Abstract
Basal Cell Carcinoma (BCC) is the most frequent skin cancer and its increasing incidence is producing a high overload in dermatology services. In this sense, it is convenient to aid physicians in detecting it soon. Thus, in this paper, we propose a tool for the detection of BCC to provide a prioritization in the teledermatology consultation. Firstly, we analyze if a previous segmentation of the lesion improves the ulterior classification of the lesion. Secondly, we analyze three deep neural networks and ensemble architectures to distinguish between BCC and nevus, and BCC and other skin lesions. The best segmentation results are obtained with a SegNet deep neural network. A 98% accuracy for distinguishing BCC from nevus and a 95% accuracy classifying BCC vs. all lesions have been obtained. The proposed algorithm outperforms the winner of the challenge ISIC 2019 in almost all the metrics. Finally, we can conclude that when deep neural networks are used to classify, a previous segmentation of the lesion does not improve the classification results. Likewise, the ensemble of different neural network configurations improves the classification performance compared with individual neural network classifiers. Regarding the segmentation step, supervised deep learning-based methods outperform unsupervised ones.
1. Introduction
Skin cancer is the most common cancer in the United States and worldwide [1]. Although the majority of the works in the literature are focused on melanoma detection, the most common malign skin lesion is the non-melanoma skin cancer (NMSC). Over 95% of NMSC cases are Basal Cell Carcinoma (BCC) and cutaneous squamous cell carcinoma (SCC) [2]. Specifically, BCC has an incidence higher than 70% [3] among all skin cancer, it has the best validated clinical criteria for its diagnosis [4], and it presents the higher variability in the presence of these dermoscopic criteria.
The detection of NMSC can be performed by visual inspection by a skilled dermatologist, but there are many benign lesions that can be confused with NMSC, leading to unnecessary biopsies, in a proportion of five biopsies versus one actual cancer case [5].
The increase in the incidence of BCC is provoking an overload for dermatologists. In the Andalusian Health System, teledermatology is being implanted. Nowadays, 315 demands of teledermatology consultation per month are received and 210 receive diagnostic criteria of BCC. Thus, a Computer Aided Diagnosis (CAD) tool that assists general practitioner physicians and provides a prioritization in the teledermatology consultation would have great utility.
Different kinds of images have been traditionally used in order to classify NMSC automatically (spectroscopy, optical coherence tomography, etc.). However, the simplest one and most used is the digital dermoscopy, that is, a digital color photograph, enhanced by a dermoscope.
Lately, and due to the availability of databases due to the challenges proposed by ISIC [6], the use of artificial intelligence methods, and in particular, the use of deep learning neural networks, have become very popular in dermatology. In this sense, this paper is focused on machine learning algorithms, in particular, deep learning ones, and using dermatoscopic images from ISIC challenges [6].
Most of the works published have been focused on melanoma segmentation and classification [7,8]. On the contrary, much less work has been devoted to NMSC detection. Marka et al. performed a systematic analysis of existing methods for automatic detection of NMSC in 2019. They came to the conclusion that, although most of the methods attain an accuracy similar to the reported diagnostic accuracy of a dermatologist, all the methods require a clinical study to assess the validity of the methods in a real clinical scenario [9]. There are three methods that attain the best classification metrics according to this study. Wahba et al., in 2017, reached 100% in all the metrics but the test set was only 10 images [10]. The same authors, in 2018, tested their methods with an extended database, obtaining the same results [11]. Møllersen et al. also achieved 100% sensitivity, but their specificity was 12% [12]. Sarkar et al. applied deep neural networks to differentiate between BCC, SCC and benign lesions. They achieved an AUROC score of 0.997, 1 and 0.998, respectively [13]. Pangti et al. analyzed the performance of a deep learning-based application for the diagnosis of BCC, as compared to dermatologist and non-dermatologist physicians [14].
In Han et al., 12 skin diseases were classified, employing a deep learning algorithm. They used three databases and concluded that the tested algorithm performance is comparable to that obtained by 16 dermatologists. One of these skin diseases is BCC [15]. Following this comparison, Carcagni et al. [16] and Zhou et al. [17] also proposed methods based on deep learning to perform a multiclassification of skin diseases. Carcagni et al. proposed an ensemble approach and compared it with the original Densenet-121, obtaining a better performance.
Sies et al. [18] tested two market-approved tools, one employed a Machine Learning (ML) technique and one is based on Convolutional Neural Networks (CNN). Although they tested 1981 skin lesions, only 28 lesions were BCC. The ML algorithm detected only 5 in 28 BCC lesions, whereas the CNN-based algorithm detected 27 in 28 [18]. Dorj et al. use a pre-trained AlexNet convolutional network to extract the features that feed an SVM classifier, in order to classify among four kinds of cancers, including BCC [19].
Recent advances in the field of histopathological and microscopic image analysis, dedicated to the BCC detection, can be found at [20,21,22], where the authors use deep learning techniques to detect, classify and identify its patterns. However, our approach covers BCC classification, focusing on distinguishing BCC from Nevus, and employing dermoscopic images. From a clinical point of view, it is very interesting to differentiate BCC from nevus, because both represent the most frequent skin lesions appearing at primary health centers, and a good detection of these types of lesions could lead to a more efficient clinical management, performing a first prioritization of the images that arrive from the Primary Health Center by teledermatology.
The main contribution of the paper is that it performs a thorough analysis of deep learning techniques, applied to BCC segmentation and classification.
In addition, to the best of our knowledge, there are no previous works that evaluate the influence of a previous segmentation in the classification of skin lesions with a deep neural network.
2. Materials and Methods
In order to segment and classify the lesion, several experiments have been conducted that try to evaluate how important the segmentation is for an ulterior classification. A comparison between deep learning methods and classical segmentation algorithms is presented.
Regarding the classification task, different deep learning architectures in the following two different classification scenarios are tested: BCC vs. Nevus, BCC vs. All lesions.
2.1. Lesion Segmentation
Skin lesion segmentation becomes a challenging task due to the presence of hair, bubbles, different illumination conditions, blurry boundaries, blood vessels, scars or different skin colors, thus, the segmentation step turns into a very delicate and complex process.
Over the years, many techniques that successfully overcome the segmentation challenges have been developed. Unsupervised segmentation methods, such as thresholding, edge-based, region-based or energy minimization-based ones, and supervised methods, such as support vector machines (SVM), Bayes-based, or deep learning-based segmentation methods (DLBSM) have been successfully tested over any kind of images [23,24].
Lately, regarding dermoscopic images, many works have been focused on deep learning-based methodologies [25]. This kind of segmentation technique combines low-level feature information with high-level semantic information [26] and takes the advantage of its learning capacities, focusing on its learning properties to identify structures that allow us to segment the image. DLBSM allow us to segment images with low contrast, different intensity distribution or images with artifacts [27].
In this paper, we compare unsupervised with supervised segmentation techniques. In fact, we compare the performance of one unsupervised method based on energy minimization, and two segmentation methods based on deep learning. More specifically, the two supervised methods consist of the following: (1) A CNN as feature extractor combined with a classic segmentation method (thresholding), and (2) Semantic neural network (SegNet). These three methods were tested over ISIC-2017 database [28].
2.1.1. Unsupervised Method: Energy Minimization Based Algorithm
Unsupervised methods do not require a labelled training dataset. One of the main advantages they have is the low computational cost [29]. Another one is that they do not need a large database as no training is performed. In contrast, its performance could not be robust for low quality images, or some interaction with the user is needed to achieve a good performance.
There are many state-of-the-art unsupervised algorithms in the literature and some of them have been explored in this paper (edge-based active contour, region-based active contours, segmentation based on convex optimization). However, for the final analysis an energy minimization method was chosen because it is less dependent on the parameter setting. In this kind of algorithm, an energy measure, which includes region and boundary information, is minimized to solve the segmentation problem. Over recent years, energy minimization algorithms based on convex relaxation have been developed [29,30]. This paper presents an algorithm using convex relaxation based on a previous work by the authors [31,32]. The original idea was proposed by Papadakis and Rabin [33]. It consists of posing the problem of segmentation as a problem of minimization of a convex energy function. In this energy function, the distance between the histograms of each region within the image and histogram models is minimized.
