MDA-Unet: A Multi-Scale Dilated Attention U-Net for Medical Image Segmentation
Abstract
:1. Introduction
- Based on potential scopes of improvement, we propose a multi-scale deep learning segmentation model with a slight increase in the number of trainable parameters in comparison with the basic U-Net model.
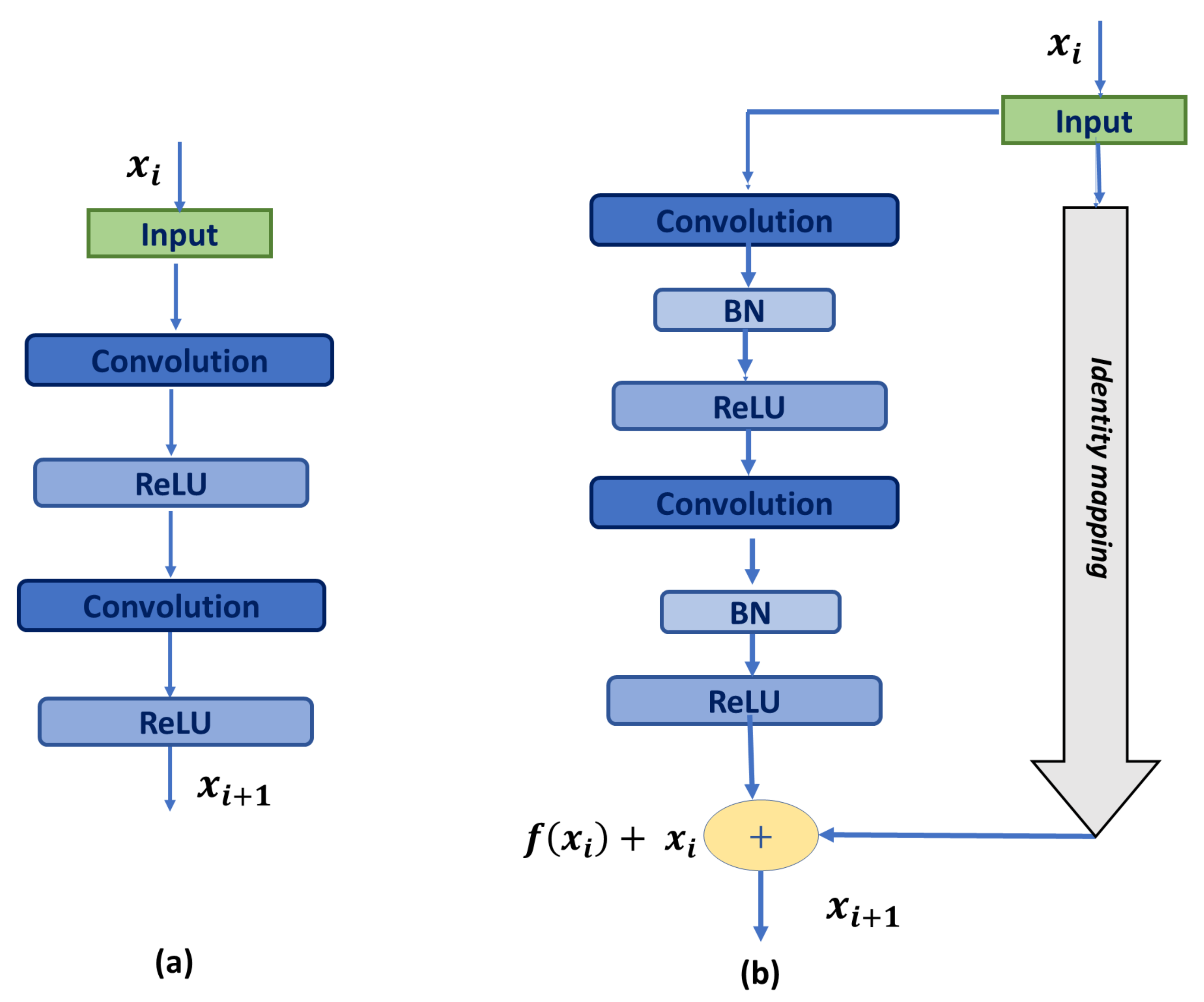
- We increase the network depth by utilizing residual blocks instead of the basic U-Net blocks, which deepens the network using identity shortcut connections. Those connections help propagate the low fine details, ease the training process, and reduce the gradient vanishing problem.
