Abstract
This study aims at developing a clinically oriented automated diagnostic tool for distinguishing malignant melanocytic lesions from benign melanocytic nevi in diverse image databases. Due to the presence of artifacts, smooth lesion boundaries, and subtlety in diagnostic features, the accuracy of such systems gets hampered. Thus, the proposed framework improves the accuracy of melanoma detection by combining the clinical aspects of dermoscopy. Two methods have been adopted for achieving the aforementioned objective. Firstly, artifact removal and lesion localization are performed. In the second step, various clinically significant features such as shape, color, texture, and pigment network are detected. Features are further reduced by checking their individual significance (i.e., hypothesis testing). These reduced feature vectors are then classified using SVM classifier. Features specific to the domain have been used for this design as opposed to features of the abstract images. The domain knowledge of an expert gets enhanced by this methodology. The proposed approach is implemented on a multi-source dataset (PH2 + ISBI 2016 and 2017) of 515 annotated images, thereby resulting in sensitivity, specificity and accuracy of 83.8%, 88.3%, and 86%, respectively. The experimental results are promising, and can be applied to detect asymmetry, pigment network, colors, and texture of the lesions.
Keywords:
color; classification; dermoscopy; hair shafts; melanoma; segmentation; shape; texture; pigment network 1. Introduction
1.1. Motivation
Melanoma is one of the worst forms of skin cancer resulting in increased morbidity and huge medical expenditure almost up to $3.3 billion [1]. Although, simple observation aids in detection of the changes in the melanocytic nevi, the deadly disease can spread to other parts of the body by metastasizing. Skin tumor thickness mainly determines the spread of the disease. Melanoma prognosis is inversely proportional to the tumor thickness, since the survival rate relies on the tumor thickness. The greater the thickness, the lesser the survival rate. However, to measure the spread and thickness of the tumor biopsy is required, which is a painful experience to the patient. Additionally, careful observation of the melanoma characteristics can be performed, as the lesion is visible on the skin. However, it is further liable to metastasize and spread to lymph nodes thus incrementing the level of malignancy. According to the Fitzpatrick Skin classification there are six skin types [2]. The type I and type II are more prone to melanoma compared to the other skin types. Melanocytes produce a pigment termed as melanin, which gives the natural color to the skin. There are two kinds of melanin, eumelanin and pheomelanin present in the dark skinned and lighter skinned population respectively. Eumelanin is insoluble and thus the skin darkening effects produced by eumelanin last relatively longer compared to the skin-reddening effects produced by pheomelanin. A Dermoscope aids the dermatologists in the primitive analysis of melanocytic skin lesions [2]. Owing to a dearth of experience and differences in visual perceptions, the prognosis of melanoma is still subjective, in spite of the availability of well-established medical methods. This fosters the need for an objective evaluation tool. Computer Aided Diagnostic (CAD), tools were introduced for dermoscopic images to provide quantitative and target assessment of the skin lesion to help clinicians in demonstrative and prognostic undertakings. Due to the inter and intra-observer variabilities, determination of melanoma is innately subjective. Thus, a CAD tool institutively eliminates the subjectivity in the diagnosis and prognosis of melanoma, and aids in early detection of melanoma in situ, thereby improving the accuracy of detection and reducing the mortality rate. This paper describes a clinical framework that can significantly identify the lesion properties and provide a diagnosis. The system incorporates the knowledge of an experienced dermatologist to co-relate the features extracted with their histopathological relevance. Additionally, it further incorporates certain statistical features to achieve promising results.
1.2. Related Work
The literature reports numerous studies for designing CAD systems used for the diagnosis of melanoma. Based on the features used for the prediction of the lesion, approaches for the diagnosis of melanoma have been broadly classified into three types: (i) methods inspired by the dermoscopic criteria (ABCD), that take into account the global and local lesion features [3,4,5]; (ii) methods based on the characteristic properties of images [6,7,8]; and (iii) combination of the aforementioned methods. These methods can be used to either develop a cumulative score [9] or can be used to develop trained models that make predictions. The proposed approach in this paper, belongs to the third category. Nevertheless, the literature also indicates a few approaches that combine the two categories [10,11]. Celebi et al. [10] used color, texture and presence of blue-white veil for classification of skin lesions. However, the lesions were segmented manually, to separate issue of feature extraction from automated border detection. The smooth boundary between the lesion and surrounding skin, poses difficulties in automated border detection. Abuzaghleh et al. [11] classified the lesions using abstract image features and two dermoscopic features. However, the method is computationally complex due extraction of large feature sets. The method proposed in [5] concentrates mainly on the clinical aspects of color in dermoscopic images and texture features, while missing out the important role of shape features. Several studies also report the use of complex deep learning architecture for segmentation and classification [12,13]. However, these methods need to tackle the vanishing gradient and degradation problem.
