COMMA: Propagating Complementary Multi-Level Aggregation Network for Polyp Segmentation
Abstract
:1. Introduction
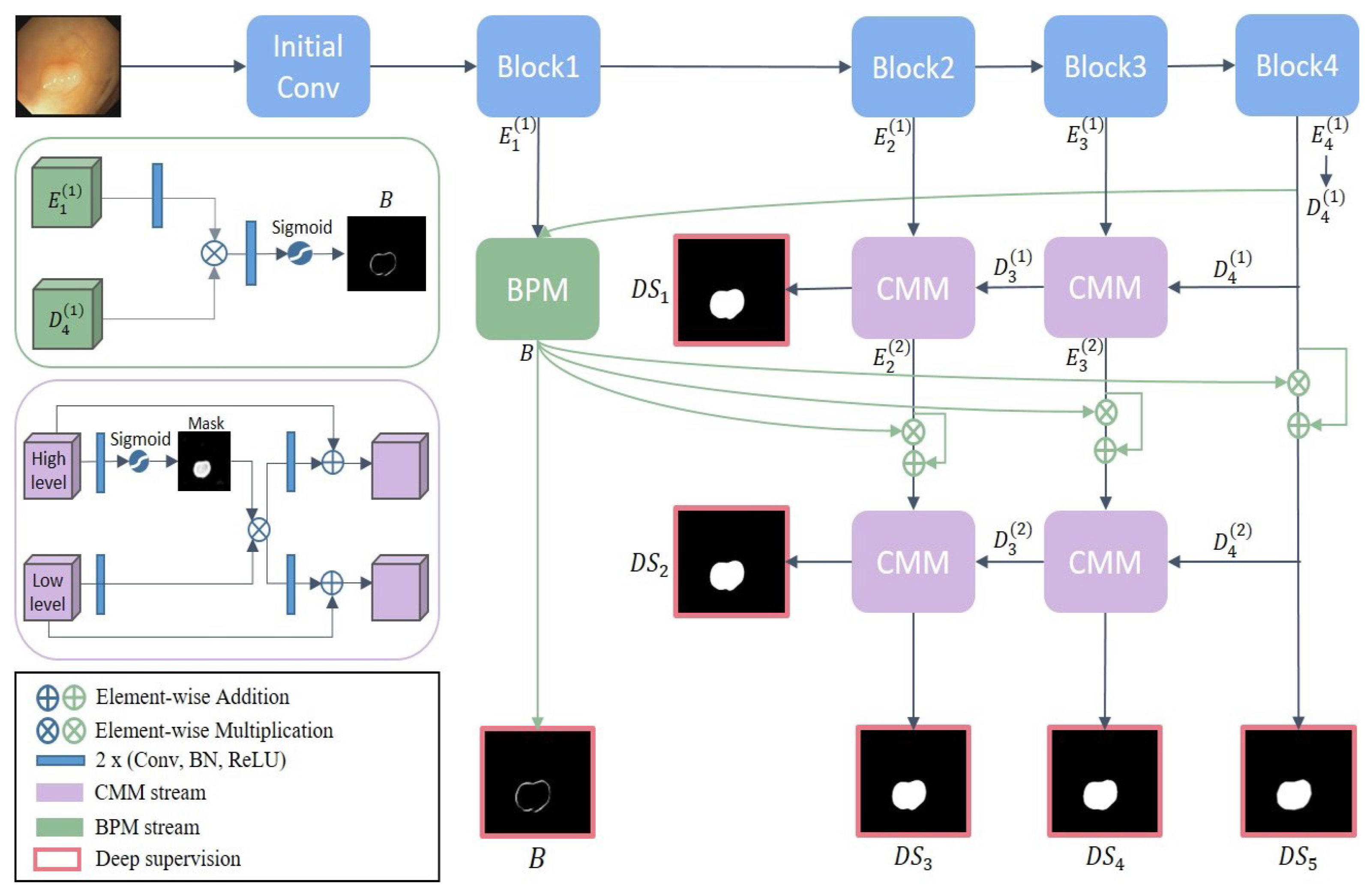
2. Materials and Methods
2.1. Complementary Masking Module
2.2. Boundary Propagation Module
2.3. Hybrid Loss Function
3. Results
3.1. Dataset
3.2. Experimental Setup
3.2.1. Evaluation Metrics
3.2.2. Implementation Details
3.3. Experimental Results
3.3.1. Comparison with State-of-the-Art Methods
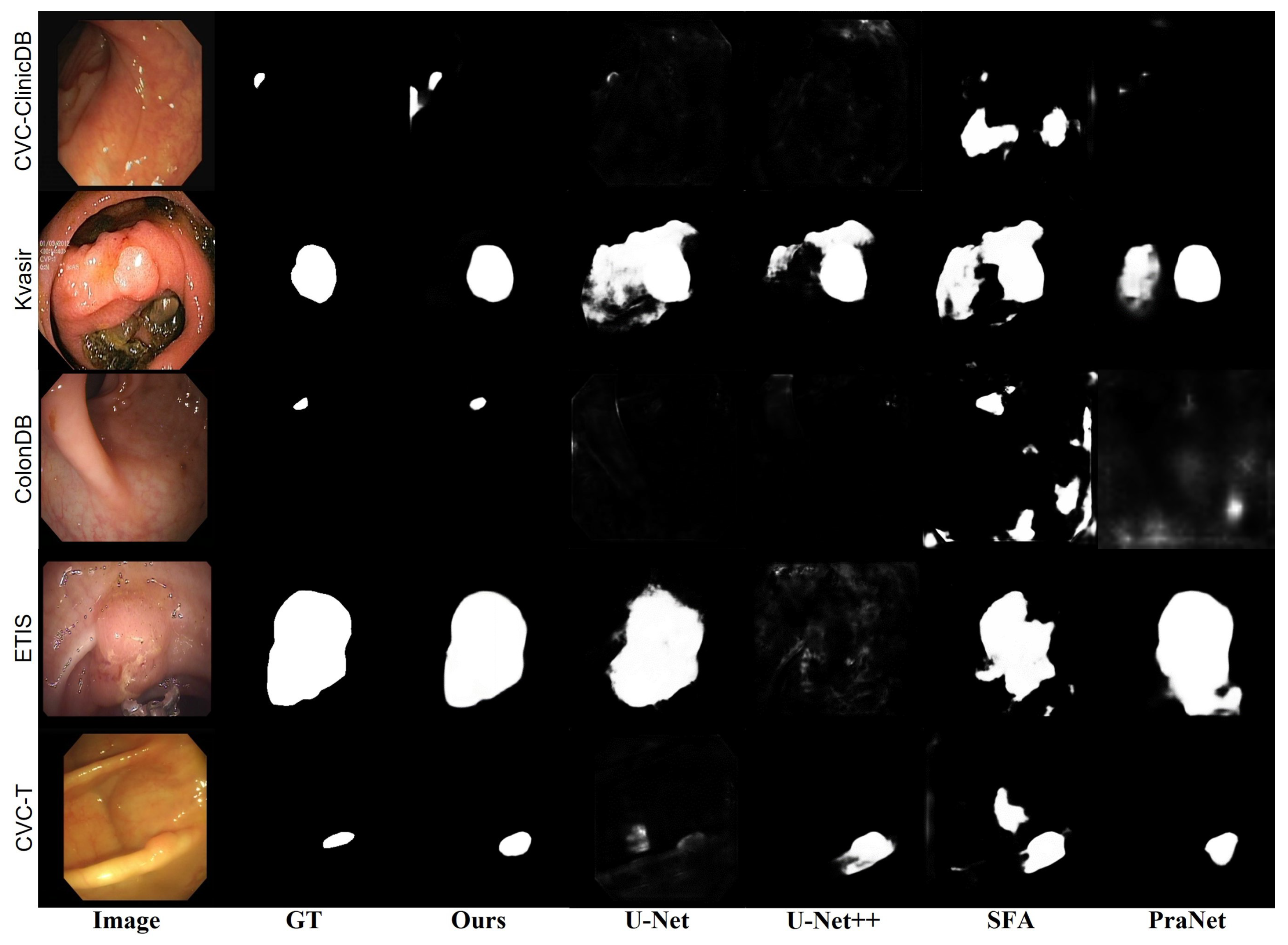
3.3.2. Qualitative Comparison
3.3.3. Inference Analysis
3.4. Further Experiments
3.4.1. Effectiveness of Multi-Decoder Structure
3.4.2. Individual Learning
3.4.3. Comparison of Interpolation Method of Up/Down-Sampling
4. Discussion
4.1. Effectiveness of Proposed Modules
4.2. Effectiveness of BPM Combinations
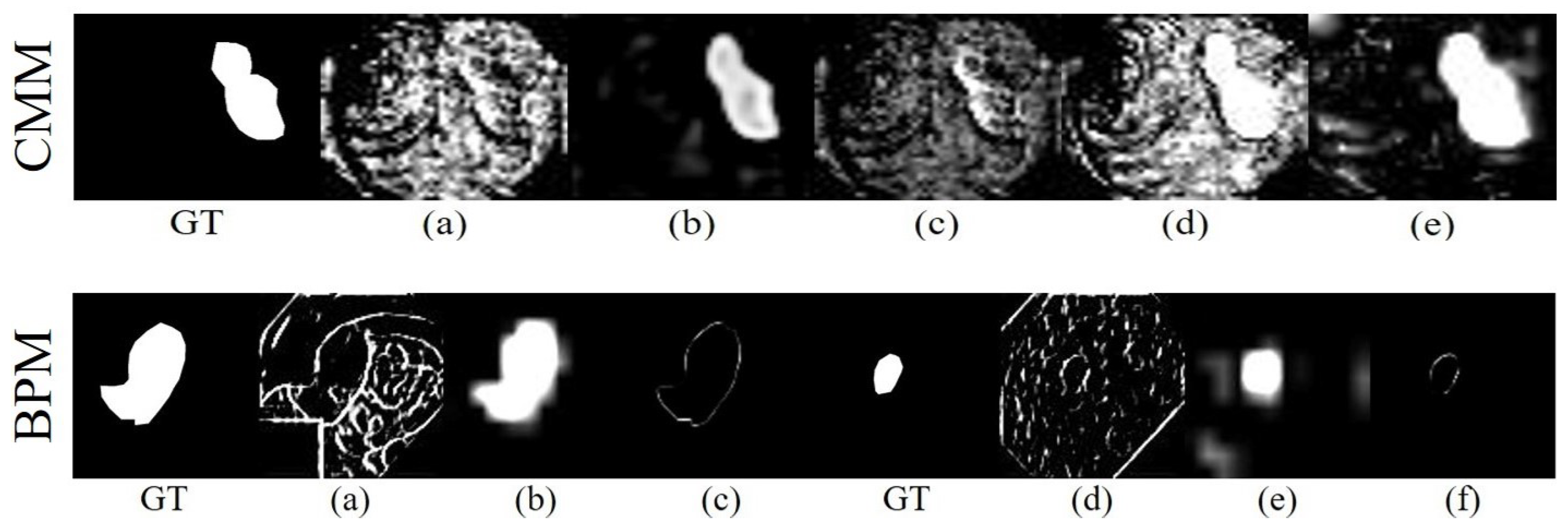
4.3. CMM and BPM Visualization
4.3.1. Complementary Masking Visualization
4.3.2. Explicit Boundary Visualization
4.4. Analysis of Loss Functions
4.4.1. Comparison of Loss Functions
4.4.2. Weighted Loss Combination
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Granados-Romero, J.J.; Valderrama-Treviño, A.I.; Contreras-Flores, E.H.; Barrera-Mera, B.; Herrera Enríquez, M.; Uriarte-Ruíz, K.; Ceballos-Villalba, J.; Estrada-Mata, A.G.; Alvarado Rodríguez, C.; Arauz-Peña, G. Colorectal cancer: A review. Int. J. Res. Med. Sci. 2017, 5, 4667–4676. [Google Scholar] [CrossRef] [Green Version]
- Schreuders, E.H.; Ruco, A.; Rabeneck, L.; Schoen, R.E.; Sung, J.J.; Young, G.P.; Kuipers, E.J. Colorectal cancer screening: A global overview of existing programmes. Gut 2015, 64, 1637–1649. [Google Scholar] [CrossRef]
- Mármol, I.; Sánchez-de Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamonov, A.V.; Figueiredo, I.N.; Figueiredo, P.N.; Tsai, Y.H.R. Automated polyp detection in colon capsule endoscopy. IEEE Trans. Med. Imaging 2014, 33, 1488–1502. [Google Scholar] [CrossRef] [Green Version]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 2015, 35, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Integrating online and offline three-dimensional deep learning for automated polyp detection in colonoscopy videos. IEEE J. Biomed. Health Inform. 2016, 21, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Brandao, P.; Mazomenos, E.; Ciuti, G.; Caliò, R.; Bianchi, F.; Menciassi, A.; Dario, P.; Koulaouzidis, A.; Arezzo, A.; Stoyanov, D. Fully convolutional neural networks for polyp segmentation in colonoscopy. In Medical Imaging 2017: Computer-Aided Diagnosis; SPIE: Orlando, FL, USA, 2017; Volume 10134, pp. 101–107. [Google Scholar]