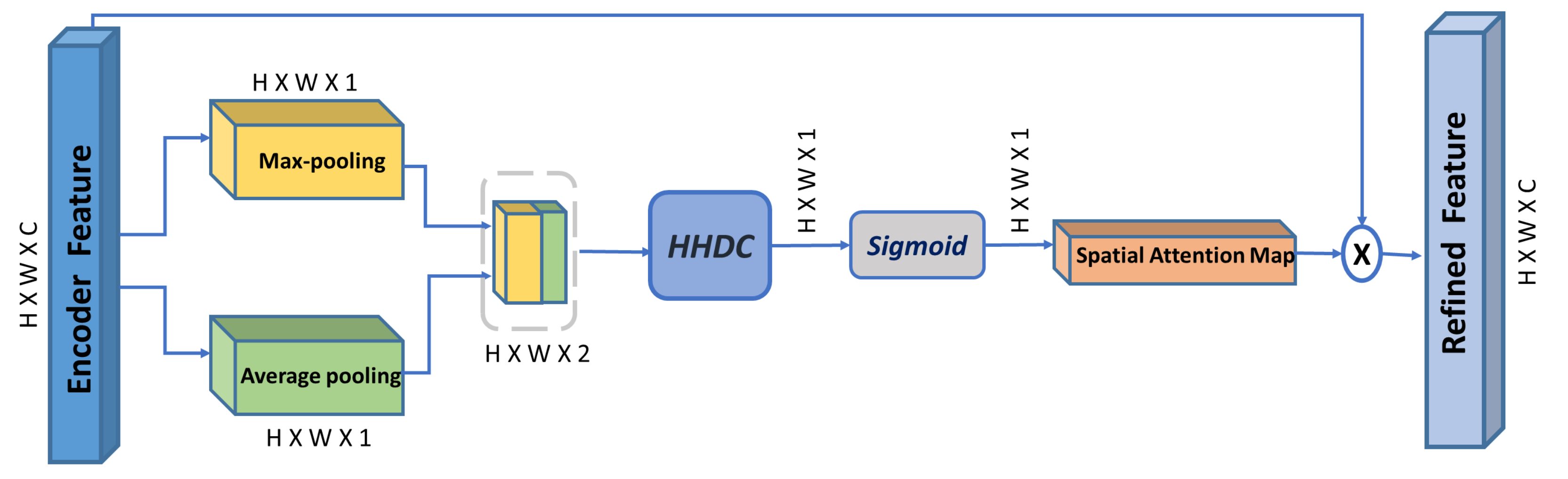
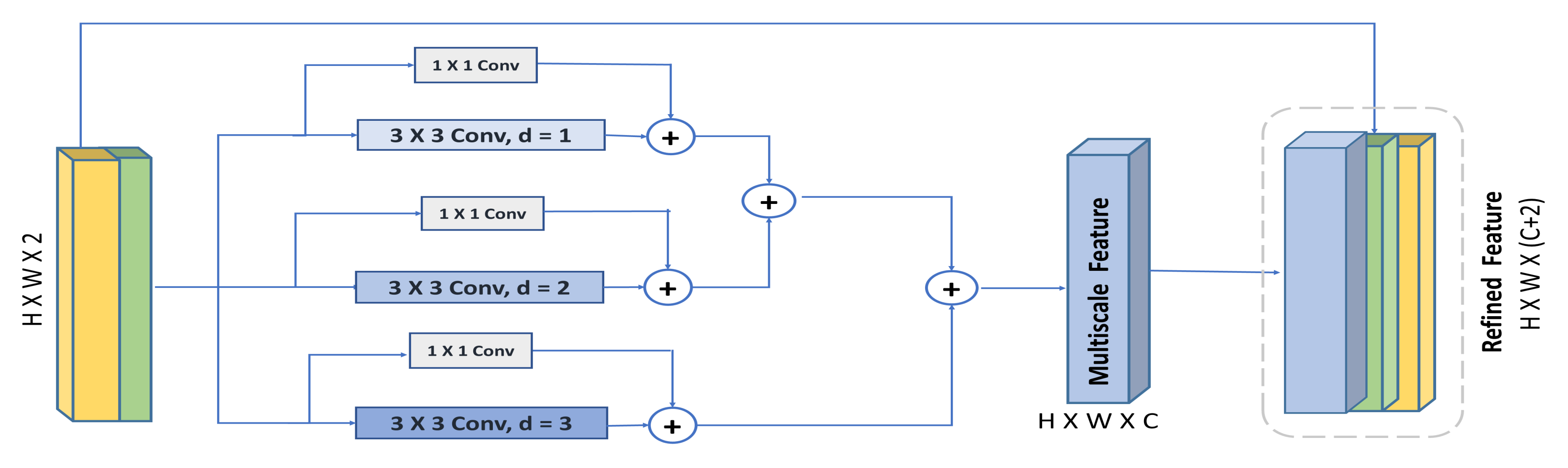
- We propose a multi-scale spatial attention module, which further enhances the capability of the spatial attention mechanism to emphasize multi-scale spatial features driven from feature maps captured using a hybrid hierarchical dilated convolution module, which additionally overcomes the gridding artifacts problem induced by increasing the receptive field using dilated convolution.