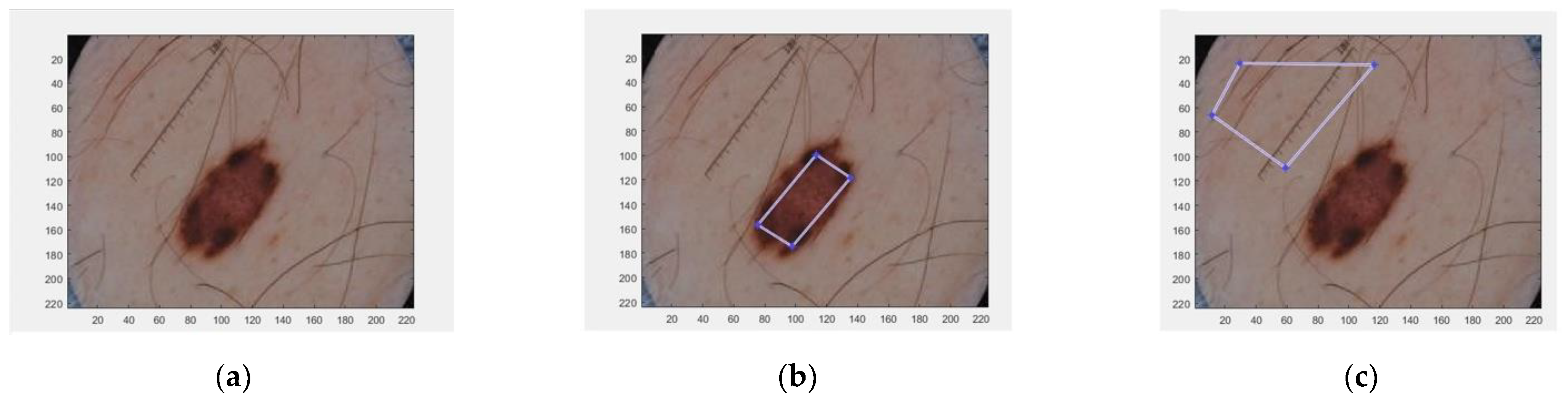
Two histogram models were defined for each dermoscopic image, one for the foreground (the lesion) and the other for the background (the skin). To generate these two histogram models, the algorithm requires a manual selection of a partial part of each region (Figure 1).
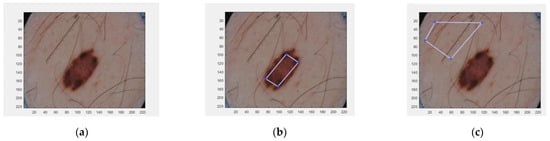
Figure 1.
Selected areas of dermoscopic images for calculating the histograms. (a) Original image; (b) Selected region inside the lesion; (c) Selected region in the healthy skin.
2.1.2. Supervised Methods
Supervised methods need a training data set in order to fix the parameters of the classifier. Some of these segmentation methods are based on SVMs, Bayes classifier, decision trees (DTs) or artificial neural networks (ANN) [34].
As supervised algorithms, two different methods were chosen. The first method has been chosen because it segments by employing the information provided by the deep features of a CNN. This fact has the advantage that a small training database is required and even a pre-trained CNN may be utilized. The second supervised segmentation is a fully convolutional neural network, which has been demonstrated to be effective in medical image segmentation. More specifically, SegNet has been chosen because it is state of the art in the field.
Segmentation from Feature Images of a CNN
A CNN possesses convolutional layers that provide a wide information about global and local features of an image. Hence, the deepest convolutional layers contain information of the global, abstract and conceptual features, whereas the lower convolutional layers give information about the local structure, which is relevant for the segmentation process [35]. Likewise, convolutional layers can be used to obtain the image features [36].
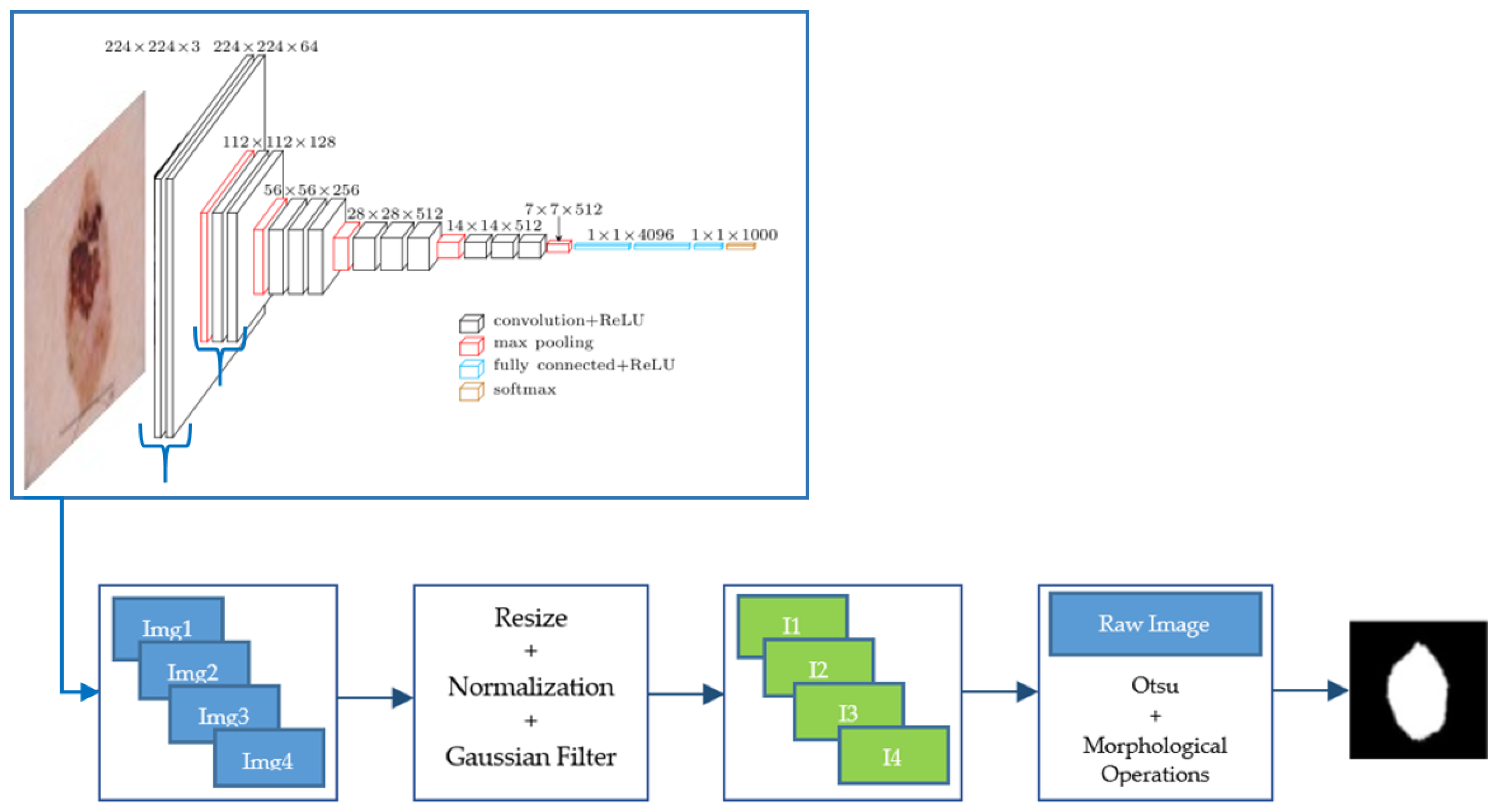
We used a VGG-16 pre-trained with ImageNet database. A set of images from the fourth convolutional layer of VGG-16 network was extracted. These images were normalized and filtered by applying a Gaussian filter with standard deviation equal to 2, before being added. Finally, a threshold using Otsu’s method and morphological operations (dilation and hole filling) was applied to obtain the final segmentation result. A scheme of this segmentation algorithm is presented in Figure 2.
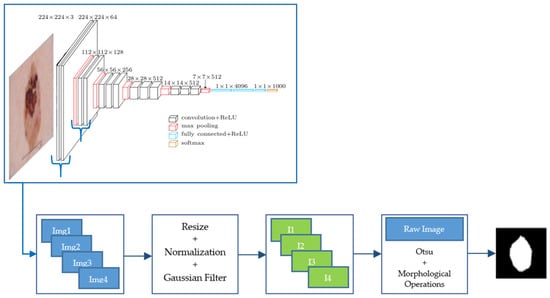
Figure 2.
Scheme of this segmentation algorithm from feature images of a CNN.
Semantic Segmentation with SegNet Deep Neural Network
Semantic Segmentation allows us to identify an object in an image by classifying each pixel into a labeled class, which is called pixel-wise labeling. As is described by Badrinarayanan et al. [37], SegNet is a Fully Convolutional Network (FCN) architecture whose encoder is topologically similar to the convolutional layers from VGG-16, but without its fully connected layers. The convolutions are performed with a filter bank. The last layer of the decoder works as a soft-max classifier, which allows us to obtain the predicted segmentation labels for each pixel as output, where each label is associated with an existing class. SegNet admits as input a map of features or an image.
2.2. Lesion Classification
The classification part of the paper will try to differentiate between different types of skin lesions, being the motivation of the paper, the detection of BCC.