1.3. Problem Statement
Based on the literature, a clinical framework for the diagnosis of melanoma should encompass the requirements mentioned below.
- Provide automated localization of the lesions.
- The features extracted need to hold clinical significance.
- Result in a balance between sensitivity and specificity for distinguishing the lesion classes.
The aforementioned issues are addressed in this work.
1.4. Contributions
The proposed system describes the development of a clinically inspired framework for diagnosis of melanocytic lesions, such that the system is informative from the perspective of a dermatologist. In contrast to the methods proposed in literature [3,4,5,6,7,8,9,10,11,12,13,14], the proposed system considers lesion specific properties in order to distinguish benign and malignant melanocytic lesions. Additionally, rather than extracting abstract image features, algorithms are developed for the extraction of melanocytic specific features.
1.5. Summary
The manuscript has been organized in the following manner: section two provides the technical depths pertaining to the methodologies developed for the extraction of hair shafts, segmentation of lesion masks, dermoscopic feature extraction and classification. The section three reports the quantitative results obtained. The manuscript concludes with the discussion and conclusion section that provide key aspects with respect to the methodologies developed and future research perspectives.
2. Methods
This section describes the proposed framework. The proposed framework’s overview is illustrated in Figure 1.
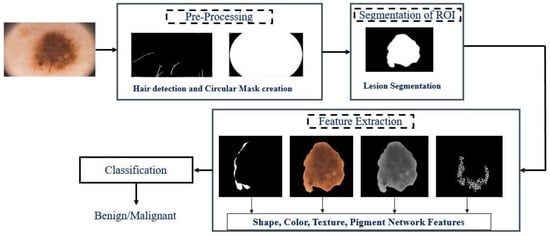
Figure 1.
Overview of the proposed system.
Initially, preprocessing of the dermoscopic images is performed for eliminating artifacts viz., dark corners, ruler markings, hair and dark frames. For the elimination of dark corners, masks of circular shape are created with a radius and centroid co-ordinates computed as given in (1).
where, correspond to the rows and columns of the dermoscopic image. The Figure 2 illustrates the circular mask created for the corresponding dermoscopic image Figure 2A.
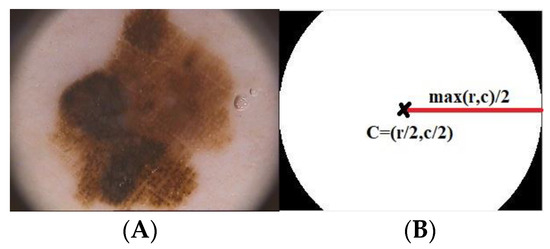
Figure 2.
Mask created (A) Dermoscopic Image (B) Mask for image (A).
The hair masks are multiplied with the initial contour prior to curve deformation to eliminate the dark corners.
2.1. Detection and Removal of Hair
Numerous techniques have been proposed for the detection and exclusion of hair [11,15,16,17,18]. These techniques have been designed assuming that hair color is much darker than the skin and lesion. Additionally, the properties of dermoscopic hair shaft were not considered to detect the dermoscopic hair. Owing to the localization of melanin in the upper and lower epidermis, most skin lesions are either brown or black in color. Therefore, consideration of color variation between hair and surrounding skin could be erroneous leading to an overlap between the attributes of the lesion and hair. This signifies that, for a hair detection algorithm, a need exists for the inclusion of attributes specific to dermoscopic hair.
Geometric deformable models and their success depends on the initial conditions and speed function evolution. As the color of melanin is dependent on the extent of localization in the skin, the attribute of color is vital in the creation of this framework [19,20,21,22]. Therefore, the approach of segmentation has been adopted in this study by giving consideration to the chroma component as opposed to the RGB channels used in conventional systems. The Figure 3 illustrates the results of dermoscopic hair detection, for the corresponding dermoscopic images of Figure 3A and Figure 3B, the hair masks obtained are Figure 3C and Figure 3D, and the Figure 3E and Figure 3F, corresponds to dermoscopic images after hair inpainting.
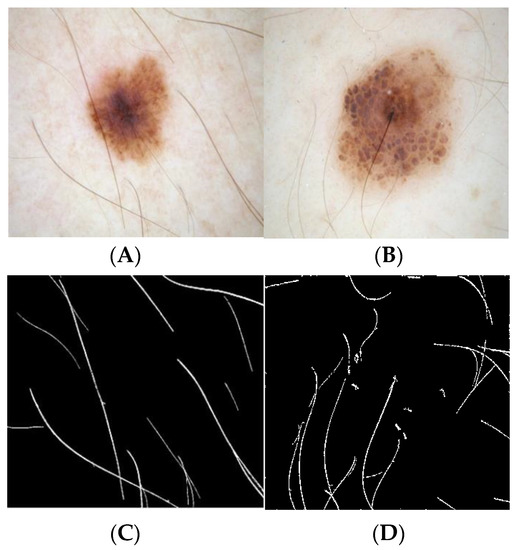
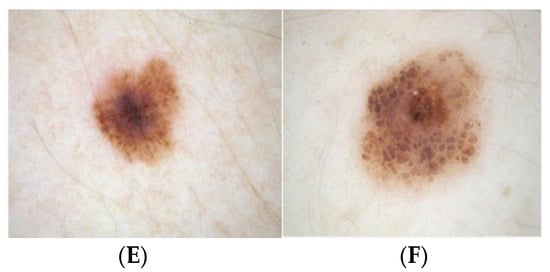
Figure 3.