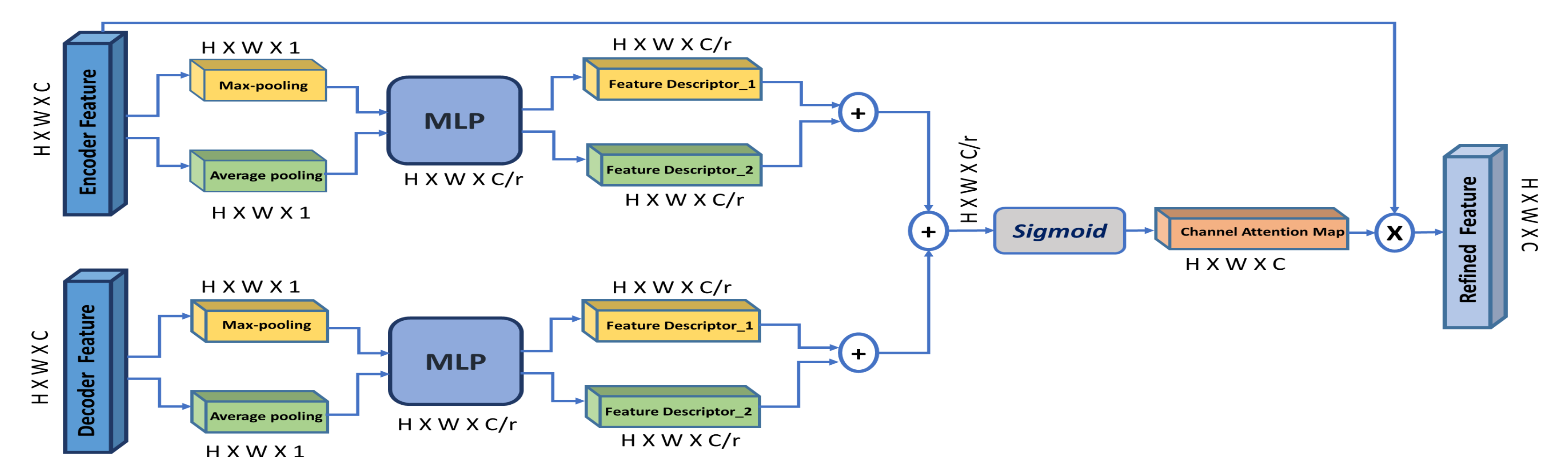
- Through a channel attention mechanism, we use the high-level decoder features to guide and enhance the low-level encoder features, thus improving the encoder–decoder concatenation process.
- We evaluate our model on two different public datasets with different modalities and show significant improvement over U-Net and its variants.