We present the following two types of classifications:
- BCC vs. Nevus;
- BCC vs. All, where the term “All” groups the following skin lesions: nevus, benign keratosis, dermatofibroma, melanoma, SCC, actinic keratosis and vascular lesion.
From a clinical point of view, it is very interesting to differentiate BCC from nevus, because both represent the most frequent skin lesions appearing at primary health centers, and a good detection of these types of lesions could lead to a more efficient clinical management.
In the two classification experiments, we have tested how the introduction of previously segmented images could affect the classification.
A wide number of experiments are conducted in order to check which configuration could be better to solve this difficult problem. To this purpose several classification approaches have been proposed, as follows:
- The use of a VGG-16 neural network. VGG-16 consists of 16 convolutional layers and is very appealing because of its very uniform architecture [38].
- The use of a ResNet50 neural network. It is a convolutional neural network with 50 layers. It is a type of Residual Network and it first introduced the concept of skip connection [39].
- The use of an InceptionV3 neural network. InceptionV3 is another type of CNN developed by Google. It is 48 layers deep [40].
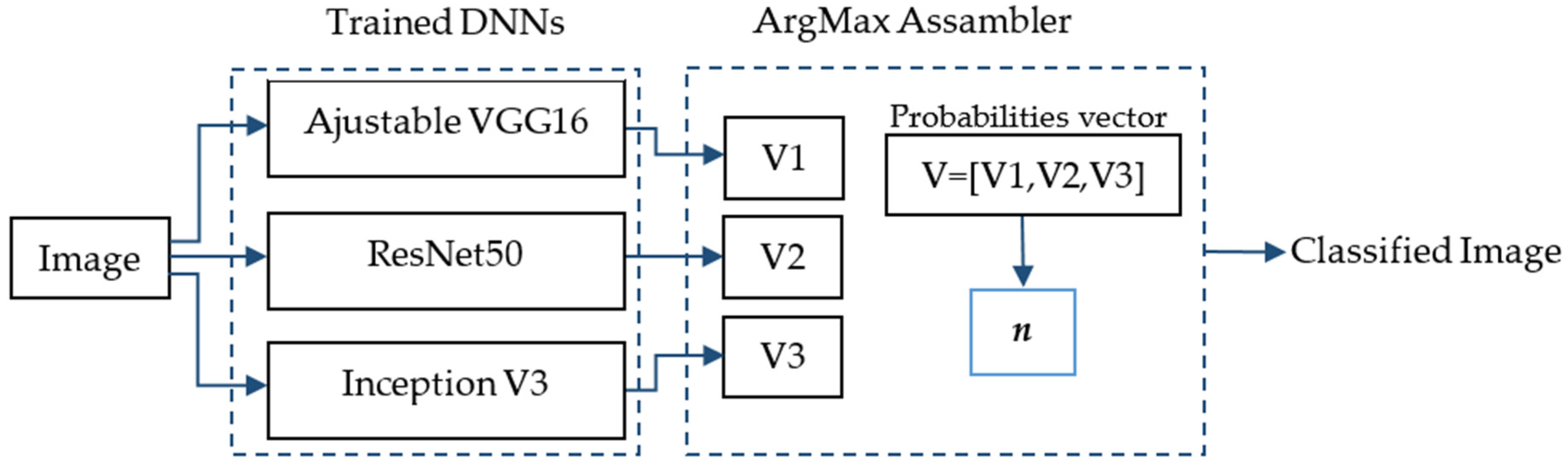
- The use of an ensemble of the three neural networks using the maximum argument. The ArgMax ensemble calculates, for each image, the probability of each class from each neural network, and it selects as the output class the one with highest probability among all the neural networks.
- The use of an ensemble of the three neural networks using the mean. In this case the average of the three probabilities for each class belonging to each neural network is calculated. The output class selected is the one with the maximum average value.
In Figure 3, the ArgMax ensemble configuration is shown. After training the three DNNs mentioned above, a vector with the probabilities belonging to each class for each DNN is obtained (Vi, i = 1,2,3). Each in this vector has dimension m × 1, where m is the number of classes. Finally, a new vector is formed V = [V1,V2,V3] of dimensions 3m × 1. The class with the highest probability, denoted by n, is chosen as the predicted class of the lesion.
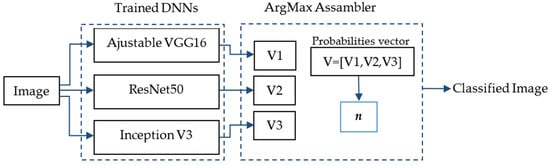
Figure 3.
ArgMax ensemble configuration for the skin lesion classification.
3. Results
3.1. Segmentation Results
3.1.1. Database
To perform the comparison among the different segmentation algorithms, the ISIC 2017 database has been used [6,26]. This database provides 2000 images for training and 600 images for testing, with their corresponding ground truth masks. The ISIC 2018 database for the task “Lesion segmentation” does not provide the ground truth masks for the test set, that is why ISIC 2017 database has been chosen for the part of segmentation. The ISIC 2019 challenge [6] does not include a “Lesion segmentation” task.
For the segmentation algorithms based on convolutional neural networks, a data augmentation process was applied. The data augmentation step consists of random rotations between −30 and 30 degrees; random translations on axes x and y within −10 and 10 interval; random horizontal and vertical reflections; scaling with a random scale factor between 0.9 and 1.1. Finally, all images were randomly sheared in horizontal and vertical angles, specified between 0 and 45 degrees. All these operations picked their random values from a continuous uniform distribution.