Hair shaft detection and exclusion method (A,B) dermoscopic images, (C,D) hair shafts detected, and (E,F) dermoscopic images after inpainting.
The Figure 4 illustrates the process of segmentation. Figure 4D illustrates the segmented border obtained by the proposed approach and the boundary of the ground truth.
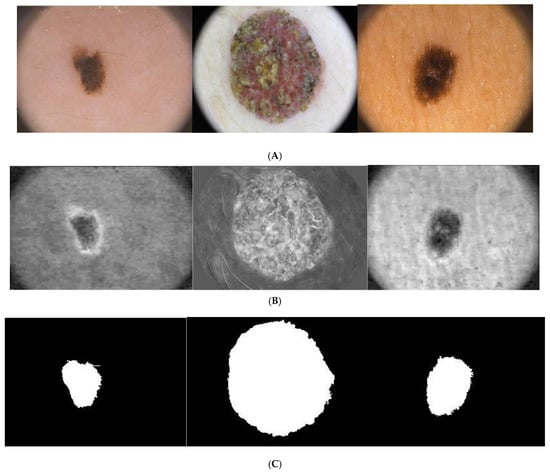
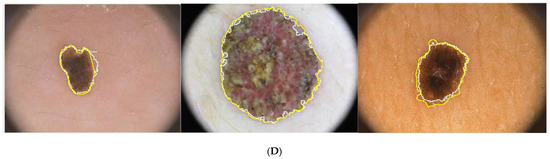
Figure 4.
Illustration of the proposed segmentation approach: (A) original Images, (B) chroma component, (C) segmented images, (D) boundary of ground truth and segmented region overlapped on the original images (yellow corresponds to ground truth, white corresponds to segmented output.
2.2. Extraction and Classification of Features
2.2.1. Color Features
The role of color in dermoscopy is inevitable. The most important chromophore of the skin is melanin. Lesions which are benign exhibit one or two colors. Since the malignant lesions are localized within the deeper structures of the skin, they tend to exhibit three or more colors. To study the color properties of the lesions, six groups of features were computed namely, color asymmetry, color similarity index, color entropy and statistical color features (i.e., color co-relation coefficient, principal component analysis and color entropy). The statistical color features are derived specifically for two categories: (i) region of interest (ROI) (ii) between ROI and non-ROI (NROI). The color features are delineated as follows:
The color asymmetry and color similarity index draw their inspiration from the ABCD rule of dermoscopy. The color asymmetry is quantified by the difference between the opposite halves of the lesion along x-axis ( and y-axis (. The perceived color difference is calculated in the CIE L*a*b color space as given in (9). The four halves are divided as indicated in Figure 5. Correspondingly, four asymmetry indices are computed as given in (2).
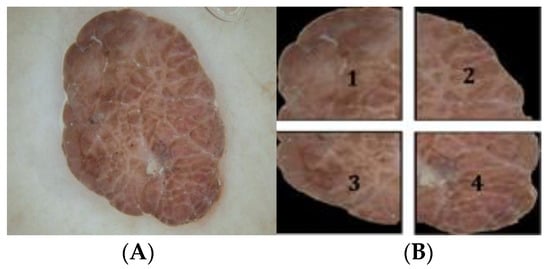
Figure 5.
Color asymmetry calculation: (A) dermoscopic image; (B) the four halves of ROI.
A set of four color asymmetry indices were computed.
Color similarity index indicates the presence of six suspicious colors in a lesion (light-brown (LB), dark-brown (DB), black (K), white (W), red (R) and blue-gray (BG)). The color similarity index is computed by computation of the Euclidean distance between the lesion RGB values and corresponding six suspicious colors. The color of the lesion pixels is used to determine the color similarity index and hence the lesion masks are used for computing the color similarity index. The color similarity index and Euclidean distance are inversely related as given in (3).
