Featured Application
The results of the current study aid in developing a potent mouthwash formulation to maintain and improve oral health in healthy people.
Abstract
This study analyzed the alteration of oral microbial composition in healthy subjects after using dextranase-containing mouthwash (DMW; Mouthwash formulation I) and dextranase-and-nisin-containing mouthwash (DNMW; Mouthwash formulation II). Eighteen participants were recruited and were randomly allocated to two groups: G1 (DMW user; n = 8) and G2 (DNMW user; n = 10). The subjects were instructed to use the provided mouthwash regularly twice a day for 30 days. The bleeding on probing (BOP), plaque index (PI), probing depth (PBD), and gingival index (GI) were analyzed, and saliva samples were collected before (day 0) and after (day 30) the use of mouthwashes. The saliva metagenomic DNA was extracted and sequenced (next-generation sequencing, Miseq paired-end Illumina 2 × 250 bp platform). The oral microbial community in the pre-and post-treated samples were annotated using QIIME 2™. The results showed the PI and PBD values were significantly reduced in G2 samples. The BOP and GI values of both groups were not significantly altered. The post-treated samples of both groups yielded a reduced amount of microbial DNA. The computed phylogenetic diversity, species richness, and evenness were reduced significantly in the post-treated samples of G2 compared to the post-treated G1 samples. The mouthwash formulations also supported some pathogens’ growth, which indicated that formulations required further improvement. The study needs further experiments to conclude the results. The study suggested that the improved DNMW could be an adjuvant product to improve oral hygiene.
1. Introduction
Oral health is important in daily life since it reflects general physical health [1,2,3]. Oral disease affects teeth and causes gut microbial dysbiosis, cardiovascular illness, urinary tract infections, arthritis, cystic fibrosis, and brain disease [4,5,6,7,8,9]. Even though brushing and flossing can be effective methods of removing plaque, toothbrushes can operate as reservoirs for harmful germs that later inoculate the oral cavity, highlighting the need to properly sanitize and store personal dental hygiene devices [10,11,12,13].
The mouth rinsing custom has been a part of human culture for more than 200 decades. Kamal et al. reported that natural mouthwashes could serve as substitutes to chemical agents such as chlorhexidine, providing effective caries control and safe long-term use [14]. Mouth rinses containing dextranase were superior to 0.12% chlorhexidine mouthwash in preventing plaque accumulation caused by Lipomyces starkeyi KSM 22 and Catenovulum agarivorans MNH15 in human experimental gingivitis [15,16].
Likewise, nisin-containing mouth rinse was proven to be a strong plaque inhibitor. Nisin inhibits the growth of oral bacteria and the development of biofilms by preventing cell attachment and proliferation. Nisin either binds to nonspecific lipids on cytoplasmic membranes or to specific Lipid II, a crucial component of the cell membranes of Gram-positive and Gram-negative bacteria such as Prevotella intermedia (P. intermedia), Porphyromonas gingivalis (P. gingivalis), and Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans). The antibacterial effect of ethanolic extract of licorice root, Triphala, and garlic on Streptococcus mutans (S. mutans) was less effective when compared with chlorhexidine mouthwash [17,18].
The collective genome of microbes in the oral cavity is the oral microbiome linked to taste thresholds such as sweet, sour, salty, and bitter, used to maintain its microbial membership and permanence in the oral cavity [19,20]. Additionally, gingival-related microbiota such as S. mutans, Streptococcus gordonii, Streptococcus oralis, Lacticaseibacillus casei, and P. gingivalis have been discovered to have a critical role in governing human health and illness. Additionally, when it grows abundantly, it substantially supports the growth of Gram-negative bacteria such as P. gingivalis, Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans), P. intermedia, and Tannerella forsythia (T. forsythia), resulting in periodontal organ inflammation [21,22].