After data augmentation, the number of training images was 18,000.
3.1.2. Implementation Details
The three methodologies were implemented on a system with an Intel Core I9-3.6 GHz processor, 32 GB of RAM, and NVIDIA TITAN RTX card.
The SegNet neural network was pre-initialized with layers and weights from a VGG-16 pretrained network, with an ImageNet database from the ImageNet Large Scale Visual Recognition Challenge (ILSVRC). The stochastic gradient descent with momentum (SGDM) optimizer was applied and the training parameters were set as follows: momentum of 0.9, mini-batch size of 5, initial learning rate of 0.001, and weight decay (L2Regularization) of 0.005. The model was trained for 200 epochs.
3.1.3. Results
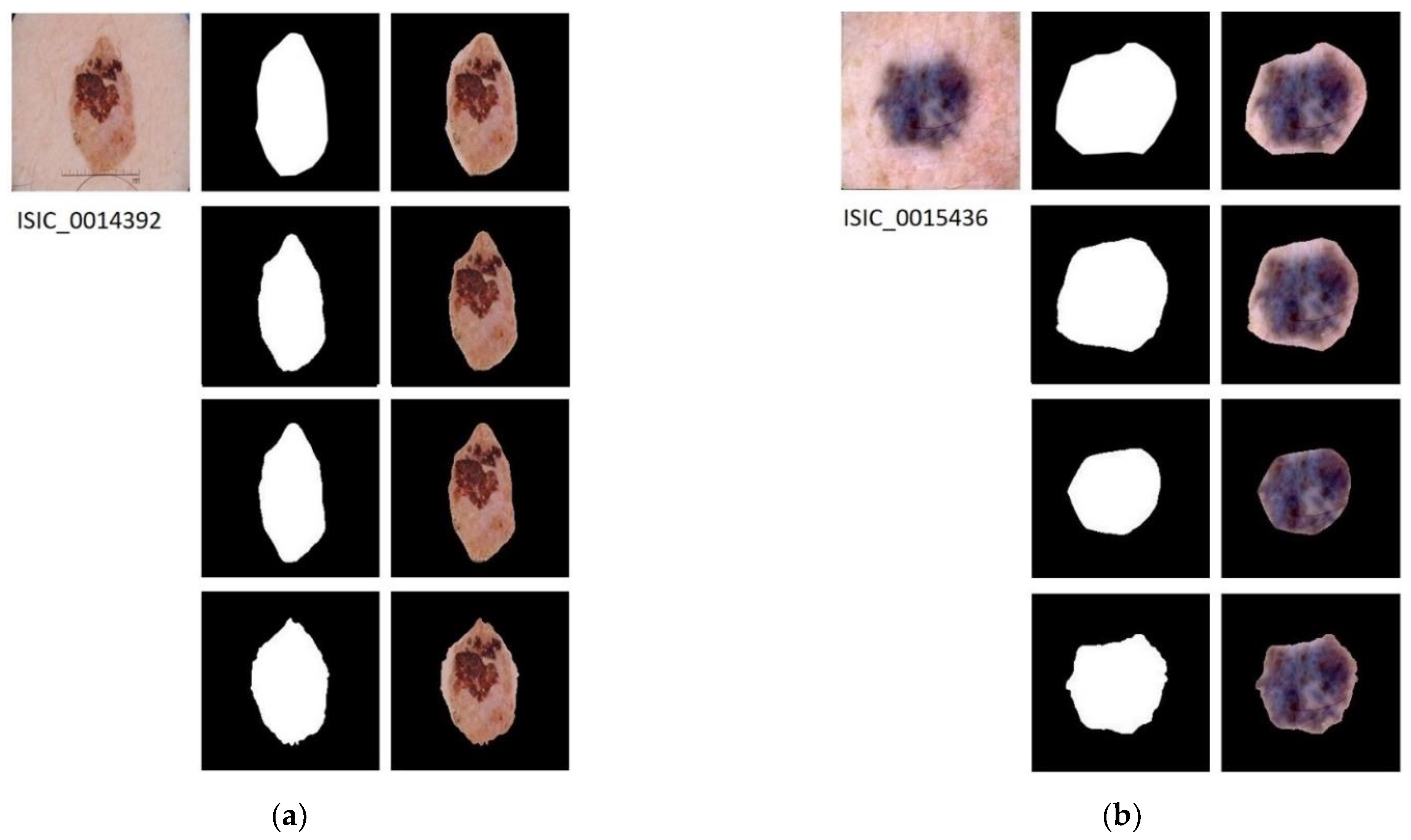
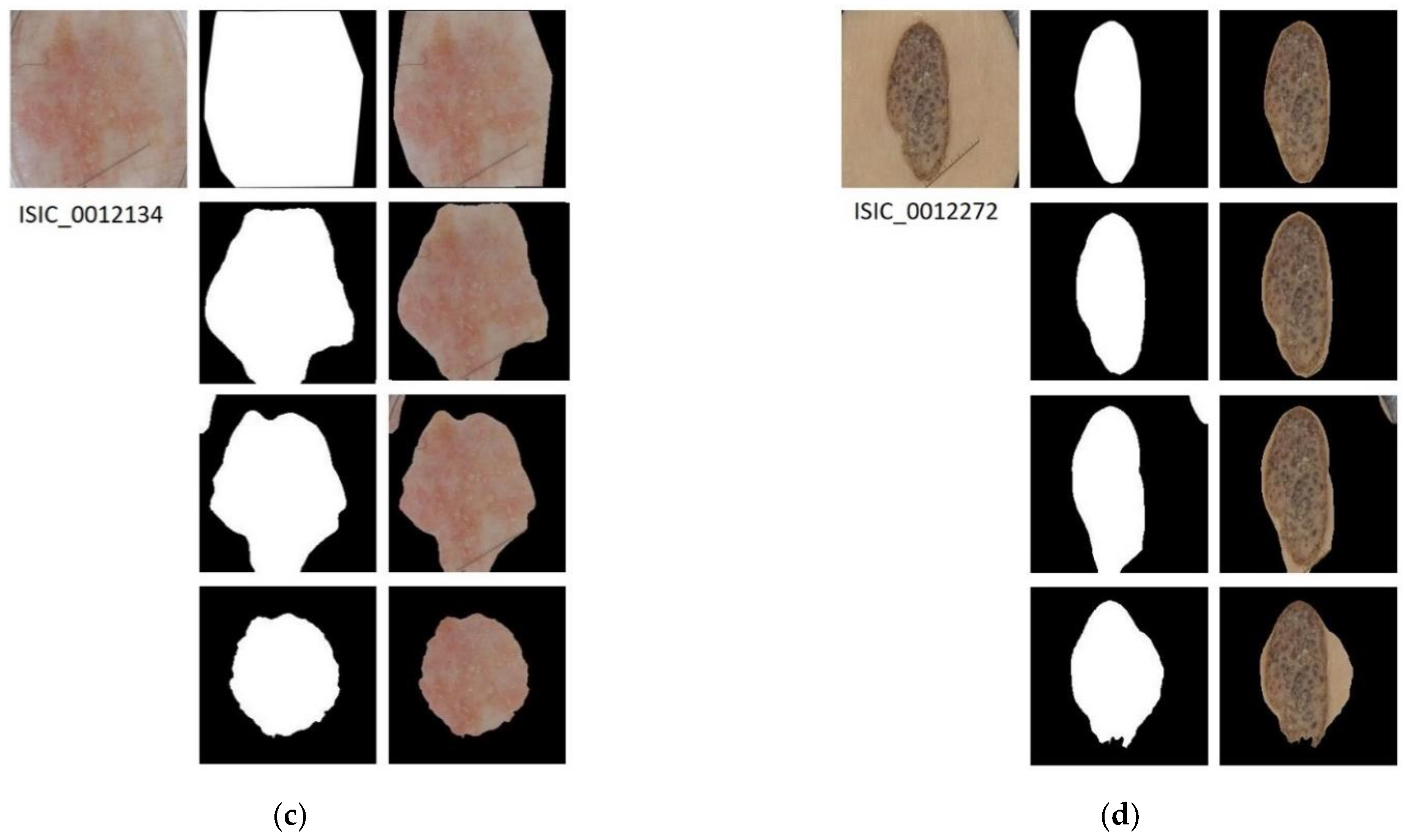
In Figure 4, the results applying the three segmentation methods for four example images are shown.
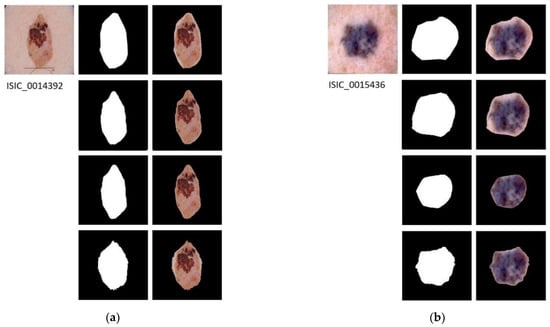
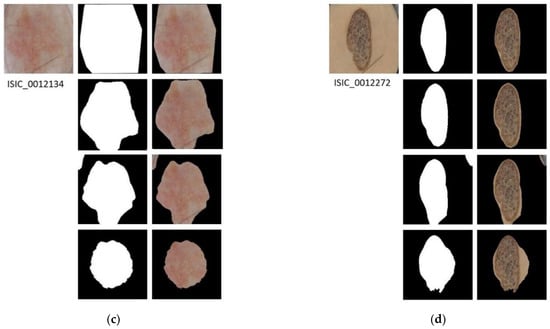
Figure 4.
Segmented Images. For images (a–d): first row shows the original image with its ground truth mask and its corresponding segmentation result; second row shows the results of applying semantic segmentation with SegNet; third row shows the result of energy minimization algorithm via convex optimization; fourth row shows the result of segmentation based on VGG16 feature images.
Table 1 presents the performance parameters for each method, as follows: Dice coefficient (DICE), Jaccard index (JACC), Sensitivity (Se), Specificity (Sp) and Accuracy (Acc). This table presents the results of applying the three methods to the test set of the database, ISIC 2017 (600 images). The best performance is achieved by the SegNet neural network in four out of five parameters.

Table 1.
Segmentation results of the tested methods over the Test set (600 images). Numbers represent the average values of the different segmentation performance parameters calculated over the 600 test images.
Beyond the evaluation parameters, it is relevant to discuss the advantages and drawbacks of each methodology. Although the SegNet neural network obtains the highest performance, the accuracy obtained by the energy minimization algorithm is acceptable, taking into account the resources needed. Nevertheless, SegNet requires a high computational effort for the training but, once the training process has been done, the computational cost of the segmentation process is comparable to the cost required by the other two methods. On the other hand, semantic segmentation via SegNet is an automatic process, which allows us to obtain the segmented lesion without human supervision, as well as the segmentation based on feature images from VGG16. In contrast, energy minimization segmentation requires human intervention, slowing down the segmentation process or making it difficult to use, in the case of large databases.
In Table 2, a comparison with other methods published in the literature is presented. This table shows that the segmentation with SegNet attains competitive results. This justifies that this technique can be chosen as a good segmentation method, in order to evaluate the convenience of including a segmentation step before the classification of the lesions.

Table 2.
Segmentation results of benchmark methods over the Test set (600 images). The results obtained with the SegNet neural network have also been included to facilitate the comparison.