2. Related Work
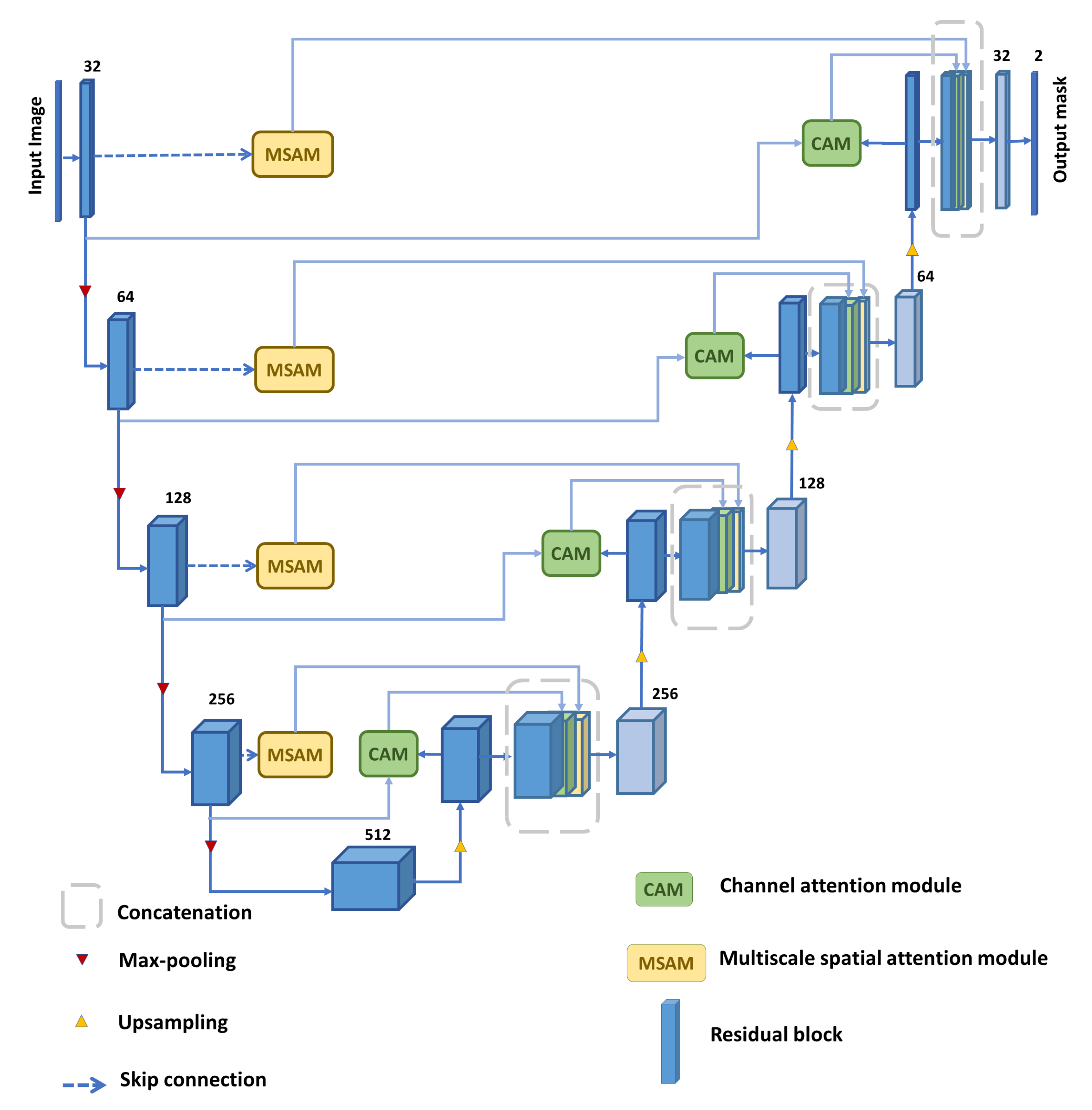
3. Proposed Model
3.1. Residual Block
3.2. Multi-Scale Spatial Attention Module (MSAM)
3.3. Hybrid Hierarchical Dilated Convolution Module (HHDC)
3.4. Channel Attention Module (CAM)
4. Materials and Evaluation Metrics
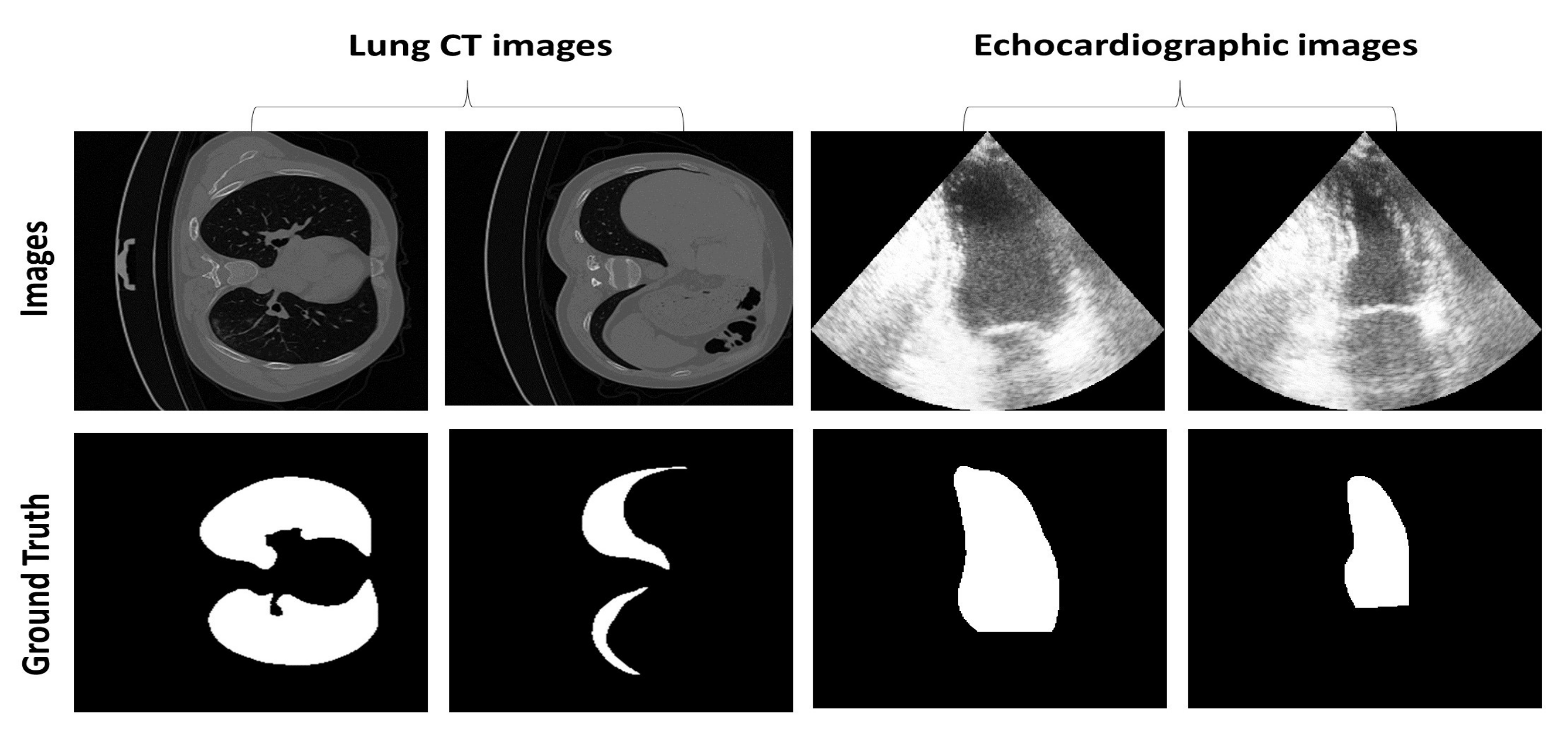
4.1. Dataset
4.2. Implementation Details
4.3. Evaluation Measures
5. Evaluation Results and Discussion
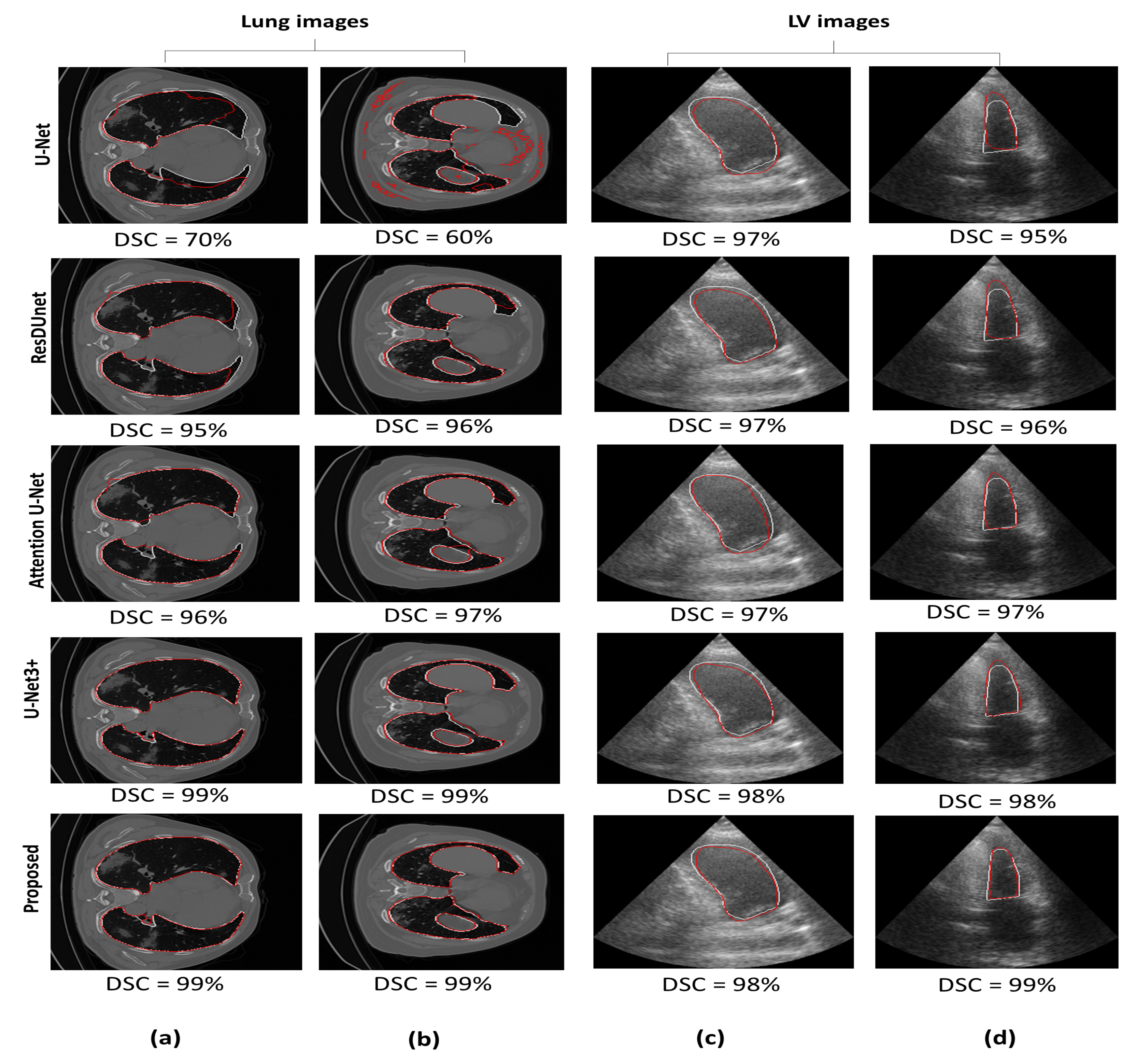
5.1. Comparison with Other Segmentation Models
5.2. Ablation Experiments
5.2.1. Adding Attention Modules
5.2.2. Adding Hierarchical Hybrid Dilated Convolution Module
5.2.3. Changing the Reduction Ratio
5.2.4. Changing the Dilation Rate
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McGuinness, K.; O’connor, N.E. A comparative evaluation of interactive segmentation algorithms. Pattern Recognit. 2010, 43, 434–444. [Google Scholar] [CrossRef] [Green Version]
- Naik, S.; Doyle, S.; Agner, S.; Madabhushi, A.; Feldman, M.; Tomaszewski, J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In Proceedings of the 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Paris, France, 14–17 May 2008; pp. 284–287. [Google Scholar]
- Rouhi, R.; Jafari, M.; Kasaei, S.; Keshavarzian, P. Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Syst. Appl. 2015, 42, 990–1002. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin, Germany, 2018; pp. 3–11. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent Residual Convolutional Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Guo, C.; Szemenyei, M.; Pei, Y.; Yi, Y.; Zhou, W. SD-UNet: A structured dropout U-Net for retinal vessel segmentation. In Proceedings of the 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering (BIBE), Athens, Greece, 28–30 October 2019; pp. 439–444. [Google Scholar]
