaccone, G.; Atkins, M.B.; Sausville, E.A. Capítulo 66: Fármacos Citotóxicos; McGraw Hill: New York, NY, USA, 2022. [Google Scholar]
- Bailly, C. Irinotecan: 25 years of cancer treatment. Pharmacol. Res. 2019, 148, 104398. [Google Scholar] [CrossRef] [PubMed]
- Wellstein, A.; Giaccone, G.; Atkins, M.B.; Sausville, E.A. Fármacos citotóxicos. In Goodman & Gilman: Las Bases Farmacológicas De La Terapéutic, 13th ed.; Brunton, L.L., Chabner, B.A., Knollmann, B.C., Eds.; McGraw-Hill Education: New York, NY, USA, 2019. [Google Scholar]
- American Cancer Society. Colorrectal Cancer. Available online: https://www.cancer.org/cancer/colon-rectal-cancer.html (accessed on 15 January 2023).
- Laroui, H.; Wilson, D.S.; Dalmasso, G.; Salaita, K.; Murthy, N.; Sitaraman, S.V.; Merlin, D. Nanomedicine in GI. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, 371–383. [Google Scholar] [CrossRef]
- Chakravarty, K.; Dalal, D.C. Mathematical modelling of liposomal drug release to tumour. Math. Biosci. 2018, 306, 82–96. [Google Scholar] [CrossRef]
- Sahai, N.; Gogoi, M.; Ahmad, N. Mathematical Modeling and Simulations for Developing Nanoparticle-Based Cancer Drug Delivery Systems: A Review. Curr. Pathobiol. Rep. 2021, 9, 1–8. [Google Scholar] [CrossRef]
- Ebrahimnejad, P.; Sodagar Taleghani, A.; Asare-Addo, K.; Nokhodchi, A. An updated review of folate-functionalized nanocarriers: A promising ligand in cancer. Drug Discov. Today 2021, 27, 471–489. [Google Scholar] [CrossRef]
- Moosmann, N.; Von Weikersthal, L.F.; Vehling-Kaiser, U.; Stauch, M.; Hass, H.G.; Dietzfelbinger, H.; Oruzio, D.; Klein, S.; Zellmann, K.; Decker, T.; et al. Cetuximab plus capecitabine and irinotecan compared with cetuximab plus capecitabine and oxaliplatin as first-line treatment for patients with metastatic colorectal cancer: AIO KRK-0104—A randomized trial of the German AIO CRC study group. J. Clin. Oncol. 2011, 29, 1050–1058. [Google Scholar] [CrossRef]
- Shah, S. Numerical Simulation of Particle Adhesion Dynamics for Applications in Nanomedicine and Biosensing; The University of Texas at Arlington: Arlington, TX, USA, 2009. [Google Scholar]
- Chandran, S.P.; Natarajan, S.B.; Chandraseharan, S.; Mohd Shahimi, M.S.B. Nano drug delivery strategy of 5-fluorouracil for the treatment of colorectal cancer. J. Cancer Res. Pract. 2017, 4, 45–48. [Google Scholar] [CrossRef]
- Hani, U.; Honnavalli, Y.K.; Begum, M.Y.; Yasmin, S.; Osmani, R.A.M.; Ansari, M.Y. Colorectal cancer: A comprehensive review based on the novel drug delivery systems approach and its management. J. Drug Deliv. Sci. Technol. 2021, 63, 102532. [Google Scholar] [CrossRef]
- Pérez-Herrero, E.; Fernández-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef] [PubMed]
- Bitounis, D.; Fanciullino, R.; Iliadis, A.; Ciccolini, J. Optimizing Druggability through Liposomal Formulations: New Approaches to an Old Concept. ISRN Pharm. 2012, 2012, 738432. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pi, C.; Feng, X.; Hou, Y.; Zhao, L.; Wei, Y. The influence of nanoparticle properties on oral bioavailability of drugs. Int. J. Nanomed. 2020, 15, 6295–6310. [Google Scholar] [CrossRef] [PubMed]
- Yamazoe, E.; Fang, J.Y.; Tahara, K. Oral mucus-penetrating PEGylated liposomes to improve drug absorption: Differences in the interaction mechanisms of a mucoadhesive liposome. Int. J. Pharm. 2021, 593, 120148. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K. Liposomes for enhanced bioavailability of water-insoluble drugs: In vivo evidence and recent approaches. Pharmaceutics 2020, 12, 264. [Google Scholar] [CrossRef]
- Hua, S. Orally administered liposomal formulations for colon targeted drug delivery. Front. Pharmacol. 2014, 5, 138. [Google Scholar] [CrossRef]
- Ying, K.; Bai, B.; Gao, X.; Xu, Y.; Wang, H.; Xie, B. Orally Administrable Therapeutic Nanoparticles for the Treatment of Colorectal Cancer. Front. Bioeng. Biotechnol. 2021, 9, 670124. [Google Scholar] [CrossRef]