3.2. Classification Results
3.2.1. Database
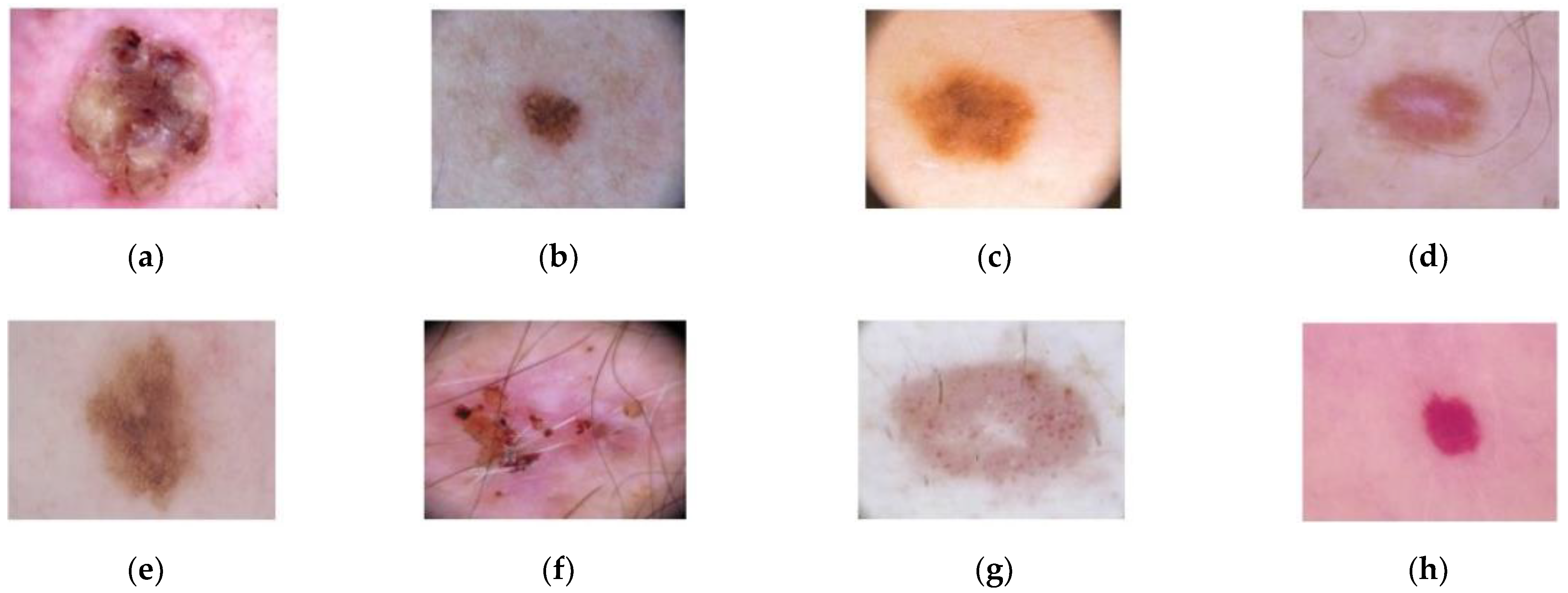
For the classification step, the ISIC-2019 database has been used [44,45], which contains 25,331 dermoscopic images. This database consists of the following lesions: actinic keratosis (867 images), Basal Cell Carcinoma (3323 images), benign keratosis (2624 images), dermatofibroma (239 images), melanoma (4522 images), nevus (12,875 images), squamous cell carcinoma (628 images), and vascular lesion (253 images). An example of these dermoscopic images is shown in Figure 5.
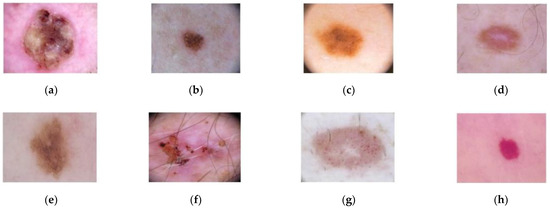
Figure 5.
Dermoscopic images from ISIC-2019 database. (a) Squamous cell carcinoma; (b) Nevus; (c) Melanoma; (d) Dermofibroma; (e) Benign keratosis; (f) Basal Cell Carcinoma; (g) Actinic keratosis; (h) Vascular lesion.
In order to train the convolutional neural networks used to classify the lesions, the database was balanced by a data augmentation process. Data augmentation was carried out by mirror operations and rotations of 36 degrees over each image. The test images were not modified.
As mentioned above, two different experiments were carried out, hence, the database was balanced in two different ways.
For the classification of BCC vs. Nevus, after data augmentation, the following number of images were obtained: 8982 images for training and 1287 for validation, for each class. In this sense, for all the classes, except nevus, the number of images was artificially augmented by the data augmentation process described above. For the nevus class, a downsampling process has been carried out, randomly selecting 10,269 images in total (8982 training + 1287 validation) out of the 12,875 nevus images that the ISIC provides.
For BCC vs. All lesions, the balanced dataset is constituted as follows: 15,419 training images and 2199 validation images, for each of the two classes (BCC and the rest of the types). In this case, data augmentation was applied for the BCC class, increasing the number of nevus images up to 17,618 in total (training and validation set). For each class belonging to the remaining seven classes, the number of images was fixed to 2517 images in total for each class.
3.2.2. Classification Results of BCC vs. Nevus
The database consists of 23,780 images in total, after the data augmentation. The training set consists of 17,964 images, the validation set consists of 2574 images and there are 3242 images for the test process. The results are described in Table 3 and Table 4.

Table 3.
Performance parameters for the different classifiers without the previous segmentation of the image when classifying BCC vs. Nevus. Se: Sensitivity, Sp: Specificity, Pre: Precision, FPR: False Positive Rate, Acc: Accuracy. The highest values are shown in bold numbers.

Table 4.
Performance parameters for the different classifiers with the previous segmentation of the image by using a SegNet neural network when classifying BCC vs. Nevus. Se: Sensitivity, Sp: Specificity, Pre: Precision, FPR: False Positive Rate, Acc: Accuracy. The highest values are shown in bold numbers.
Transfer learning has been applied to the pre-trained neural network configurations.
If we denote true positives as TP, true negatives as TN, false positives as FP and false negatives as FN, the classification performance parameters we have used are defined as follows:
Sensitivity, which represents the proportion of people who test positive among all those who actually have the disease: Se = TP/(TP + FN).
Specificity, which is the proportion of people who test negative among all those who actually do not have that disease: Sp = TN/(TN + FP).
Precision, which represents the probability that following a positive test result, that individual will truly have that specific disease: Pre = TP/(TP + FP).
False positive rate, which is calculated as the ratio between the number of negative events wrongly categorized as positive (false positives) and the total number of actual negative events: FPR = FP/(FP + TN).
Accuracy, which represents the proportion of true positive results (both true positive and true negative) in the selected population: Acc = (TN + TP)/(TN + TP + FN + FP).
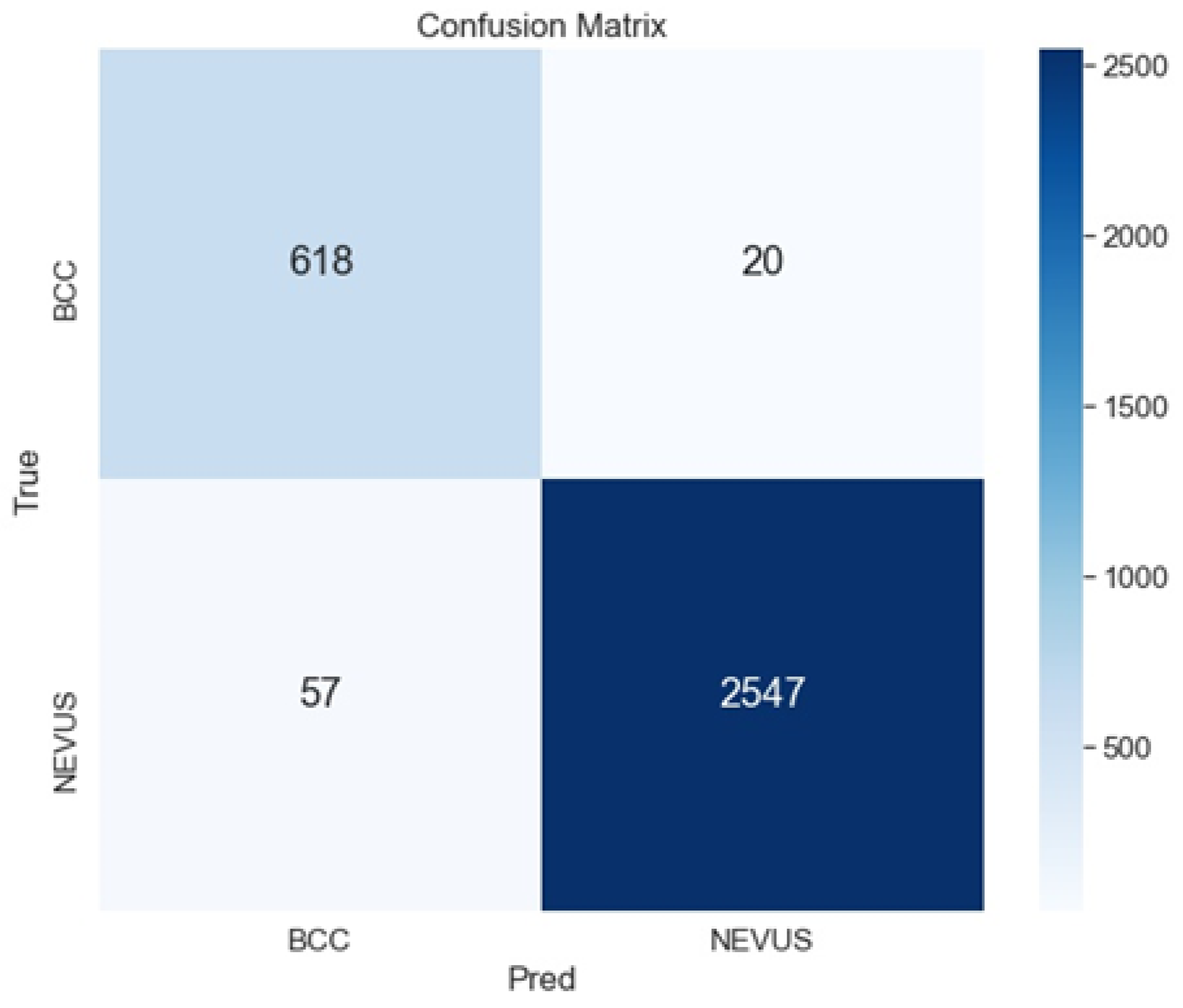
As shown in Table 3 and Table 4, the best configuration is the ensemble ArgMax, without previous segmentation of the image. For this case, the confusion matrix is shown in Figure 6. The confusion matrix shows the classification performed by the specialists (named True), versus the predicted classification performed by the ensemble ArgMax classification tool. The larger the numbers in the diagonal are, the better the classification results are.