The most common oral pathogens, such as S. mutans, P. gingivalis, Staphylococcus species, and Lactobacillus species, are present in dental plaque and cavities [23,24,25]. The Lactobacillus ferments sugar and produces a large amount of lactic acid, which can easily cause caries [26]. Oral bacteria reach the digestive tract rapidly through the airway, generating an imbalance in the intestinal microflora and harming the digestive system, particularly pathogenic periodontitis bacteria [27,28,29,30]. Likewise, Fusobacterium nucleatum colonizes and governs the gastrointestinal tract via the blood flow system, as outlined in a prior study on colorectal cancer [31]. Additionally, P. gingivalis can target the complement C5a receptor 1 (C5aR1) and the Toll-like receptor 2 (TLR2) [32]. P. gingivalis and Helicobacter pylori can activate the PI3K signaling pathway, inhibiting internalization and increasing inflammation, targeting these two receptors and leading to pancreatic cancer [33,34]. The prevalence of Neisseria and Haemophilus differs between hunter–gatherers and westerners, with rural farmers falling somewhere in the middle. Some oral pathogens have been discovered among hunter–gatherers, indicating that consuming too much meat increases the risk of dental degenerative diseases [35]. It is necessary to elucidate the complete microbiome within the human mouth. The dynamics of the oral microbial community are intricate, and we are still in a long way off properly comprehending it. New high-throughput approaches will most likely help us explore microbial ecology in healthy and oral diseased volunteers [36,37,38]. In this study, we implemented high-throughput sequencing technology to elucidate the composition of the whole microbial community in healthy subjects before and after the treatment of dextranase-containing mouthwash (DMW; Formula I) and dextranase-and-nisin-containing mouthwash (DNMW; Formula II).
2. Materials and Methods
The experimental procedure of the current study is illustrated in Figure 1.
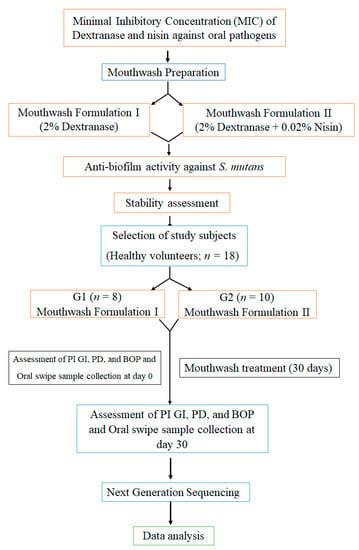
Figure 1.
Follow chart describes the experimental procedure of the study.
2.1. Preparation of Mouthwashes
The 2% dextranase-containing mouthwash (DMW; Formulation I) was prepared as detailed in the previous report [39]. A concentration of 0.02% nisin was added to DMW to formulate the dextranase-and-nisin-containing mouthwash (DNMW; Formulation II).
2.2. Minimal Inhibitory Concentration (MIC) of Dextranase and Nisin
Streptococcus mutans, Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Tannerella forsythia were cultured on a suitable medium at 37 ± 2 °C in an anaerobic chamber for 24–72 h. The microbial cell suspensions (1.5 × 106 cells/mL) were prepared in phosphate-buffered saline (PBS). The cells were mixed with serially diluted nisin (1 mg/mL) and dextranase (144 to 0.07 units/g) in a 96-well plate and incubated. After the incubation, the microbial growth was measured via spectrophotometric analysis at 600 nm; then, the MIC values were derived [40].
2.3. Antibiofilm Activity
The effect of DMW and DNMW against S. mutans biofilm was tested [41]. S. mutans biofilms were allowed to form on 12 mm glass coverslips placed in 24-well titer plates with 2 mL of DMW and DNMW separately, then incubated at 37 °C and 5% CO2 for 24 h. After 24 h of incubation, the medium was removed and the slides were rinsed with PBS. Finally, fixative glutaraldehyde fixed the biofilm and was observed using scanning electron microscopy (SEM). The efficacy of mouthwash formulations was compared with a positive control.
2.4. Estimation of Mouthwash Stability
The guidelines of the Thai industrial standard (TIS 2342-2550) were followed to evaluate the stability of mouthwashes. The mouthwashes were stored at 4 °C, room temperature, and 45 °C for three months. The stability of the mouthwash formulations was determined by the changes in their enzyme activity and MIC values. As detailed previously, dextranase activity was measured using the dinitrosalicylic acid (DNS) method [42].
2.5. Selection and Characteristics of the Study Population
Four weeks of experimental design were employed using 18 volunteers. The study was approved (No. 145/2562) by the Research Ethics Committee, Mae Fah Luang University, Thailand. Inclusion criteria were: volunteers had to be more than 18 years old with a medically healthy oral condition, with no antibiotic use for at least the last one month at the time of the experiment, a non-smoker, and the volunteer had to give informed consent. A pre-study medical questionnaire was used to examine general health conditions. The potential participants were screened out to fulfil the following criteria: volunteers with systemic diseases such as diabetes, heart diseases, blood-associated diseases, tissue disease in teeth or dead teeth, pregnant and breastfeeding women, and damage/loss of periodontal organs. Initially, we allowed 10 subjects in each group; later, 2 subjects from group 2 withdrew from the study due to personal reasons. The demographic data of the study subjects are detailed in Table 1. The subjects in group 1 (G1) and group 2 (G2) were recommended to use mouthwash formulation I and II, respectively, at least twice a day for four weeks (the study duration was selected based on our preliminary study). The plaque index (PI), probing depth (PBD), gingival index (GI), and bleeding on probing (BOP) scores of the subjects before and after four weeks of experiments were recorded in detail previously [43,44,45,46]. The saliva samples were collected before and after the treatment to assess the changes in oral microbiota. Furthermore, the gingival inflammation was assessed according to the GI by Loe and Silness [47].