- Matsuzawa, F.; Ohki, S.Y.; Aikawa, S.I.; Eto, M. Computational simulation for interactions of nano-molecules: The phospho-pivot modeling algorithm for prediction of interactions between a phospho-protein and its receptor. Sci. Technol. Adv. Mater. 2005, 6, 463–467. [Google Scholar] [CrossRef]
- Yang, W.; Lai, L. Computational design of ligand-binding proteins. Curr. Opin. Struct. Biol. 2017, 45, 67–68. [Google Scholar] [CrossRef]
- Kang, G.; Márquez, C.; Barat, A.; Byrne, A.T.; Prehn, J.H.M.; Sorribes, J.; César, E. Colorectal tumour simulation using agent based modelling and high performance computing. Futur. Gener. Comput. Syst. 2017, 67, 397–408. [Google Scholar] [CrossRef]
- Mustapha, K.; Gilli, Q.; Frayret, J.M.; Lahrichi, N.; Karimi, E. Agent-based Simulation Patient Model for Colon and Colorectal Cancer Care Trajectory. Procedia Comput. Sci. 2016, 100, 188–197. [Google Scholar] [CrossRef]
- Bethge, A.; Schumacher, U.; Wedemann, G. Simulation of metastatic progression using a computer model including chemotherapy and radiation therapy. J. Biomed. Inform. 2015, 57, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Wedemann, G.; Bethge, A.; Haustein, V.; Schumacher, U. Computer simulation of the metastatic progression. In Methods in Molecular Biology; Dwek, M., Schumacher, U., Brooks, S.A., Eds.; Springer: New York, NY, USA, 2014; Volume 1070, pp. 107–116. [Google Scholar]
- Mollazadeh, S.; Sahebkar, A.; Shahlaei, M.; Moradi, S. Nano drug delivery systems: Molecular dynamic simulation. J. Mol. Liq. 2021, 332, 115823. [Google Scholar] [CrossRef]
- Dhanavel, S.; Praveena, P.; Narayanan, V.; Stephen, A. Chitosan/reduced graphene oxide/Pd nanocomposites for co-delivery of 5-fluorouracil and curcumin towards HT-29 colon cancer cells. Polym. Bull. 2020, 77, 5681–5696. [Google Scholar] [CrossRef]
- Osorio, M.; Martinez, E.; Naranjo, T.; Castro, C. Recent advances in polymer nanomaterials for drug delivery of adjuvants in colorectal cancer treatment: A scientific-technological analysis and review. Molecules 2020, 25, 2270. [Google Scholar] [CrossRef]
- Soltani, M.; Souri, M.; Moradi Kashkooli, F. Effects of hypoxia and nanocarrier size on pH-responsive nano-delivery system to solid tumors. Sci. Rep. 2021, 11, 19350. [Google Scholar] [CrossRef]
- Wang, J.; Shi, X. Molecular dynamics simulation of diffusion of nanoparticles in mucus. Acta Mech. Solida Sin. 2017, 30, 241–247. [Google Scholar] [CrossRef]
- Rendón, J.P.; Cañas, A.I.; Correa, E.; Bedoya-Betancur, V.; Osorio, M.; Castro, C.; Naranjo, T.W. Evaluation of the Effects of Genistein In Vitro as a Chemopreventive Agent for Colorectal Cancer—Strategy to Improve Its Efficiency When Administered Orally. Molecules 2022, 27, 7042. [Google Scholar] [CrossRef]
- Sohrabi, S.; Zheng, J.; Finol, E.A.; Liu, Y. Numerical Simulation of Particle Transport and Deposition in the Pulmonary Vasculature. J. Biomech. Eng. 2014, 136, 121010. [Google Scholar] [CrossRef]
- Longest, P.W.; Xi, J. Effectiveness of direct Lagrangian tracking models for simulating nanoparticle deposition in the upper airways. Aerosol Sci. Technol. 2007, 41, 380–397. [Google Scholar] [CrossRef]
- Rahman, M.M.; Zhao, M.; Islam, M.S.; Dong, K.; Saha, S.C. Numerical study of nanoscale and microscale particle transport in realistic lung models with and without stenosis. Int. J. Multiph. Flow 2021, 145, 103842. [Google Scholar] [CrossRef]
- Graham, S.; Chen, H.; Gamper, J.; Dou, Q.; Heng, P.A.; Snead, D.; Tsang, Y.W.; Rajpoot, N. MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Med. Image Anal. 2019, 52, 199–211. [Google Scholar] [CrossRef]
- Kather, J.N.; Weis, C.A.; Bianconi, F.; Melchers, S.M.; Schad, L.R.; Gaiser, T.; Marx, A.; Zöllner, F.G. Multi-class texture analysis in colorectal cancer histology. Sci. Rep. 2016, 6, 27988. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Zöllner, F.G.; Bianconi, F.; Melchers, S.M.; Schad, L.R.; Gaiser, T.; Marx, A.; Weis, C.-A. Collection of textures in colorectal cancer histology. Zenodo 2016. [Google Scholar] [CrossRef]