The Algorithm 1 summarizes the steps used for calculating the color similarity score. The threshold is determined from the RGB values of two opposite colors [9]. The two opposite colors are black and white. Further, a score of 1 is assigned if more than two percent of the lesion area has suspicious color.
| Algorithm 1. Color Similarity Index Calculation |
| |
The color variance is computed considering the red, green, blue and gray-scale values for the lesion along with the entire image, thus resulting in computation of eight features (VR, VG, VB, and VK). The degree of randomness quantified by color entropy (E) is computed similarly for the red and blue values of the lesion and entire image, leading to computation of four features (ER and EB). The correlation co-efficient signifying the direction and degree of closeness of intra and inter linear relations between the RGB channels and grayscale images resulted in computation of twelve features (CRG, CGB, CBR, CRK, CGK, and CBK). Along with these, the lesion RGB values are projected on the three principal components (PC). Therefore, a total of 37 color features were computed.
2.2.2. Texture Features
According to the opinion of expert dermatologists malignant melanocytic lesions are characterized by coarse texture with a contrast which is inhomogeneous and irregular patterns. Since Tamura’s et al. [23] texture features are based on the visual perception, a set of three texture features, namely coarseness (T1), contrast (T2) and directionality (T3) were computed. A larger value of coarseness specifies a greater degree of roughness. Coarseness is calculated as the average of the best size that gives the maximum difference between the non-overlapping neighborhoods in horizontal and vertical directions. Directionality is computed by taking the gradient of the neighboring pixels given by (4).
where, is the edge strength, () and () indicate the horizontal and vertical change in direction. Further, the contrast is calculated as the statistical distribution of pixel gray values.
2.2.3. Shape Features
The computation of the shape symmetry index is performed using the lesion mark. The lesion centroid is positioned at the centroid of the image by using the difference in centroid positions method showcased in (5). This is carried out since the lesions are not aligned with the center of the image.
and indicate the image and lesion centroids respectively. The image is divided into two halves with respect to x-axis and y-axis as illustrated in Figure 6 to determine the asymmetry along and y axis. The maximum possible asymmetry index of the lesion is given in (6).
where, and are the two halves with respect to the x-axis and and are the two halves with respect to the y-axis. Since, asymmetry quantifies malignancy the proposed method takes into account the maximum possible asymmetry to minimize the classification error.

Figure 6.
AI calculation: (A) dermoscopic image, (B) left half of the image, (C) right half of the image, (D) asymmetric region over y-axis.
A multi-scale method termed as fractal dimension (FD) is used to quantify border irregularity. It is computed by dividing the image in small grids of size as given in (7) [9].
gives the contour length, indicates the scaling constant. The circularity of the lesion is measured using the metric of Compactness (8) [24].
indicates the perimeter of the lesion and indicates the area of the lesion.
2.2.4. Detection of the Pigment Network
Pigment network is a honeycomb-like structure characterized by linear strokes and directional shapes. The presence of pigment network histopathologically, indicates the melanin presence in keratinocytes and melanocytes at the dermal and epidermal junction [25]. Additionally, the lines in the pigment network have diverse orientation. Thus, the proposed approach for detection of pigment network is similar to the hair detection method proposed in Section 2.1 with a few additional steps and change in Gabor parameter . The green channel is used for processing due to relatively greater contrast. A second order derivative Laplacian operator [ mask] is used for enhancing the finer details in the image after median filtering. The enhanced image is convolved with the 2D Gabor filter defined in (3). The empirically determined values for and are same. However, the value of thickness parameter is set to 3.3, since the lines of the pigment network are comparably thinner than the hair shafts.
For post processing the Adaptive Histogram Equalization (AHE) is followed by determination of the threshold to extract the pigment network efficiently. The threshold is computed by fitting a fourth degree polynomial as given in (9) to the contrast enhanced image.
, and are three different points to fit a curve with distinct co-ordinates. The pigment network mask serves to calculate the five distinct features () as given in [25]. The Figure 6 illustrates the pigment network detection process. A comparison of the proposed pigment network detection method with the method proposed by Barata et al. [15] is illustrated in Figure 7. It can be observed by from Figure 8B,C that, the proposed method accurately identifies honeycomb-like pigment network structures in comparison to the method in [15]. An overview of the features extracted is summarized in Table 1.
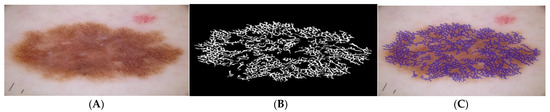
Figure 7.
Detection of pigment network: (A) dermoscopic image, (B) pigment network mask detected, (C) corresponding mask ((A) overlaid on (B)).
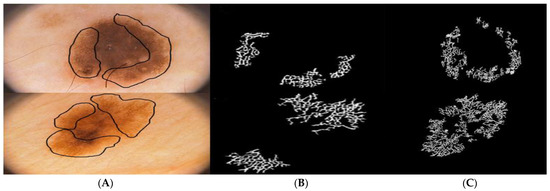
Figure 8.