Table 1.
The demographic data of the study subjects.
2.6. Oral Microbiome Assay
According to the manufacturer protocol, we extracted bacterial genomic DNA from healthy subjects using a QIAmp UCP DNA Micro Kit (Catalog no. 56204, QIAGEN, Hilden, Germany). The metagenomic analysis was performed by Omics Sciences and Bioinformatics Center, Faculty of Science, Chulalongkorn University, Thailand. This method obtained 16s RNA from a total of 36 metagenomic DNA samples (pre and post samples from G1 (n = 16); pre and post samples from G2 (n = 20)) covering the variable regions V3–V4 were generated using forward and reverse primers (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′ and 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′) on the Illumina MiSeq device (5200 Illumina Way, San Diego, CA, USA). MOTHURv.1.43.0 software (The University of Michigan, State Street, Ann Arbor, MI 48109, United States) was used in sequence filtration and in the analysis of sequenced data [48]. After checking the quality of pair-end reads with the information in FASTQ files using DADA2, the poor-quality reads were filtered out as per the default QIIME 2™ threshold values (minimum quality score = 25, minimum/maximum length = 200/1000, no ambiguous bases allowed, and no mismatches allowed in the primer sequence) [49].
From the total of 36 samples, we obtained 1,013,094 non-chimeric 16S rRNA gene sequence reads. The resulting high-quality sequences were clustered into de novo operational taxonomic units (OTUs) with 97% identity and assigned taxonomy for each OUT sequence. The resulting QIIME artifacts data were visualized and analyzed using QIIME 2 View. Applying microbiomes composition analyses, we found the substantially prevalent bacterial taxonomic classifications at the species level.
2.7. Statistical Analysis
Analysis of weighted and unweighted UniFrac distances [50] performed using the QIIME 2™ workflow was used to compare microbial richness in pre-treated and post-treated samples in G1 and G2 [51]. The first three main coordinates were used to create principal coordinate analysis plots, labeled according to their variance. The OTU table of raw counts was normalized to an OTU table of relative abundances. Taxa of the same type were agglomerated at the phylum, class, order, family, and genus levels. Our analysis included taxa from the major phylum of the pre-treated and post-treated G1 and G2 microbiome (proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Spirochaetes, TM7, and Synergistetes). Phylogenetic diversity (PD) was annotated from the resulting rarefaction curve. The relation between samples was identified using principal coordinate analysis (PcoA) and visualized using QIIME 2 View.
3. Results
3.1. Minimal Inhibitory Concentration (MIC)
The MIC of nisin against P. gingivalis, A. actinomycetemcomitans, T. forsythia, and S. mutans was 0.031 mg/mL, 0.50 mg/mL, 0.25 mg/mL, and 0.50 mg/mL, respectively. The MIC of dextranase against S. mutans was 4.5 units/g (Table 2). The mouthwashes showed anti-biofilm activity against S. mutans (a key etiologic factor leading to gingivitis, dental caries, periodontal, and other oral-related diseases). The antibiofilm activity of the mouth with regard to formulations against S. mutans was observed under SEM (Figure 2).

Table 2.
The MIC of the nisin and dextranase against the representative pathogens. NA: no activity.
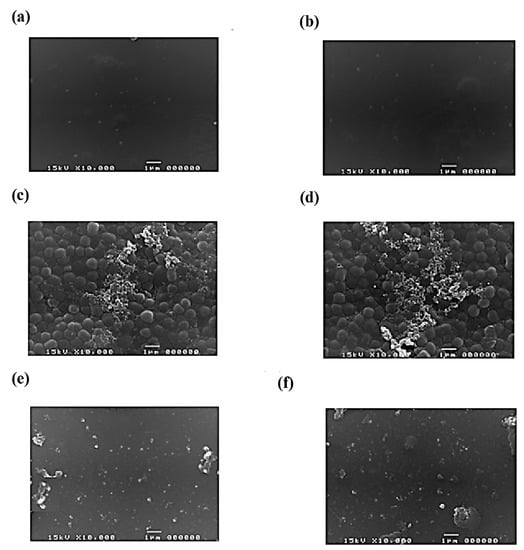
Figure 2.