- Ji, G.P.; Chou, Y.C.; Fan, D.P.; Chen, G.; Fu, H.; Jha, D.; Shao, L. Progressively normalized self-attention network for video polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 142–152. [Google Scholar]
- Wei, J.; Hu, Y.; Zhang, R.; Li, Z.; Zhou, S.K.; Cui, S. Shallow attention network for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 699–708. [Google Scholar]
- Wang, T.; Zhang, L.; Wang, S.; Lu, H.; Yang, G.; Ruan, X.; Borji, A. Detect globally, refine locally: A novel approach to saliency detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3127–3135. [Google Scholar]
- Wu, Z.; Su, L.; Huang, Q. Cascaded partial decoder for fast and accurate salient object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 12–20 June 2019; pp. 3907–3916. [Google Scholar]
- Zhang, P.; Wang, D.; Lu, H.; Wang, H.; Ruan, X. Amulet: Aggregating multi-level convolutional features for salient object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 202–211. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Johansen, D.; de Lange, T.; Johansen, H.D.; Halvorsen, P.; Riegler, M.A. A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation. IEEE J. Biomed. Health Inform. 2021, 25, 2029–2040. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. Resunet++: An advanced architecture for medical image segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; pp. 225–2255. [Google Scholar]
- Fang, Y.; Chen, C.; Yuan, Y.; Tong, K.y. Selective feature aggregation network with area-boundary constraints for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 302–310. [Google Scholar]
- Zhang, R.; Li, G.; Li, Z.; Cui, S.; Qian, D.; Yu, Y. Adaptive context selection for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 253–262. [Google Scholar]
- Patel, K.; Bur, A.M.; Wang, G. Enhanced u-net: A feature enhancement network for polyp segmentation. In Proceedings of the 2021 18th Conference on Robots and Vision (CRV), Burnaby, BC, Canada, 26–28 May 2021; pp. 181–188. [Google Scholar]
- Murugesan, B.; Sarveswaran, K.; Shankaranarayana, S.M.; Ram, K.; Joseph, J.; Sivaprakasam, M. Psi-Net: Shape and boundary aware joint multi-task deep network for medical image segmentation. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 7223–7226. [Google Scholar]