Comparison of pigment network detection: (A) dermoscopic images with pigment network marked, (B) pigment network masks detected by Barata et al. [15], (C) pigment network masks detected by the proposed method.

Table 1.
Overview of the features extracted.
2.2.5. Classification and Diagnosis
The features selected from the groups of are concatenated to benefit from the complimentary information captured by the feature types. The classification of the observations into two classes classifier (benign and malignant) yielding the largest probability which is performed using a probabilistic SVM, as given in (10).
indicates the cumulative probabilities of shape, color, texture and pigment network features. The features are concatenated and used to train a single SVM classifier. Platt’s method is used for the computation of posterior probabilities [26,27]. Linear kernel is used to map the scores.
3. Results
3.1. Dataset and Evaluation Metrics
A multi-source dataset of 515 images which was taken from PH2 [28], ISBI 2016 and ISBI 2017 [29,30], has been used for experimentation in this article. The dataset consists of 304 benign and 211 melanoma lesions with annotated ground truths. In the initial stage, pre-processing of the images is performed for the removal of dermoscopic hair and dark corners. The algorithms have been implemented using MATLAB 2016®. Three metrics viz., Sensitivity (SE), Specificity (SP) and Accuracy (ACC) have been used for the detection of ROI and lesion classification. In addition to this the overlap error (between the ground truth and segmented mask) is also computed for evaluation of lesion segmentation. The null hypothesis testing has been performed using the Wilcoxon Rank Sum statistics which is a non-parametric test. The null hypothesis is stated below:
H0.
The extracted features for benign and malignant lesions have equal medians.
The testing of the null hypothesis against the alternative hypothesis is performed. The alternate hypothesis states that the features extracted do not have equal medians and hence are statistically significant at 5% significance level. Among the 48 p-values, 34 p-values satisfied the alternative hypothesis and hence were used for classification. A hold-out set of 25% is used for testing. The classification metrics are computed by repeating the training and test procedures ten times by stratifying the training and test sets.
3.2. Evaluation of Hair Detection and Lesion Segmentation: Results
Hair detection and exclusion is performed prior to lesion segmentation to eliminate the artifacts thereby increasing the accuracy of segmentation. A positive effect of pre-processing (hair detection + black frame removal) on the segmentation accuracy for the combined dataset can be observed from the Figure 9. The overlap error after applying hair detection algorithm prior to segmentation was 0.07, and the overlap error obtained without applying hair detection algorithm prior to segmentation was 0.15. This proves that hair detection improved the accuracy of segmentation.
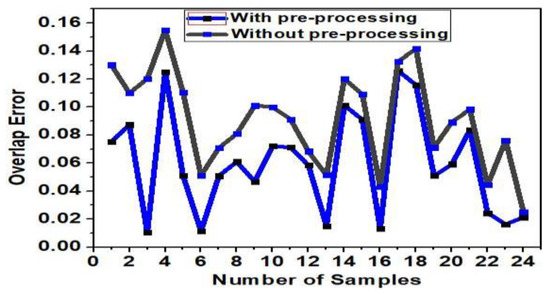
Figure 9.
Effect of pre-processing on segmentation accuracy.
The proposed segmentation method resulted in sensitivity, specificity, accuracy and overlap error of 92.5%, 96.7%, 95.7%, and 8.2%, respectively, for the combined datasets. The overlap segmentation error for modified Chan-Vese (proposed method) and Chan-Vese for combined dataset is illustrated in Figure 10. The Overlap Error (OE) is calculated as given in (11).
where G is the ground truth and S is the segmented binary image.
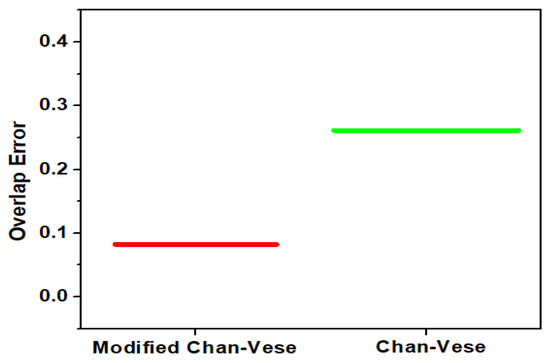
Figure 10.
Overlap segmentation error for Modified Chan-Vese and Chan-Vese Algorithm.
3.3. Evaluation of Features Extracted and Lesion Classification: Results
Various experiments were conducted to deduce the classifier with a major goal to assess the best subset of features and compare the performance of training the SVM with single set of features against training with concatenation of features. The Figure 11 illustrates the plot of features for combined dataset versus the p-values. A good score has a p-values less than or equal to 0.05 whereas is considered a bad score. It is seen from Figure 11, that the shape and pigment network features have good scores. However, the performance scores for color asymmetry index (), indicates comparably lower scores. Insignificant p-values for color similarity index for colors red, light brown and dark brown can be observed. This justifies the fact that presence of red is due to vascularization of blood vessels, irrespective of the lesion class and shades of brown are common to both the lesion types (benign and malignant). Regarding texture features, T2 (contrast) performed comparably poor then T1 and T3. Similarly, the role of statistical color features for ROI and NROI can be interpreted from Figure 11. The Table 2 provides the mean and standard deviation values of the features with significant p-values for combined datasets.