The scanning electron microscopy pictures show the efficacy of mouthwash formulation I (a,c,e) and II (b,d,f) against S. mutans biofilm. Positive control (a,b), negative control (c,d), and test (e,f). The treatment of mouthwash formulations effectively suppressed the formation of biofilm. Scale bar = 1 µm.
3.2. Stability of the Mouthwashes
The result showed that both mouthwash formulations are clear, light brown, have no precipitation, no cracking, and passed the Thai standards after three months of storage at different temperatures. The MIC values (Table 3) and dextranase activities (Table 4) were not affected significantly after the storage.

Table 3.
The stability of mouthwash formulation I and II on S. mutans biofilm formation based on MIC value.

Table 4.
The stability of mouthwash formulations I and II are based on enzyme activity. RT: room temperature.
3.3. Plaque Index, Probing Depth Gingival Index, and Bleeding on Probing
The PI, PBD, GI, and BOP values were estimated in the G1 and G2 subjects before and after the use of mouthwash formulations (Table 5 and Table 6).

Table 5.
The comparison of the plaque index, probing depth, gingival index, and bleeding on probe values of pre-and post-treatment samples of G1 and G2. * indicates the statistical significance.

Table 6.
The Mann–Whitney test compares the plaque index, probing depth, gingival index, and bleeding on probe between G1 and G2.
The results revealed that the PI values significantly differed among the pre-and post-treated samples of G1 and G2 (Table 5). However, the Mann–Whitney-test-based investigation showed no significant differences between G1 and G2 (p = 0.884) (Table 6).
3.4. Microbiome Analysis
3.4.1. Taxonomy Assignment
Proteobacteria (μ = 86.909%) was found majorly before treatment, whereas the amount of Firmicutes (μ = 36.012%) was increased after treatment in G1 samples. Bacteroidetes, Actinobacteria, Fusobacteria, Spirochaetes, TM7, Synergistetes, SR1, Tenericutes, GN02, Verrucomicrobia, and cyanobacteria phylum were also found in G1 samples (Figure 3a). Similarly, Proteobacteria (μ = 81.711%) was found majorly before treatment, whereas the amount of Firmicutes (μ = 41.440%) was increased after treatment in G2 samples. Bacteroidetes, Actinobacteria, Fusobacteria, TM7, SR1, Spirochaetes, GN02, Synergistetes, Tenericutes, and Verrucomicrobia phylum were also found in G2 samples (Figure 3b).
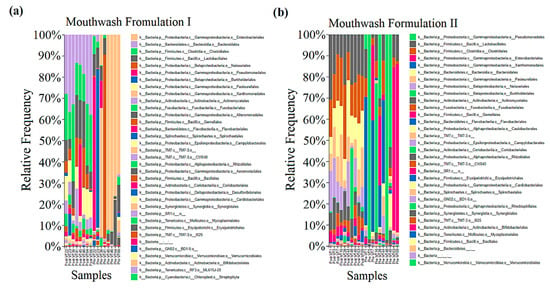
Figure 3.
Taxonomy was assigned to the samples (pre-and post-treatment) collected from G1 (a) and G2 (b), and its relative frequency was compared.
The OTUs can be divided into several categories according to their occurrences among different samples in groups before and after the mouthwash treatment. The relative frequency of the phyla can be used to identify the efficacy of the DMW. The relative frequencies of Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Spirochaetes, TM7, and Synergistetes were found to be μ = 86.91%, μ = 10.37%, μ = 1.23%, μ = 0.81%, μ = 0.2%, μ = 0.21%, μ = 0.17%, and μ = 0.03%, respectively, in G1 samples before treatment. Likewise, the relative frequency of Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Spirochaetes, TM7, and Synergistetes were μ = 22.52%, μ = 36.01%, μ = 28.03%, μ = 6.24%, μ = 3.62%, μ = 1.43%, μ = 1.27%, and μ = 0.18%, respectively, in G1 samples after treatment with DMW (Figure 4).