- Lee, C.Y.; Xie, S.; Gallagher, P.; Zhang, Z.; Tu, Z. Deeply-supervised nets. In Proceedings of the Artificial Intelligence and Statistics, San Diego, CA, USA, 9–12 May 2015; pp. 562–570. [Google Scholar]
- Wang, D.; Hao, M.; Xia, R.; Zhu, J.; Li, S.; He, X. MSB-Net: Multi-Scale Boundary Net for Polyp Segmentation. In Proceedings of the 2021 IEEE 10th Data Driven Control and Learning Systems Conference (DDCLS), Suzhou, China, 14–16 May 2021; pp. 88–93. [Google Scholar]
- Cao, F.; Gao, C.; Ye, H. A novel method for image segmentation: Two-stage decoding network with boundary attention. Int. J. Mach. Learn. Cybern. 2021, 1–13. [Google Scholar] [CrossRef]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 263–273. [Google Scholar]
- Chen, S.; Tan, X.; Wang, B.; Hu, X. Reverse attention for salient object detection. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 234–250. [Google Scholar]
- Wei, J.; Wang, S.; Huang, Q. F3Net: Fusion, Feedback and Focus for Salient Object Detection. In Proceedings of the AAAI Conference on Artificial Intelligence, Hilton, NY, USA, 7–12 February 2020; Volume 34, pp. 12321–12328. [Google Scholar]
- Ghosh, A.; Kumar, H.; Sastry, P. Robust loss functions under label noise for deep neural networks. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31. [Google Scholar]
- Wang, G.; Liu, X.; Li, C.; Xu, Z.; Ruan, J.; Zhu, H.; Meng, T.; Li, K.; Huang, N.; Zhang, S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging 2020, 39, 2653–2663. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Gao, S.; Cheng, M.M.; Zhao, K.; Zhang, X.Y.; Yang, M.H.; Torr, P.H. Res2net: A new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 652–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 630–645. [Google Scholar]
- Jain, V.; Seung, S. Natural image denoising with convolutional networks. Adv. Neural Inf. Process. Syst. 2008, 21, 769–776. [Google Scholar]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Image super-resolution using deep convolutional networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 295–307. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.X.; Liu, J.J.; Fan, D.P.; Cao, Y.; Yang, J.; Cheng, M.M. EGNet: Edge guidance network for salient object detection. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019; pp. 8779–8788. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; Lange, T.d.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the International Conference on Multimedia Modeling, Daejeon, Korea, 5–8 January 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 451–462. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef]