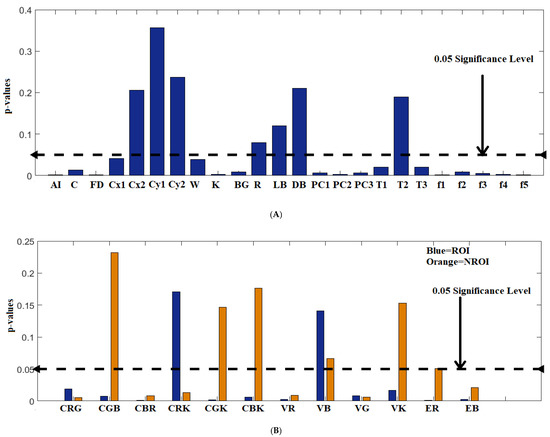
Figure 11.
Plot of features extracted versus the p-values: (A) lesion specific features, (B) statistical color features (CRG, CGB, CBR, CGK, and CBK indicate correlation between red (R), green (G), blue (B), gray values (K)), V indicates color variance, and E indicates entropy).

Table 2.
Mean and standard deviation values of the features with significant p-values.
Based on the p-values the feature set was reduced from 48 to 34 features. The role of single and combined features in lesion diagnosis is given in Table 3. It can be inferred from the Table 3 that, the role of color features is significantly pre-dominant for lesion diagnosis, followed by pigment network features. Interestingly, the best overall results were obtained for a combination of the features. Table 4, shows the results of applying the proposed framework on diverse datasets, correspondingly the plot of ROC for the same is illustrated in Figure 12. It can be observed from the Table 4, acceptable results were obtained for diverse datasets, with PH2 dataset yielding the greatest accuracy. The generalization ability of the features extracted is tested by training on ISBI dataset and testing on PH2 dataset and vice-versa, the results are depicted in Table 5.

Table 3.
Contribution of features for lesion diagnosis (PH2 Dataset).

Table 4.
Classifier Performance for different datasets.
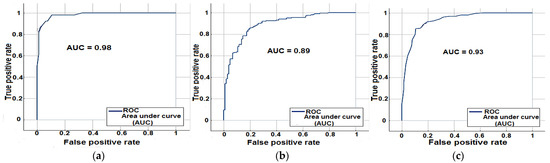
Figure 12.
ROC curves (a) For PH2 data (b) For ISBI data (c) For Combined datasets.

Table 5.
Classifier performance depicting the classifier generalization ability.
4. Discussion
The major objective of this study is the development of an automated computer aided melanoma diagnostic system using clinical aspects of dermoscopy on a diverse dataset. In this regard, the diagnosis system was built using a sequence of algorithms for pre-processing, ROI extraction, feature extraction and classification. Since these steps are sequential in nature, the accuracy of classification mainly relies on the efficiency of the preceding steps. The hair detection algorithm considers the dermoscopic knowledge of hair shafts thereby neglecting a overlap of the attributes of lesion and hair. Such an algorithm prevents loss of lesion specific information and efficiently eliminates the light and dark hair that subsequently aids in improving the lesion segmentation accuracy. Border irregularity is a major indication of malignancy of melanocytic lesions. Thus, while localizing the ROI appropriate care has to be taken to prevent the loss of lesion border details. Geometric deformable models incorporating color information have provided promising segmentation accuracy even in the presence of background noise and poor contrast. The segmentation process was followed by extraction of a set of 48 features specific to shape, color, texture and pigment network from the segmented ROI’s to facilitate identification of benign and malignant lesions. The non-parametric Wilcoxon Rank Sum statistics was used to obtain the p-values for the features extracted with the goal of finding the best features.