- Kather, J.N.; Halama, N.; Marx, A. 100,000 histological images of human colorectal cancer and healthy tissue. Zenodo 2018. [Google Scholar] [CrossRef]
- Awan, R.; Sirinukunwattana, K.; Epstein, D.; Jefferyes, S.; Qidwai, U.; Aftab, Z.; Mujeeb, I.; Snead, D.; Rajpoot, N. Glandular Morphometrics for Objective Grading of Colorectal Adenocarcinoma Histology Images. Sci. Rep. 2017, 7, 2220–2243. [Google Scholar] [CrossRef]
- Derwinger, K.; Kodeda, K.; Bexe-Lindskog, E.; Taflin, H. Tumour differentiation grade is associated with TNM staging and the risk of node metastasis in colorectal cancer. Acta Oncol. 2010, 49, 57–62. [Google Scholar] [CrossRef]
- Helander, H.F.; Fändriks, L. Surface area of the digestive tract-revisited. Scand. J. Gastroenterol. 2014, 49, 681–689. [Google Scholar] [CrossRef]
- Devadason, D.; Goldman, D.A. 90—Management of Diarrhea. In Pediatric Gastrointestinal and Liver Disease, 6th ed.; Wyllie, R., Hyams, J.S., Kay, M., Eds.; Elsevier: Philadelphia, PA, USA, 2021; pp. 1012–1022.e3. [Google Scholar]
- Yao, W.; Shen, Z.; Ding, G. Simulation of interstitial fluid flow in ligaments: Comparison among Stokes, Brinkman and Darcy models. Int. J. Biol. Sci. 2013, 9, 1050–1056. [Google Scholar] [CrossRef]
- Pedersen, J.A.; Boschetti, F.; Swartz, M.A. Effects of extracellular fiber architecture on cell membrane shear stress in a 3D fibrous matrix. J. Biomech. 2007, 40, 1484–1492. [Google Scholar] [CrossRef]
- Deshpande, S.; Birnie, A.; Dekker, C. On-chip density-based purification of liposomes. Biomicrofluidics 2017, 11, 034106. [Google Scholar] [CrossRef] [PubMed]
- ANSYS Inc. ANSYS CFX-Solver Theory Guide; ANSYS Inc.: Canonsburg, PA, USA, 2019. [Google Scholar]
- Peters, M.J.; Stinstra, J.G.; Leveles, I. The Electrical Conductivity of Living Tissue: A Parameter in the Bioelectrical Inverse Problem. In Modeling and Imaging of Bioelectrical Activity: Principles and Applications; Springer: New York, NY, USA, 2004; pp. 281–319. [Google Scholar] [CrossRef]
- Chan, K.L.; Gascoyne, P.R.; Becker, F.F.; Pethig, R. Electrorotation of liposomes: Verification of dielectric multi-shell model for cells. Bone 1997, 1349, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Riedl, S.; Leber, R.; Rinner, B.; Schaider, H.; Lohner, K.; Zweytick, D. Human lactoferricin derived di-peptides deploying loop structures induce apoptosis specifically in cancer cells through targeting membranous phosphatidylserine. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2918–2931. [Google Scholar] [CrossRef] [PubMed]
- Dan, N. Effect of liposome charge and PEG polymer layer thickness on cell-liposome electrostatic interactions. Biochim. Biophys. Acta Biomembr. 2002, 1564, 343–348. [Google Scholar] [CrossRef]



| Description | Units | Symbol |
|---|---|---|
| Fluid mobility | m/s | |
| Vectorized deposition efficiency | % | |
| Maximum depth reached by the particles | m | d |
| Time | s | t |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, E.V.; Guerrero, G.S.; Hoyos Palacio, L.M.; Maday, Y. Computational Analysis of Nanocarriers in the Tumor Microenvironment for the Treatment of Colorectal Cancer. Appl. Sci. 2023, 13, 6248. https://doi.org/10.3390/app13106248
Morales EV, Guerrero GS, Hoyos Palacio LM, Maday Y. Computational Analysis of Nanocarriers in the Tumor Microenvironment for the Treatment of Colorectal Cancer. Applied Sciences. 2023; 13(10):6248. https://doi.org/10.3390/app13106248
Chicago/Turabian StyleMorales, Esteban Vallejo, Gustavo Suárez Guerrero, Lina M. Hoyos Palacio, and Yvon Maday. 2023. "Computational Analysis of Nanocarriers in the Tumor Microenvironment for the Treatment of Colorectal Cancer" Applied Sciences 13, no. 10: 6248. https://doi.org/10.3390/app13106248
APA StyleMorales, E. V., Guerrero, G. S., Hoyos Palacio, L. M., & Maday, Y. (2023). Computational Analysis of Nanocarriers in the Tumor Microenvironment for the Treatment of Colorectal Cancer. Applied Sciences, 13(10), 6248. https://doi.org/10.3390/app13106248