- Bernal, J.; Sánchez, J.; Vilarino, F. Towards automatic polyp detection with a polyp appearance model. Pattern Recognit. 2012, 45, 3166–3182. [Google Scholar] [CrossRef]
- Silva, J.; Histace, A.; Romain, O.; Dray, X.; Granado, B. Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, D.; Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; López, A.M.; Romero, A.; Drozdzal, M.; Courville, A. A benchmark for endoluminal scene segmentation of colonoscopy images. J. Healthc. Eng. 2017, 2017, 4037190. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, Q.; Cong, R.; Huang, Q. Global context-aware progressive aggregation network for salient object detection. In Proceedings of the AAAI Conference on Artificial Intelligence, Hilton, NY, USA, 7–12 February 2020; Volume 34, pp. 10599–10606. [Google Scholar]
- Fan, D.P.; Cheng, M.M.; Liu, Y.; Li, T.; Borji, A. Structure-measure: A new way to evaluate foreground maps. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 4548–4557. [Google Scholar]
- Fan, D.P.; Gong, C.; Cao, Y.; Ren, B.; Cheng, M.M.; Borji, A. Enhanced-alignment measure for binary foreground map evaluation. arXiv 2018, arXiv:1805.10421. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef] [Green Version]
| Model | CVC-ClinicDB | Kvasir | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | MAE | mDice | mIoU | MAE | |||||
| U-Net [9] | 0.823 | 0.760 | 0.890 | 0.953 | 0.019 | 0.818 | 0.750 | 0.858 | 0.893 | 0.055 |
| U-Net++ [17] | 0.794 | 0.733 | 0.873 | 0.931 | 0.022 | 0.821 | 0.747 | 0.862 | 0.909 | 0.048 |
| ResUNet-mod [46] | 0.779 | 0.455 | n/a | n/a | n/a | 0.791 | 0.429 | n/a | n/a | n/a |
| ResUNet++ [19] | 0.796 | 0.796 | n/a | n/a | n/a | 0.813 | 0.793 | n/a | n/a | n/a |
| SFA [20] | 0.702 | 0.611 | 0.793 | 0.885 | 0.042 | 0.725 | 0.614 | 0.781 | 0.849 | 0.075 |
| PraNet [27] | 0.899 | 0.853 | 0.937 | 0.979 | 0.009 | 0.897 | 0.844 | 0.915 | 0.948 | 0.030 |
| COMMA * | 0.916 | 0.871 | 0.947 | 0.979 | 0.008 | 0.904 | 0.860 | 0.925 | 0.963 | 0.024 |
| COMMA | 0.933 | 0.891 | 0.956 | 0.985 | 0.007 | 0.901 | 0.852 | 0.919 | 0.951 | 0.027 |
| Dataset | Model | mDice | mIoU | MAE | ||
|---|---|---|---|---|---|---|
| ColonDB | U-Net [9] | 0.504 | 0.440 | 0.710 | 0.781 | 0.059 |
| U-Net++ [17] | 0.482 | 0.412 | 0.693 | 0.764 | 0.061 | |
| SFA [20] | 0.457 | 0.341 | 0.628 | 0.753 | 0.094 | |
| PraNet [27] | 0.711 | 0.644 | 0.820 | 0.872 | 0.043 | |
| E: U-Net [22] | 0.740 | 0.663 | - | - | - | |