Of late, deep learning techniques have been extensively used in skin lesion classification [12,13,31]. In spite of the fact that these architectures have increase the accuracy of classification using large data for learning, the optimization of network parameters for reducing computational complexity is unexplored. A quantitative comparison of the proposed method and the state of art methods reported above may be tenuous due to the diversity of the datasets involved, however a comparative analysis of the studies carried out using the same datasets is given in Table 6. Sensitivity indicates the accurate rate of classification of melanoma lesions. Specificity indicates the accurate rate of classification of benign lesions, whereas accuracy gives a cumulative score of classification of benign and malignant lesions. In [5], the sensitivity was higher in contrast to specificity, since the main focus was on color feature of the lesions. An imbalance in sensitivity and specificity was obtained by Yu et al. [31], by employing a deep learning-based architecture. A methodological approach to detect pigmented skin lesions was proposed in [32]. Pennisi et al. [33], have used standard color, shape and texture feature for classification after applying Delaunay Triangulation based segmentation acc Nonetheless, the comparison provides us with relevant information about the significance of the proposed method. Nonetheless, the proposed method employs domain specific features, thereby improving the accuracy in classification of benign and malignant lesions. However, the study did not consider the thickness feature of the lesion due to lack of third dimensional image data and ground truth. The thickness feature would be an important parameter to rate the stage of malignancy once, the lesion malignancy has been detected by the classification model. Another limitation of the study if the processing time, since it approximately takes 90 s, on an average system of 8 GB RAM, and clock frequency of 1.60 GHz to provide the diagnosis once, the dermoscopic image is given as the input to the system The trained system can be employed in a clinical scenario, by using a dermoscopic based image capturing system, since a dermo scope would enhance the resolution of the lesions that would aid in better analysis, rather than a normal image capturing device.

Table 6.
Comparative analysis of lesion classification methods with the state-of art.
5. Conclusions
This paper presents the development of a clinically oriented framework for melanoma diagnosis. On the basis of the color characteristics of the lesion, the regions are segmented. It can be observed from the Table 4 that the role of color is evident in melanoma detection relative to other features. However, the color features could be severely affected due to variations in image acquisition modalities. Hence, while acquiring real-time images, appropriate illumination correction techniques should be employed to eliminate the effects of non-uniform illuminations.
The experimental results are promising and can be applied to detect asymmetry, pigment network, colors and texture of the lesions. Finally, the detected criteria are combined to develop a cumulative model which exhibits sensitivity, specificity and accuracy of 83.8%, 88.3%, and 86%, respectively.
Author Contributions
Conceptualization, S.P., T.A., S.V., Y.N., R.M.D. and O.P.K.; Formal analysis, S.P., T.A., R.M.D. and O.P.K.; Investigation, S.P. and S.V.; Methodology, S.P., Y.N. and O.P.K.; Software, S.P.; Visualization, T.A. and S.V.; Writing—original draft, S.P.; Writing—review & editing, T.A., Y.N., R.M.D. and O.P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- The Medical Futurist. Amazing Technologies Changing the Future of Dermatology—The Medical Futurist. 2017. Available online: http://medicalfuturist.com/future-of-dermatology/ (accessed on 24 September 2017).
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P.C. Techniques and algorithms for computer aided diagnosis of pigmented skin lesions—A review. Biomed. Signal Process. Control 2018, 39, 237–262. [Google Scholar] [CrossRef]
- Abbas, Q.; Celebi, M.E.; García, I.F. Skin tumor area extraction using an improved dynamic programming approach. Ski. Res. Technol. 2011, 18, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Q.; Celebi, M.E.; Garcia, I.F.; Ahmad, W. Melanoma recognition framework based on expert definition of ABCD for dermoscopic images. Ski. Res. Technol. 2013, 19, e93–e102. [Google Scholar] [CrossRef] [PubMed]
- Barata, C.; Celebi, M.E.; Marques, J. Development of a clinically oriented system for melanoma diagnosis. Pattern Recognit. 2017, 69, 270–285. [Google Scholar] [CrossRef]
- Garnavi, R.; Aldeen, M.; Bailey, J. Computer-Aided Diagnosis of Melanoma Using Border- and Wavelet-Based Texture Analysis. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 1239–1252. [Google Scholar] [CrossRef] [PubMed]