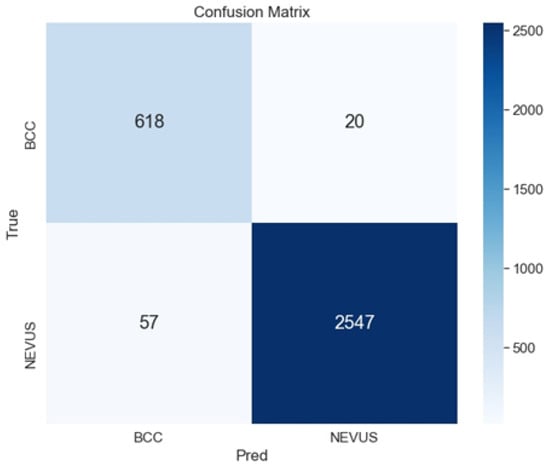
Figure 6.
Confusion matrix for the classification of BCC vs. Nevus using the ensemble ArgMax and without previous segmentation.
3.2.3. Classification Results of BCC vs. All Lesions
The total number of images of the database, after data augmentation, was 40,302. The training set was composed of 30,838 images, the validation set, 4398 images, and the test set, 5066 images. The different classes were balanced after applying the data augmentation step. Results are summarized in Table 5 and Table 6.

Table 5.
Performance metrics for the different classifiers without the previous segmentation of the image when classifying BCC vs. All lesions. Se: Sensitivity, Sp: Specificity, Pre: Precision, FPR: False Positive Rate, Acc: Accuracy. The highest values are shown in bold numbers.

Table 6.
Performance parameters for the different classifiers with the previous segmentation of the image with a SegNet when classifying BCC vs. All lesions. Se: Sensitivity, Sp: Specificity, Pre: Precision, FPR: False Positive Rate, Acc: Accuracy. The highest values are shown in bold numbers.
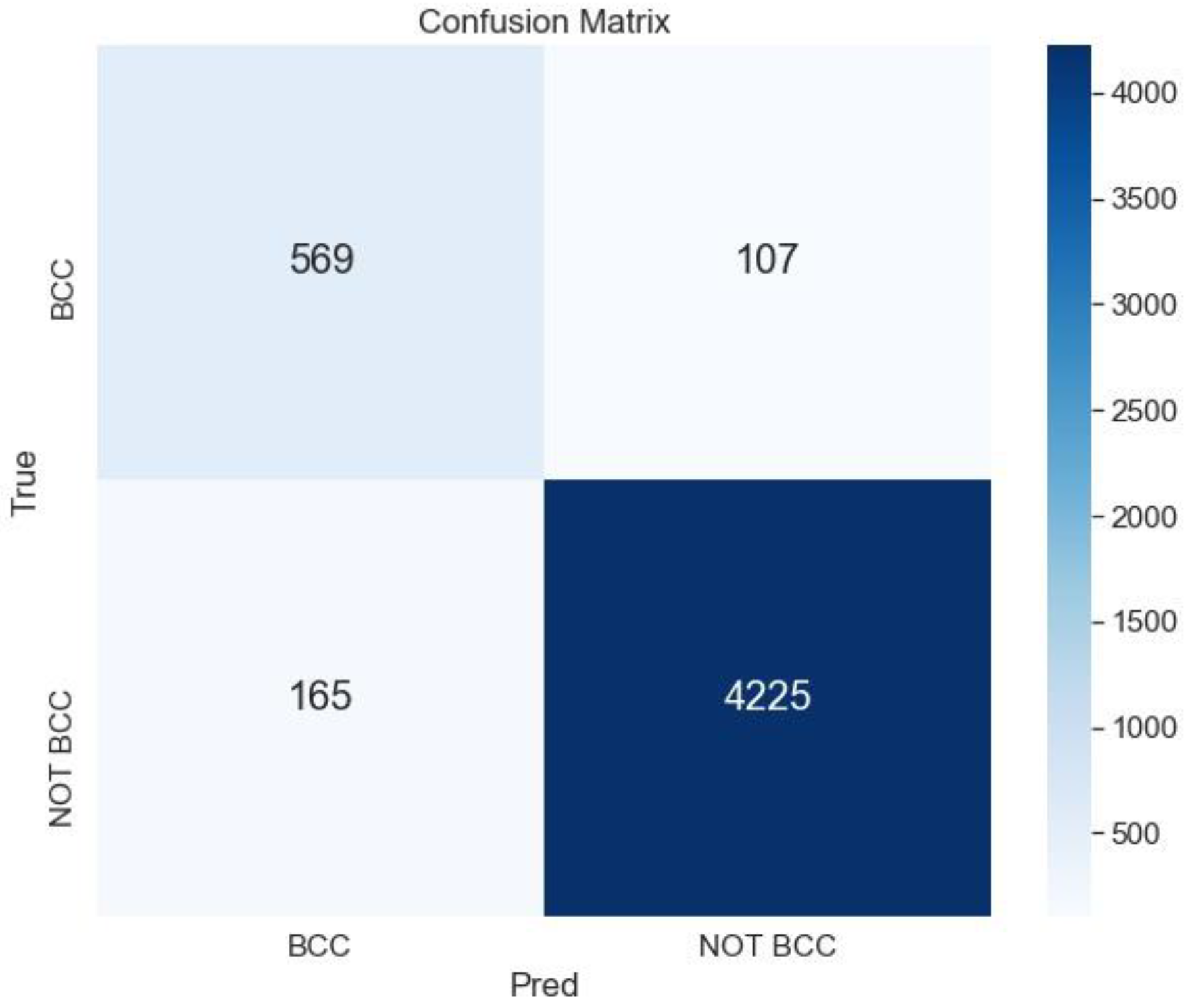
The best result is obtained for the ensemble ArgMax without previous segmentation; the confusion matrix for this case is shown in Figure 7.
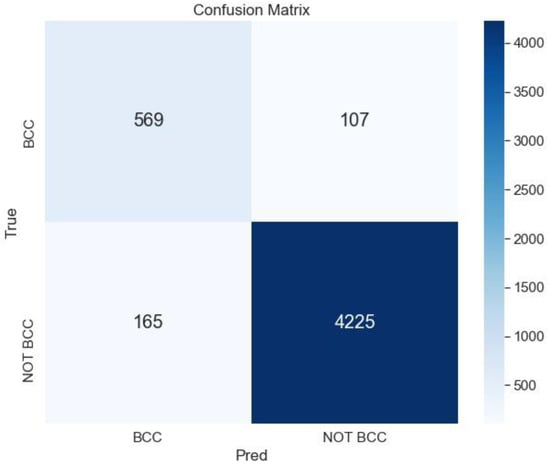
Figure 7.
Confusion matrix for the classification BCC vs. All lesions using the ensemble ArgMax without previous segmentation of the lesion.
To the best of our knowledge, there are no published works devoted to classifying BCC vs. non-BCC lesions or BCC vs. nevus. Thus, in order to compare the proposed method with the state of the art, we have performed the classification proposed in the ISIC 2019 Challenge, where eight different lesions are classified. Results are shown in Table 7. We have used as a benchmark algorithm, the winner of the ISIC 2019 Challenge [6,46]. As can be observed, the proposed method outperforms the benchmark algorithm in almost all the metrics.

Table 7.