| MSBNet [25] | 0.741 | - | 0.826 | 0.875 | 0.040 | |
| COMMA * | 0.712 | 0.645 | 0.823 | 0.864 | 0.045 | |
| COMMA | 0.754 | 0.689 | 0.849 | 0.897 | 0.037 | |
| ETIS | U-Net [9] | 0.399 | 0.340 | 0.684 | 0.740 | 0.036 |
| U-Net++ [17] | 0.401 | 0.348 | 0.683 | 0.776 | 0.035 | |
| SFA [20] | 0.298 | 0.221 | 0.557 | 0.632 | 0.109 | |
| PraNet [27] | 0.628 | 0.571 | 0.794 | 0.841 | 0.031 | |
| E: U-Net [22] | 0.651 | 0.582 | - | - | - | |
| MSBNet [25] | 0.606 | - | 0.772 | 0.841 | 0.023 | |
| COMMA * | 0.709 | 0.643 | 0.845 | 0.887 | 0.018 | |
| COMMA | 0.711 | 0.648 | 0.844 | 0.887 | 0.015 | |
| CVC-T | U-Net [9] | 0.711 | 0.631 | 0.843 | 0.875 | 0.022 |
| U-Net++ [17] | 0.708 | 0.629 | 0.839 | 0.898 | 0.018 | |
| SFA [20] | 0.468 | 0.334 | 0.641 | 0.817 | 0.065 | |
| PraNet [27] | 0.871 | 0.801 | 0.925 | 0.972 | 0.010 | |
| E: U-Net [22] | 0.886 | 0.813 | - | - | - | |
| MSBNet [25] | 0.866 | - | 0.917 | 0.966 | 0.010 | |
| COMMA * | 0.871 | 0.801 | 0.924 | 0.980 | 0.011 | |
| COMMA | 0.906 | 0.843 | 0.945 | 0.988 | 0.006 |
| Models | Batch. | #Params | CVC-ClinicDB | Kvasir | ColonDB | ETIS | Mean FPS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mDice | MAE | FPS | mDice | MAE | FPS | mDice | MAE | FPS | mDice | MAE | FPS | ||||
| PraNet | 1 | 32.55 M | 0.899 | 0.009 | 32.99 | 0.897 | 0.030 | 31.71 | 0.711 | 0.043 | 37.10 | 0.628 | 0.031 | 19.56 | 30.34 |
| COMMA | 1 | 31.10 M | 0.933 | 0.007 | 42.32 | 0.901 | 0.027 | 31.90 | 0.754 | 0.037 | 51.96 | 0.711 | 0.015 | 42.31 | 42.12 |
| COMMA | 8 | 31.10 M | 0.933 | 0.007 | 71.30 | 0.901 | 0.027 | 46.47 | 0.754 | 0.037 | 110.00 | 0.711 | 0.015 | 71.07 | 74.70 |
| Dataset | #Decoder (d) | #Params | mDice | mIoU | MAE | ||
|---|---|---|---|---|---|---|---|
| CVC-ClinicDB | 1 | 28.43 M | 0.919 | 0.875 | 0.944 | 0.978 | 0.0072 |
| 2 | 31.10 M | 0.933 | 0.891 | 0.956 | 0.985 | 0.0066 | |
| 3 | 33.76 M | 0.925 | 0.882 | 0.945 | 0.981 | 0.0074 | |
| 4 | 36.42 M | 0.931 | 0.889 | 0.953 | 0.982 | 0.0068 | |
| 5 | 39.09 M | 0.921 | 0.878 | 0.950 | 0.979 | 0.0074 | |
| Kvasir | 1 | 28.43 M | 0.870 | 0.823 | 0.898 | 0.920 | 0.032 |
| 2 | 31.10 M | 0.901 | 0.852 | 0.919 | 0.951 | 0.027 | |
| 3 | 33.76 M | 0.898 | 0.849 | 0.919 | 0.953 | 0.028 | |
| 4 | 36.42 M | 0.897 | 0.848 | 0.918 | 0.957 | 0.028 | |
| 5 | 39.09 M | 0.901 | 0.851 | 0.919 | 0.950 | 0.027 | |
| ColonDB | 1 | 28.43 M | 0.701 | 0.665 | 0.817 | 0.844 | 0.051 |
| 2 | 31.10 M | 0.754 | 0.689 | 0.849 | 0.897 | 0.037 | |