- Kostopoulos, S.A.; Asvestas, P.A.; Kalatzis, I.K.; Sakellaropoulos, G.C.; Sakkis, T.H.; Cavouras, D.A.; Glotsos, D.T. Adaptable pattern recognition system for discriminating Melanocytic Nevi from Malignant Melanomas using plain photography images from different image databases. Int. J. Med. Inform. 2017, 105, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ferris, L.K.; Harkes, J.A.; Gilbert, B.; Winger, D.G.; Golubets, K.; Akilov, O.; Satyanarayanan, M. Computer-aided classification of melanocytic lesions using dermoscopic images. J. Am. Acad. Dermatol. 2015, 73, 769–776. [Google Scholar] [CrossRef]
- Kasmi, R.; Mokrani, K. Classification of malignant melanoma and benign skin lesions: Implementation of automatic ABCD rule. IET Image Process. 2016, 10, 448–455. [Google Scholar] [CrossRef]
- Celebi, M.E.; Kingravi, H.A.; Uddin, B.; Iyatomi, H.; Aslandogan, Y.A.; Stoecker, W.V.; Moss, R.H. A methodological approach to the classification of dermoscopy images. Comput. Med. Imaging Graph. 2007, 31, 362–373. [Google Scholar] [CrossRef] [Green Version]
- Abuzaghleh, O.; Barkana, B.D.; Faezipour, M. Noninvasive Real-Time Automated Skin Lesion Analysis System for Melanoma Early Detection and Prevention. IEEE J. Transl. Eng. Health Med. 2015, 3, 4300212. [Google Scholar] [CrossRef]
- Bozorgtabar, B.; Sedai, S.; Roy, P.K.; Garnavi, R. Skin lesion segmentation using deep convolution networks guided by local unsupervised learning. IBM J. Res. Dev. 2017, 61, 6:1–6:8. [Google Scholar] [CrossRef]
- Premaladha, J.; Ravichandran, K.S. Novel Approaches for Diagnosing Melanoma Skin Lesions Through Supervised and Deep Learning Algorithms. J. Med. Syst. 2016, 40, 96. [Google Scholar] [CrossRef]
- Celebi, M.E.; Iyatomi, H.; Stoecker, W.V.; Moss, R.H.; Rabinovitz, H.S.; Argenziano, G.; Soyer, H.P. Automatic detection of blue-white veil and related structures in dermoscopy images. Comput. Med. Imaging Graph. 2008, 32, 670–677. [Google Scholar] [CrossRef] [Green Version]
- Barata, C.; Marques, J.S.; Rozeira, J. A System for the Detection of Pigment Network in Dermoscopy Images Using Directional Filters. IEEE Trans. Biomed. Eng. 2012, 59, 2744–2754. [Google Scholar] [CrossRef]
- Abbas, Q.; Celebi, M.E.; García, I.F. Hair removal methods: A comparative study for dermoscopy images. Biomed. Signal Process. Control 2011, 6, 395–404. [Google Scholar] [CrossRef]
- Toossi, M.T.B.; Pourreza, H.R.; Zare, H.; Sigari, M.-H.; Layegh, P.; Azimi, A. An effective hair removal algorithm for dermoscopy images. Ski. Res. Technol. 2013, 19, 230–235. [Google Scholar] [CrossRef]
- Liu, Z.-Q.; Cai, J.-H.; Buse, R. Hand-writing Recognition: Soft Computing and Probablistic Approaches, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Rakowska, A. Trichoscopy (hair and scalp videodermoscopy) in the healthy female. Method standardization and norms for measurable parameters. J. Dermatol. Case Rep. 2019, 3, 14. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Tavares, J.M.R.S. A Novel Approach to Segment Skin Lesions in Dermoscopic Images Based on a Deformable Model. IEEE J. Biomed. Health Inform. 2016, 20, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Weatherall, I.L.; Coombs, B.D. Skin Color Measurements in Terms of CIELAB Color Space Values. J. Investig. Dermatol. 1992, 99, 468–473. [Google Scholar] [CrossRef] [Green Version]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Tamura, H.; Mori, S.; Yamawaki, T. Textural Features Corresponding to Visual Perception. IEEE Trans. Syst. Man Cybern. 1978, 8, 460–473. [Google Scholar] [CrossRef]
- Lee, T.K.; McLean, D.I.; Atkins, M.S. Irregularity index: A new border irregularity measure for cutaneous melanocytic lesions. Med. Image Anal. 2002, 7, 47–64. [Google Scholar] [CrossRef]
- Eltayef, K.; Li, Y.; Liu, X. Detection of Pigment Networks in Dermoscopy Images. J. Physics Conf. Ser. 2017, 787, 012033. [Google Scholar] [CrossRef] [Green Version]
- Platt, J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In Advances in Large Margin Classifiers; The MIT Press: Cambridge, MA, USA, 2000; pp. 61–74. [Google Scholar]
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P.C. Hair detection and lesion segmentation in dermoscopic images using domain knowledge. Med. Biol. Eng. Comput. 2018, 56, 2051–2065. [Google Scholar] [CrossRef]
- Mendonca, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.S.; Rozeira, J. PH2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar] [CrossRef]
- ISIC 2016: Skin Lesion Analysis Towards Melanoma Detec-tion. Available online: https://challenge.kitware.com/#challenge/n/ISBI_2016%3A_Skin_Lesion_Analysis_Towards_Melanoma_Detection (accessed on 24 September 2017).
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2017, arXiv:1710.05006. [Google Scholar]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.-A. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Net-works. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef]
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P. A methodological approach to classify typical and atypical pigment network patterns for melanoma diagnosis. Biomed. Signal Process. Control 2018, 44, 25–37. [Google Scholar] [CrossRef]
- Pennisi, A.; Bloisi, D.D.; Nardi, D.; Giampetruzzi, A.R.; Mondino, C.; Facchiano, A. Skin lesion image segmentation using Delaunay Triangulation for melanoma detection. Comput. Med. Imaging Graph. 2016, 52, 89–103. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).