Performance parameters for the winner of the ISIC Challenge 2019 [46] and for the proposed method (Argmax ensemble without previous segmentation) when classifying into eight types of lesions. Se: Sensitivity, Sp: Specificity, Acc: Accuracy.
The reason for these good results may be found in different factors. First of all, the three networks chosen for the ensemble are networks of proven effectiveness (VGG16, ResNet and Inceptionv3 were the winners in the ILSVRC 2014 or ILSVRC 2015 challenges). On the other hand, these networks do not have an excessive number of parameters, which would require a large training database to obtain good classification results. Finally, the ensemble of these three winning networks leads to the obtaining of better results.
4. Discussion
There are few papers devoted to the detection of BCC in the literature [10,11,12,13], and less that apply deep neural networks to solve this problem. One of the main reasons is the lack of public databases with BCC lesions, with contours delineation or with labelled dermoscopic criteria.
From a clinical point of view, it is very convenient to distinguish between BCC and nevus, due to the high incidence of these two types of lesions. Specifically, in primary health centers, it would be desirable to have an automatic tool, in order to help the non-specialist in the diagnosis and to establish a good priority in the attendance at the dermatology services. To the best of our knowledge this is the first time that a classification between BCC and nevus has been performed.
Most works devoted to segment skin lesions claim that an accurate segmentation is necessary to achieve a proper extraction of features and consequent lesion characterization [47]. However, in this paper, we demonstrated that, when using deep learning methods, it is not advantageous to include a segmentation before classifying the lesion. Actually, we get worse results when segmenting the lesion previously to the classification step, showing that, when using a large database, the previous segmentation of the lesion does not improve the classification results. This suggests that the healthy skin surrounding the lesion may contain information significant for the classification. In this sense, other works, such as the one by Teixeira et al., support this statement [48].
The main limitation of our method is the lack of explainability of the classification. An explanation of the classification, by providing the automatic detection of dermoscopic criteria of BCC, would considerably improve the utility of the method for physicians. To this purpose, we are working on developing a database with the dermoscopic criteria of BCC and a system for the automatic detection of these dermoscopic criteria.
As future research, a clinical study to assess the validity of the methods in a real clinical scenario would be desirable.
5. Conclusions
In this paper, two analyses have been performed. Firstly, a comparison between an unsupervised segmentation method and two supervised segmentation methods, based on deep learning, has been carried out. Secondly, the identification of BCC amongst other types of skin lesions has been performed in the following two different scenarios: with a previous segmentation of the lesion and without segmenting the lesion. To this second task, different deep neural networks have been tested.
Experiments to compare the different segmentation methods show that SegNet architecture has attained the best behavior, obtaining 94% accuracy.
The ISIC 2019 public database [6] has been used to carry out the classification task. A 98% accuracy, 0.84% sensitivity and 0.96% specificity, for distinguishing BCC from nevus, and a 95% accuracy, 0.68% sensitivity and 0.97% specificity, classifying BCC vs. all lesions, have been obtained. Furthermore, the proposed algorithm outperforms the winner of the ISIC 2019 challenge in almost all the metrics, when lesions are classified into eight classes.
In summary, this paper adds important comparison studies, applied to the analysis of BCC, that have not been performed previously. These studies are of interest, because BCC is the skin cancer of highest incidence. First, an analysis of the utility of BCC segmentation to improve classification is carried out, driving to the conclusion that previous segmentation does not improve the classification. Secondly, a tool for the discrimination between BCC and nevus, which is the most common pigmented lesion, is provided. Finally, we have demonstrated that an ensemble of well-known CNN can attain results that can compete with the best methods in the ISIC challenge.
Author Contributions
Conceptualization, P.V., C.S. and B.A.; methodology, P.V. and M.M.; software, P.V. and M.M.; validation, P.V., M.M., C.S. and B.A.; resources, C.S. and B.A.; writing—original draft preparation, P.V.; writing—review and editing, C.S. and B.A.; visualization, P.V.; supervision, C.S. and B.A.; funding acquisition, C.S. and B.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Project DPI2016-81103-R (“Ministerio de Economía y Competitividad”, Spanish Government) and Project US-1381640 (FEDER-US, Regional Government of Andalusia). Manuel Miranda has been hired by “Fondo Social Europeo Iniciativa de Empleo Juvenil” 2019-3-EJ3-83-1 (European Union and Andalusian Government).
Data Availability Statement
Databases employed in this study are available at https://challenge.isic-archive.com/data/ (accessed on 30 December 2021).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviation
The following abbreviations are used in this manuscript:
| Acc | Accuracy |
| AK | Actinic Keratosis |
| ANN | Artificial Neural Networks |
| AUROC | Area Under the Receiver Operating Characteristic |
| BCC | Basal Cell Carcinoma |
| BKL | Bening Keratosis |
| CAD | Computer Aid Diagnosis |
| CNN | Convolutional Neural Network |
| DF | Dermatofibroma |
| DICE | Dice Coefficient |
| DLBSM | Deep Learning-Based Segmentation Methods |
| DNN | Deep Neural Network |
| DT | Decision Trees |
| FCN | Fully Convolutional Network |
| FPR | False Positive Rate |
| ILSVRC | ImageNet Large Scale Visual Recognition Challenge |
| ISIC | International Skin Imaging Collaboration |
| JACC | Jaccard Index |
| MEL | Melanoma |
| ML | Machine Learning |
| NMSC | Non-Melanoma Skin Cancer |
| Pre | Precision |
| RAM | Random Access Memory |
| ResNet | Residual Networks |
| SCC | Squamous Cell Carcinoma |
| Se | Sensitivity |
| SegNet | Semantic Neural Network |
| SGDM | Stochastic Gradient Descent with Momentum |
| Sp | Specificity |
| SVM | Support Vector Machine |
| VASC | Vascular |
References
- Skin Cancer Foundation. Skin Cancer Facts and Statistics. Available online: https://www.skincancer.org/skin-cancer-information/skin-cancer-facts (accessed on 11 October 2021).
- Gillard, M.; Wang, T.S.; Johnson, T.M. Nonmelanoma cutaneous malignancies. In Oncology, An Evidence-Based Approach; Chang, A.E., Ganz, P.A., Hayes, D.F., Kinsella, T., Pass, H.I., Schiller, J.H., Stone, R.M., et al., Eds.; Springer: New York, NY, USA, 2006; pp. 1102–1118. [Google Scholar]
- Ciążyńska, M.; Narbutt, J.; Woźniacka, A.; Lesiak, A. Trends in basal cell carcinoma incidence rates: A 16-year retrospective study of a population in central Poland. Adv. Dermatol. Allergol. 2018, 35, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Peris, K.; Fargnoli, M.C.; Garbe, C.; Kaufman, R.; Bastholt, L.; Basset Seguin, N.; Bataille, V.; Del Marmol, V.; Dummer, R.; Harwood, C.A.; et al. Diagnosis and treatment of basal cell carcinoma: European consensus-based interdisciplinary guidelines. Eur. J. Cancer 2019, 118, 10–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitbart, E.W.; Waldmann, A.; Nolte, S.; Capellaro, M.; Greinert, R.; Volkmer, B.; Katalinic, A. Systematic skin cancer screening in northern Germany. J. Am. Acad. Dermatol. 2012, 66, 201–211. [Google Scholar] [CrossRef] [PubMed]
- International Skin Imaging Collaboration. Available online: https://www.isic-archive.com (accessed on 26 October 2020).