| 3 | 33.76 M | 0.762 | 0.697 | 0.852 | 0.874 | 0.039 | |
| 4 | 36.42 M | 0.756 | 0.689 | 0.850 | 0.875 | 0.035 | |
| 5 | 39.09 M | 0.753 | 0.689 | 0.846 | 0.876 | 0.039 | |
| ETIS | 1 | 28.43 M | 0.677 | 0.621 | 0.831 | 0.880 | 0.0160 |
| 2 | 31.10 M | 0.711 | 0.648 | 0.844 | 0.887 | 0.0151 | |
| 3 | 33.76 M | 0.708 | 0.644 | 0.844 | 0.878 | 0.0176 | |
| 4 | 36.42 M | 0.711 | 0.649 | 0.844 | 0.893 | 0.0164 | |
| 5 | 39.09 M | 0.694 | 0.633 | 0.836 | 0.874 | 0.0167 | |
| CVC-T | 1 | 28.43 M | 0.850 | 0.793 | 0.894 | 0.957 | 0.012 |
| 2 | 31.10 M | 0.906 | 0.843 | 0.945 | 0.988 | 0.006 | |
| 3 | 33.76 M | 0.892 | 0.826 | 0.935 | 0.987 | 0.007 | |
| 4 | 36.42 M | 0.870 | 0.803 | 0.927 | 0.964 | 0.008 | |
| 5 | 39.09 M | 0.888 | 0.822 | 0.936 | 0.977 | 0.009 |
| Dataset | Model | mDice | mIoU | MAE | ||
|---|---|---|---|---|---|---|
| CVC-ClinicDB | PraNet | 0.899 | 0.853 | 0.937 | 0.979 | 0.009 |
| COMMA | 0.919 | 0.877 | 0.948 | 0.984 | 0.007 | |
| COMMA | 0.933 | 0.891 | 0.956 | 0.985 | 0.007 | |
| Kvasir | PraNet | 0.897 | 0.844 | 0.915 | 0.948 | 0.030 |
| COMMA * | 0.913 | 0.867 | 0.929 | 0.965 | 0.024 | |
| COMMA | 0.901 | 0.852 | 0.919 | 0.951 | 0.027 | |
| ColonDB | PraNet | 0.711 | 0.644 | 0.820 | 0.872 | 0.043 |
| COMMA | 0.757 | 0.690 | 0.849 | 0.893 | 0.038 | |
| COMMA * | 0.676 | 0.609 | 0.803 | 0.858 | 0.045 | |
| COMMA | 0.754 | 0.689 | 0.849 | 0.897 | 0.037 | |
| ETIS | PraNet | 0.628 | 0.571 | 0.794 | 0.841 | 0.031 |
| COMMA | 0.671 | 0.593 | 0.820 | 0.830 | 0.036 | |
| COMMA * | 0.689 | 0.616 | 0.829 | 0.863 | 0.023 | |
| COMMA | 0.711 | 0.648 | 0.844 | 0.887 | 0.015 | |
| CVC-T | PraNet | 0.871 | 0.801 | 0.925 | 0.972 | 0.010 |
| COMMA | 0.850 | 0.785 | 0.914 | 0.960 | 0.014 | |
| COMMA * | 0.844 | 0.765 | 0.909 | 0.943 | 0.010 | |
| COMMA | 0.906 | 0.843 | 0.945 | 0.988 | 0.006 |
| Dataset | Model | mDice | mIoU | MAE | ||
|---|---|---|---|---|---|---|
| CVC-ClinicDB | Nearest | 0.901 | 0.842 | 0.929 | 0.978 | 0.009 |
| Bilinear | 0.933 | 0.891 | 0.956 | 0.985 | 0.007 | |
| Bicubic | 0.920 | 0.876 | 0.943 | 0.979 | 0.008 | |
| Kvasir | Nearest | 0.881 | 0.821 | 0.904 | 0.942 | 0.031 |
| Bilinear | 0.901 | 0.852 | 0.919 | 0.951 | 0.027 | |
| Bicubic | 0.890 | 0.841 | 0.914 | 0.941 | 0.028 | |
| ColonDB | Nearest | 0.750 | 0.672 | 0.842 | 0.883 | 0.037 |
| Bilinear | 0.754 | 0.689 | 0.849 | 0.897 | 0.037 | |
| Bicubic | 0.753 | 0.687 | 0.846 | 0.885 | 0.038 | |
| ETIS | Nearest | 0.681 | 0.604 | 0.821 | 0.846 | 0.014 |
| Bilinear | 0.711 | 0.648 | 0.844 | 0.887 | 0.015 | |