- Amer, A.; Ye, X.; Janan, F. ResDUnet: A Deep Learning based Left Ventricle Segmentation Method for Echocardiography. IEEE Access 2021, 9, 159755–159763. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, R.; Yang, Y.; Ye, N. Learning cascade attention for fine-grained image classification. Neural Netw. 2020, 122, 174–182. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Computer Vision—ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 3–19. [Google Scholar]
- Park, J.; Woo, S.; Lee, J.Y.; Kweon, I.S. BAM: Bottleneck Attention Module. arXiv 2018, arXiv:1807.06514. [Google Scholar]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef]
- Li, H.; Xiong, P.; An, J.; Wang, L. Pyramid attention network for semantic segmentation. arXiv 2018, arXiv:1805.10180. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Guo, C.; Szemenyei, M.; Yi, Y.; Wang, W.; Chen, B.; Fan, C. Sa-unet: Spatial attention u-net for retinal vessel segmentation. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 1236–1242. [Google Scholar]
- Huang, G.; Zhu, J.; Li, J.; Wang, Z.; Cheng, L.; Liu, L.; Li, H.; Zhou, J. Channel-attention U-Net: Channel attention mechanism for semantic segmentation of esophagus and esophageal cancer. IEEE Access 2020, 8, 122798–122810. [Google Scholar] [CrossRef]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual attention network for scene segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 3146–3154. [Google Scholar]
- Zhao, P.; Zhang, J.; Fang, W.; Deng, S. SCAU-Net: Spatial-Channel Attention U-Net for Gland Segmentation. Front. Bioeng. Biotechnol. 2020, 8, 670. [Google Scholar] [CrossRef] [PubMed]