- Kaymak, S.; Esmaili, P.; Serener, A. Deep Learning for Two-Step Classification of Malignant Pigmented Skin Lesions. In Proceedings of the 14th Symposium on Neural Networks and Applications (NEUREL 2018), Belgrade, Serbia, 20–21 November 2018. [Google Scholar] [CrossRef]
- Sultana, N.N.; Puhan, N.B. Recent Deep Learning Methods for Melanoma Detection: A Review. In Proceedings of the 4th International Conference Mathematics and Computing (ICMC 2018), Varanasi, India, 9–11 January 2018. [Google Scholar] [CrossRef]
- Marka, A.; Carter, J.B.; Toto, E.; Hassanpour, S. Automated detection of nonmelanoma skin cancer using digital images: A systematic review. BMC Med. Imaging 2019, 19, 21. [Google Scholar] [CrossRef]
- Wahba, M.A.; Ashour, A.S.; Napoleon, S.A.; Abd Elnaby, M.M.; Guo, Y. Combined empirical mode decomposition and texture features for skin lesion classification using quadratic support vector machine. Health Inf. Sci. Syst. 2017, 5, 10. [Google Scholar] [CrossRef]
- Wahba, M.A.; Ashour, A.S.; Guo, Y.; Napoleon, S.A.; Elnaby, M.M. A novel cumulative level difference mean based GLDM and modified ABCD features ranked using eigenvector centrality approach for four skin lesion types classification. Comput. Methods Programs Biomed. 2018, 165, 163–174. [Google Scholar] [CrossRef]
- Møllersen, K.; Kirchesch, H.; Zortea, M.; Schopf, T.R.; Hindberg, K.; Godtliebsen, F. Computer-aided decision support for melanoma detection applied on melanocytic and nonmelanocytic skin lesions: A comparison of two systems based on automatic analysis of Dermoscopic images. Biomed. Res. Int. 2015, 2015, 579282. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, R.; Chatterjee, C.C.; Hazra, A. A novel approach for automatic diagnosis of skin carcinoma from dermoscopic images using parallel deep residual networks. In Proceedings of the Third International Conference on Advances in Computing and Data Sciences (ICACDS 2019), Ghaziabad, India, 12–13 April 2019. [Google Scholar] [CrossRef]
- Pangti, R.; Chouhan, V.; Mathur, J.; Kumar, S.; Dixit, A.; Gupta, S.; Mahajan, S.; Gupta, A.; Gupta, S. Performance of a deep learning-based application for the diagnosis of BCC in Indian patients as compared to dermatologists and nondermatologists. Int. J. Dermatol. 2020, 60, e51–e52. [Google Scholar] [CrossRef]
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E. Classification of the Clinical Images for Benign and Malignant Cutaneous Tumors Using a Deep Learning Algorithm. J. Investig. Dermatol. 2018, 138, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Carcagni, P.; Leo, M.; Cuna, A.; Mazzeo, P.; Spagnolo, P.; Celeste, G.; Distante, C. Classification of Skin Lesions by Combining Multilevel Learnings in a DenseNet Architecture. In Proceedings of the 20th International Conference Image Analysis and Processing (ICIAP 2019), Trento, Italy, 9–13 September 2019. [Google Scholar] [CrossRef]
- Zhou, H.; Xie, F.; Jiang, Z.; Liu, J.; Wang, S.; Zhu, C. Multi-classification of skin diseases for dermoscopy images using deep learning. In Proceedings of the 2017 IEEE International Conference on Imaging Systems and Techniques (IST 2017), Beijing, China, 18–20 October 2017. [Google Scholar] [CrossRef]
- Sies, K.; Winkler, K.; Fink, C.; Bardehle, F.; Toberer, F.; Buhl, T.; Enk, A.; Blum, A.; Rosenberger, A.; Haenssle, H.A. Past and present of computer-assisted dermoscopic diagnosis: Performance of a conventional image analyser versus a convolutional neural network in a prospective data set of 1981 skin lesions. Eur. J. Cancer 2020, 135, 39–46. [Google Scholar] [CrossRef]
- Dorj, U.O.; Lee, K.K.; Choi, J.Y.; Lee, M. The skin cancer classification using deep convolutional neural network. Multimed. Tools Appl. 2018, 77, 9909–9924. [Google Scholar] [CrossRef]
- Cruz-Roa, A.A.; Arevalo Ovalle, J.E.; Madabhushi, A.; González Osorio, F.A. A Deep Learning Architecture for Image Representation, Visual Interpretability and Automated Basal-Cell Carcinoma Cancer Detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2013, Nagoya, Japan, 22–26 September 2013. [Google Scholar] [CrossRef]
- Campanella, G.; Navarrete-Dechent, C.; Liopyris, K.; Monnier, J.; Aleissa, S.; Minhas, B.; Scope, A.; Longo, C.; Guitera, P.; Pellacani, G.; et al. Deep Learning for Basal Cell Carcinoma Detection for Reflectance Confocal Microscopy. J. Investig. Dermatol. 2022, 142, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kimeswenger, S.; Tschandl, P.; Noack, P.; Hofmarcher, M.; Rumetshofer, E.; Kindermann, H.; Silye, R.; Hochreiter, S.; Kaltenbrunner, M.; Guenova, E.; et al. Artificial neural networks and pathologists recognize basal cell carcinomas based on different histological patterns. Mod. Pathol. 2011, 34, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Pérez Malla, C.U.; Valdés Hernández, M.D.C.; Rachmadi, M.F.; Komura, T. Evaluation of enhanced learning techniques for segmenting ischaemic stroke lesions in brain magnetic resonance perfusion images using a convolutional neural network scheme. Front. Neuroinformatics 2019, 13. [Google Scholar] [CrossRef] [Green Version]
- Kaur, D.; Kaur, Y. Various image segmentation techniques: A review. Int. J. Comput. Sci. Mob. Comput. 2014, 3, 809–814. [Google Scholar] [CrossRef]
- Sreelatha, T.; Subramanyam, M.V.; Giri Prasad, M.N. Early Detection of Skin Cancer Using Melanoma Segmentation technique. J. Med. Syst. 2019, 43, 190. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Jinman, K.; Ahn, E.; Kumar, A.; Feng, D.; Fulham, M. Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recognit. 2019, 85, 78–89. [Google Scholar] [CrossRef] [Green Version]
- Tang, P.; Liang, Q.; Yan, X.; Xiang, S.; Sun, W.; Zhang, D.; Coppola, G. Efficient skin lesion segmentation using separable-Unet with stochastic weight averaging. Comput. Methods Programs Biomed. 2019, 178, 289–301. [Google Scholar] [CrossRef]
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection. A challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018. [Google Scholar] [CrossRef] [Green Version]
- Punithakumar, K.; Yuan, J. A convex max-flow approach to distribution-based figure-ground separation. SIAM J. Imaging Sci. 2012, 5, 1333–1354. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Yuan, J.; Ukwatta, E.; Sun, Y.; Rajchl, M.; Fenster, A. Prostate segmentation: An efficient convex optimization approach with axial symmetry using 3-D TRUS and MR images. IEEE Trans. Med. Imaging 2014, 33, 947–960. [Google Scholar] [CrossRef]
- Pérez-Carrasco, J.A.; Acha, B.; Suárez-Mejías, C.; López-Guerra, J.L.; Serrano, C. Joint segmentation of bones and muscles using an intensity and histogram-based energy minimization approach. Comput. Methods Programs Biomed. 2018, 156, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Mejías, C.; Pérez-Carrasco, J.A.; Serrano, C.; López-Guerra, J.L.; Parra-Calderón, C.; Gómez-Cía, T.; Acha, B. Three-dimensional segmentation of retroperitoneal masses using continuous convex relaxation and accumulated gradient distance for radiotherapy planning. Med. Biol. Eng. Comput. 2017, 55, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, N.; Rabin, J. Convex histogram-based joint image segmentation with regularized optimal transport cost. J. Math. Imaging Vis. 2017, 59, 161–186. [Google Scholar] [CrossRef] [Green Version]
- Al-masni, M.A.; Al-antari, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef]
- Kwasigroch, A.; Mikolajczyk, A.; Grochowski, M. Deep convolutional neural networks as a decision support tool in medical problems–malignant melanoma case study. In Proceedings of the 19th Polish Control Conference (KKA 2014), Kraków, Poland, 12–21 June 2017. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2016), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2016), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Chao, M.; Lo, Y.-C. Automatic skin lesion segmentation with fully convolutional deconvolutional networks. IEEE J. Biomed. Health Inform. 2018, 36, 1876–1886. [Google Scholar] [CrossRef] [Green Version]
- Tschandl, P.; Sinz, C.; Kittler, H. Domain-specific classification pre-trained fully convolutional network encoders for skin lesion segmentation. Comput. Biol. Med. 2019, 104, 111–116. [Google Scholar] [CrossRef]
- Sarker, M.K.; Rashwan, H.A.; Akram, F.; Singh, V.K.; Banu, S.F.; Chowdhury, F.U.H.; Choudhury, K.A.; Chambon, S.; Radeva, P.; Puig, D.; et al. SLSNet: Skin lesion segmentation using a lightweight generative adversarial network. Expert Syst. Appl. 2021, 183, 115433. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Combalia, M.; Codella, N.F.C.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Halpern, A.C.; Puig, S.; Malvehy, J. BCN20000: Dermoscopic Lesions in the Wild. arXiv 2019, arXiv:1908.02288. [Google Scholar]
- Gessert, N.; Nielsen, M.; Shaikh, M.; Werner, R.; Schlaefer, A. Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data. MethodsX 2020, 7, 100864. [Google Scholar] [CrossRef] [PubMed]
- Barata, C.; Celebi, M.E.; Marques, J.S. A survey of feature extraction in dermoscopy image analysis of skin cancer. IEEE J. Biomed. Health Inform. 2019, 23, 1096–1109. [Google Scholar] [CrossRef]
- Teixeira, L.O.; Pereira, R.M.; Bertolini, D.; Oliveira, L.S.; Nanni, L.; Cavalcanti, G.D.C.; Costa, M.G. Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest X-ray images. Sensors 2021, 21, 7116. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).