| Bicubic | 0.699 | 0.637 | 0.837 | 0.846 | 0.013 | |
| CVC-T | Nearest | 0.865 | 0.782 | 0.916 | 0.980 | 0.009 |
| Bilinear | 0.906 | 0.843 | 0.945 | 0.988 | 0.006 | |
| Bicubic | 0.891 | 0.826 | 0.936 | 0.978 | 0.007 |
| Dataset | Model | mDice | mIoU | MAE | ||
|---|---|---|---|---|---|---|
| CVC-ClinicDB | Base | 0.898 | 0.850 | 0.933 | 0.967 | 0.010 |
| Base + CMM | 0.926 | 0.882 | 0.945 | 0.981 | 0.008 | |
| Base + BPM | 0.911 | 0.863 | 0.940 | 0.972 | 0.008 | |
| Base + CMM + BPM | 0.933 | 0.891 | 0.956 | 0.985 | 0.007 | |
| Kvasir | Base | 0.882 | 0.831 | 0.906 | 0.945 | 0.032 |
| Base + CMM | 0.897 | 0.847 | 0.915 | 0.949 | 0.029 | |
| Base + BPM | 0.890 | 0.836 | 0.912 | 0.950 | 0.034 | |
| Base + CMM + BPM | 0.901 | 0.852 | 0.919 | 0.951 | 0.027 | |
| ColonDB | Base | 0.676 | 0.615 | 0.806 | 0.810 | 0.043 |
| Base + CMM | 0.720 | 0.652 | 0.822 | 0.855 | 0.040 | |
| Base + BPM | 0.671 | 0.609 | 0.803 | 0.813 | 0.044 | |
| Base + CMM + BPM | 0.754 | 0.689 | 0.849 | 0.897 | 0.037 | |
| ETIS | Base | 0.628 | 0.567 | 0.799 | 0.769 | 0.028 |
| Base + CMM | 0.675 | 0.609 | 0.832 | 0.872 | 0.021 | |
| Base + BPM | 0.664 | 0.597 | 0.816 | 0.810 | 0.018 | |
| Base + CMM + BPM | 0.711 | 0.648 | 0.844 | 0.887 | 0.015 | |
| CVC-T | Base | 0.830 | 0.752 | 0.890 | 0.931 | 0.015 |
| Base + CMM | 0.887 | 0.819 | 0.936 | 0.985 | 0.008 | |
| Base + BPM | 0.859 | 0.784 | 0.915 | 0.961 | 0.009 | |
| Base + CMM + BPM | 0.906 | 0.843 | 0.945 | 0.988 | 0.006 |
| Dataset | BPM Combination | #Params | mDice | mIoU | MAE | ||
|---|---|---|---|---|---|---|---|
| CVC-ClinicDB | , | 31.10 M | 0.927 | 0.884 | 0.944 | 0.983 | 0.0070 |
| , | 31.10 M | 0.930 | 0.889 | 0.949 | 0.984 | 0.0068 | |
| , | 31.10 M | 0.933 | 0.891 | 0.956 | 0.985 | 0.0066 | |
| , | 33.32 M | 0.928 | 0.885 | 0.954 | 0.982 | 0.0070 | |
| Kvasir | , | 31.10 M | 0.905 | 0.855 | 0.924 | 0.955 | 0.027 |
| , | 31.10 M | 0.891 | 0.842 | 0.915 | 0.945 | 0.029 | |
| , | 31.10 M | 0.901 | 0.852 | 0.919 | 0.951 | 0.027 | |
| , | 33.32 M | 0.903 | 0.852 | 0.920 | 0.954 | 0.028 | |
| ColonDB | , | 31.10 M | 0.762 | 0.697 | 0.852 | 0.883 | 0.037 |
| , | 31.10 M | 0.738 | 0.677 | 0.840 | 0.864 | 0.039 | |
| , | 31.10 M | 0.754 | 0.689 | 0.849 | 0.897 | 0.037 | |
| , | 33.32 M | 0.753 | 0.684 | 0.849 | 0.887 | 0.037 | |
| ETIS | , | 31.10 M | 0.679 | 0.619 | 0.830 | 0.830 | 0.016 |
| , | 31.10 M | 0.714 | 0.656 | 0.845 | 0.858 | 0.015 | |
| , | 31.10 M | 0.711 | 0.648 | 0.844 | 0.887 | 0.015 | |