- Hariyani, Y.S.; Eom, H.; Park, C. DA-Capnet: Dual Attention Deep Learning Based on U-Net for Nailfold Capillary Segmentation. IEEE Access 2020, 8, 10543–10553. [Google Scholar] [CrossRef]
- Mou, L.; Chen, L.; Cheng, J.; Gu, Z.; Zhao, Y.; Liu, J. Dense dilated network with probability regularized walk for vessel detection. IEEE Trans. Med. Imaging 2019, 39, 1392–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, S.; Ding, J.; Liu, B.; Ge, X.; Ma, G. Multi-U-Net: Residual Module under Multisensory Field and Attention Mechanism Based Optimized U-Net for VHR Image Semantic Segmentation. Sensors 2021, 21, 1794. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Yang, M.; Yu, K.; Zhang, C.; Li, Z.; Yang, K. Denseaspp for semantic segmentation in street scenes. In Proceedings of the IEEE conference on computer vision and pattern recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3684–3692. [Google Scholar]
- Wu, S.; Zhong, S.; Liu, Y. Deep residual learning for image steganalysis. Multimed. Tools Appl. 2018, 77, 10437–10453. [Google Scholar] [CrossRef]
- Bi, L.; Kim, J.; Kumar, A.; Fulham, M.; Feng, D. Stacked fully convolutional networks with multi-channel learning: Application to medical image segmentation. Vis. Comput. 2017, 33, 1061–1071. [Google Scholar] [CrossRef]
- Pang, S.; Du, A.; Orgun, M.A.; Wang, Y.; Yu, Z. Tumor attention networks: Better feature selection, better tumor segmentation. Neural Netw. 2021, 140, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, P.; Yuan, Y.; Liu, D.; Huang, Z.; Hou, X.; Cottrell, G. Understanding convolution for semantic segmentation. In Proceedings of the 2018 IEEE winter conference on applications of computer vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 1451–1460. [Google Scholar]
- Amer, A.; Ye, X.; Janan, F. Residual Dilated U-Net for the Segmentation of COVID-19 Infection From CT Images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 462–470. [Google Scholar]
- Amer, A.; Ye, X.; Zolgharni, M.; Janan, F. ResDUnet: Residual dilated UNet for left ventricle segmentation from echocardiographic images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 2019–2022. [Google Scholar]
- Yu, F.; Koltun, V.; Funkhouser, T. Dilated residual networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 472–480. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. arXiv 2015, arXiv:1511.07122. [Google Scholar]
- Mehta, S.; Mercan, E.; Bartlett, J.; Weaver, D.; Elmore, J.; Shapiro, L. Learning to segment breast biopsy whole slide images. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 663–672. [Google Scholar]
- Mehta, S.; Rastegari, M.; Caspi, A.; Shapiro, L.; Hajishirzi, H. Espnet: Efficient spatial pyramid of dilated convolutions for semantic segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 552–568. [Google Scholar]
- Tong, X.; Wei, J.; Sun, B.; Su, S.; Zuo, Z.; Wu, P. ASCU-Net: Attention Gate, Spatial and Channel Attention U-Net for Skin Lesion Segmentation. Diagnostics 2021, 11, 501. [Google Scholar] [CrossRef]
- Leclerc, S.; Smistad, E.; Pedrosa, J.; Østvik, A.; Cervenansky, F.; Espinosa, F.; Espeland, T.; Berg, E.A.R.; Jodoin, P.M.; Grenier, T.; et al. Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE Trans. Med. Imaging 2019, 38, 2198–2210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- COVID-19—Medical Segmentation. Available online: http://medicalsegmentation.com/covid19/ (accessed on 26 October 2021).