| , | 33.32 M | 0.697 | 0.629 | 0.840 | 0.851 | 0.024 | |
| CVC-T | , | 31.10 M | 0.880 | 0.814 | 0.933 | 0.979 | 0.007 |
| , | 31.10 M | 0.898 | 0.835 | 0.939 | 0.981 | 0.007 | |
| , | 31.10 M | 0.906 | 0.843 | 0.945 | 0.988 | 0.006 | |
| , | 33.32 M | 0.869 | 0.800 | 0.926 | 0.984 | 0.011 |
| Loss Function | Kvasir | ColonDB | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | MAE | mDice | mIoU | MAE | |||||
| BCE | 0.868 | 0.805 | 0.913 | 0.948 | 0.036 | 0.703 | 0.632 | 0.841 | 0.862 | 0.042 |
| IoU | 0.886 | 0.838 | 0.902 | 0.937 | 0.038 | 0.729 | 0.664 | 0.829 | 0.848 | 0.045 |
| L1 | 0.886 | 0.835 | 0.903 | 0.941 | 0.032 | 0.751 | 0.681 | 0.843 | 0.869 | 0.038 |
| wBCE | 0.876 | 0.815 | 0.915 | 0.945 | 0.033 | 0.731 | 0.658 | 0.852 | 0.878 | 0.042 |
| wIoU | 0.892 | 0.842 | 0.905 | 0.940 | 0.037 | 0.762 | 0.699 | 0.845 | 0.870 | 0.041 |
| Hybrid (wBCE + wIoU + L1) | 0.901 | 0.852 | 0.919 | 0.951 | 0.027 | 0.754 | 0.689 | 0.849 | 0.897 | 0.037 |
| / / | Ver. | CVC-ClinicDB | ETIS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | MAE | mDice | mIoU | MAE | ||||||
| 1.0 / 1.0 / 0.0 | (1) | 0.929 | 0.886 | 0.956 | 0.984 | 0.008 | 0.700 | 0.639 | 0.839 | 0.880 | 0.020 |
| 1.0 / 0.5 / 0.5 | (2) | 0.933 | 0.892 | 0.945 | 0.981 | 0.008 | 0.712 | 0.649 | 0.833 | 0.863 | 0.018 |
| 0.5 / 1.0 / 0.5 | (3) | 0.926 | 0.884 | 0.959 | 0.986 | 0.009 | 0.689 | 0.630 | 0.844 | 0.886 | 0.021 |
| 0.5 / 0.5 / 1.0 | (4) | 0.925 | 0.883 | 0.947 | 0.980 | 0.007 | 0.682 | 0.624 | 0.835 | 0.860 | 0.017 |
| 1.0 / 1.0 / 1.0 | (5) | 0.933 | 0.891 | 0.956 | 0.985 | 0.007 | 0.711 | 0.648 | 0.844 | 0.887 | 0.015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, W.; Lee, M.S.; Han, S.W. COMMA: Propagating Complementary Multi-Level Aggregation Network for Polyp Segmentation. Appl. Sci. 2022, 12, 2114. https://doi.org/10.3390/app12042114
Shin W, Lee MS, Han SW. COMMA: Propagating Complementary Multi-Level Aggregation Network for Polyp Segmentation. Applied Sciences. 2022; 12(4):2114. https://doi.org/10.3390/app12042114
Chicago/Turabian StyleShin, Wooseok, Min Seok Lee, and Sung Won Han. 2022. "COMMA: Propagating Complementary Multi-Level Aggregation Network for Polyp Segmentation" Applied Sciences 12, no. 4: 2114. https://doi.org/10.3390/app12042114
APA StyleShin, W., Lee, M. S., & Han, S. W. (2022). COMMA: Propagating Complementary Multi-Level Aggregation Network for Polyp Segmentation. Applied Sciences, 12(4), 2114. https://doi.org/10.3390/app12042114