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Jaccard, P. The distribution of the flora in the alpine zone. New Phytol. 1912, 11, 37–50. [Google Scholar] [CrossRef]
- Pastor-Pellicer, J.; Zamora-Martínez, F.; España-Boquera, S.; Castro-Bleda, M.J. F-measure as the error function to train neural networks. In Proceedings of the International Work-Conference on Artificial Neural Networks, Tenerife, Spain, 12–14 June 2013; Springer: Berlin, Germany, 2013; pp. 376–384. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin, Germany, 2015; pp. 234–241. [Google Scholar]
| Model | Dataset | DSC | JI | F-Score | Parameters | Training Time |
|---|---|---|---|---|---|---|
| U-Net | Lung | 94.2 ± 6.2 | 92.3 ± 3.5 | 96.5 ± 4.3 | 7,760,069 | 7 |
| LV | 93.9 ± 3.8 | 91.9 ± 2.5 | 95.2 ± 6.1 | |||
| ResDunet | Lung | 95.8 ± 5.3 | 92.3 ± 3.5 | 96.5 ± 4.3 | 11,850,069 | 10 |
| LV | 95.1 ± 0.3 | 91.9 ± 2.5 | 95.2 ± 6.1 | |||
| Attention-Unet | Lung | 96.5 ±5.4 | 95.3 ± 3.6 | 97.3 ± 5.1 | 31,901,542 | 28 |
| LV | 95.6 ± 3.4 | 94.4 ± 4.2 | 96.5 ± 4.2 | |||
| U-Net3+ | Lung | 97.9 ± 1.5 | 95.9 ± 2.6 | 98.7 ± 6.2 | 26,971,000 | 24 |
| LV | 96.1 ± 2.5 | 93.5 ± 4.3 | 97.1 ± 5.1 | |||
| Proposed | Lung | 98.3 ± 2.4 | 97.3 ± 5.4 | 98.9 ± 5.3 | 7,830,793 | 7 |
| LV | 96.7 ± 3.5 | 94.5 ± 6.1 | 97.5 ± 4.3 |
| Method | DSC | Parameters |
|---|---|---|
| Basic U-Net | 93.9 ± 3.8 | 7,760069 |
| CAM only | 95.3 ± 6.2 | 7,804,669 |
| MSAM only | 96.8 ± 3.1 | 7,779,473 |
| CAM + MSAM | 98.3 ± 2.4 | 7,830,793 |
| Method | DSC | Parameters |
|---|---|---|
| CAM only | 95.3 ± 6.2 | 7,804,669 |
| CAM + CBAM | 96.2 ± 2.5 | 7,805,065 |
| CAM + BAM | 97.4 ± 2.5 | 7,804,777 |
| CAM + MSAM | 98.3 ± 2.4 | 7,830,793 |
| Reduction Ratio (r) | DSC | Parameters |
|---|---|---|
| r = 4 | 97.8 ± 2.3 | 7,874,433 |
| r = 8 | 98.3 ± 2.4 | 7,830,793 |
| r = 16 | 97.6 ± 3.1 | 7,808,973 |
| r = 32 | 95.2 ± 1.2 | 7,798,063 |
| Dilation Rate (d) | DSC | Parameters |
|---|---|---|
| d = 1,2 | 95.3 ± 4.2 | 7,828,681 |
| d = 1,2,3 | 98.3 ± 2.4 | 7,830,793 |
| d = 1,2,5 | 97.2 ± 3.1 | 7,830,793 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amer, A.; Lambrou, T.; Ye, X. MDA-Unet: A Multi-Scale Dilated Attention U-Net for Medical Image Segmentation. Appl. Sci. 2022, 12, 3676. https://doi.org/10.3390/app12073676
Amer A, Lambrou T, Ye X. MDA-Unet: A Multi-Scale Dilated Attention U-Net for Medical Image Segmentation. Applied Sciences. 2022; 12(7):3676. https://doi.org/10.3390/app12073676
Chicago/Turabian StyleAmer, Alyaa, Tryphon Lambrou, and Xujiong Ye. 2022. "MDA-Unet: A Multi-Scale Dilated Attention U-Net for Medical Image Segmentation" Applied Sciences 12, no. 7: 3676. https://doi.org/10.3390/app12073676
APA StyleAmer, A., Lambrou, T., & Ye, X. (2022). MDA-Unet: A Multi-Scale Dilated Attention U-Net for Medical Image Segmentation. Applied Sciences, 12(7), 3676. https://doi.org/10.3390/app12073676