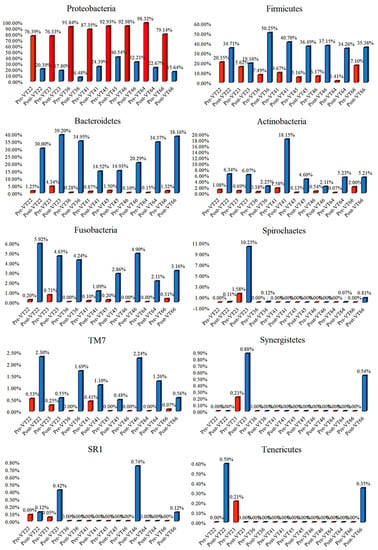
Figure 4.
The abundant phyla of the G1 (mouthwash formulation I) samples were compared before and after mouthwash to identify the formulation’s efficacy. Red and blue bars indicate the pre-and post-treatment samples, respectively.
Similarly, the relative frequency of phyla was changed in G2 samples after treatment compared to pre-treated samples, as follows: Proteobacteria (from μ = 81.71 to μ = 26.98%), Firmicutes (from μ = 14.30 to μ = 41.44%), Bacteroidetes (from μ = 1.94 to μ = 17.72%), Actinobacteria (from μ = 1.29 to μ = 8.49%), Fusobacteria (from μ = 0.33 to μ = 3.90%), TM7 (from μ = 0.36 to μ = 1.04%), SR1 (from μ = 0.02 to μ = 0.32%), and Spirochaetes (from μ = 0.05% to μ = 0.07%) (Figure 5). Other minor changes in the microbial abundances for DMW and DNMW are detailed in Figure S1a,b (Supplementary File S1).
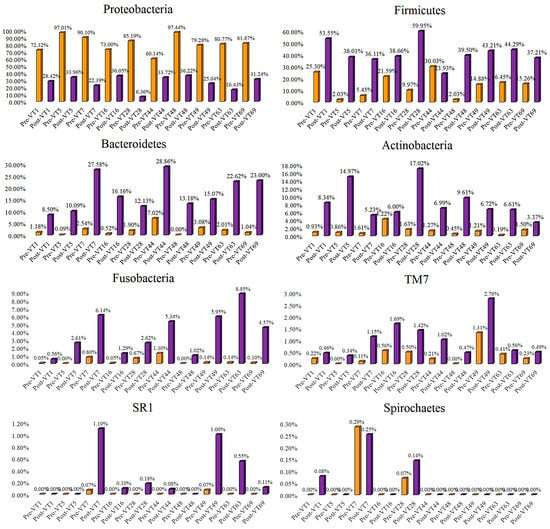
Figure 5.
The abundant phyla of the G2 (mouthwash formulation II) samples were compared before and after mouthwash to identify the formulation’s efficacy. Purple and orange bars indicate the pre-and post-treatment samples, respectively.
Klebsiella (20.733%), Citrobacter (11.615%), Chromobacterium (10.849%), Achromobacter (9.381%), Stenotrophomonas (7.601%), Serratia (7.304%), Shewanella (2.723%), Morganella (0.591%), Agrobacterium (0.502%), Staphylococcus (0.328%), Desulfovibrio (0.300%), Lactococcus (0.296%), Delftia (0.251%), and Corynebacterium (0.106%) were found in pre-treatment G1 samples (Figure S2) (Supplementary File S1).
Pseudomonas (22.881%), Stenotrophomonas (17.370%), Achromobacter (7.020%), Serratia (3.552%), Brevundimonas (1.587%), Klebsiella (0.737%), Agrobacterium (0.634%), Lactobacillus (0.127%), and Treponema (0.048%) were found in pre-treatment G2 samples (Figure S3) (Supplementary File S1).
After the use of the mouthwashes, the genera mentioned above were completely eradicated in both G1 and G2 samples.
3.4.2. Rarefaction Curve Analysis
A total of 509,199 and 656,392 16S rDNA V3-V4 sequencing reads were yielded for the pre-and post-treated G1 samples, respectively. After a series of quality control processing (where they were filtered, de-noised, and merged), 443,046 (87.00%) and 570,048 (86.84%) high-quality non-chimera reads were obtained for the pre-and post-treated G1 samples, respectively, and were clustered into OTUs. On average, each pre-and post-treated sample had 69 and 232 OTUs, respectively (Table S1) (Supplementary File S2) (Figure 6a). Similarly, a total of 756,857 and 863,350 16S rDNA V3-V4 sequencing reads were yielded for the pre-and post-treated G2 samples. After a series of quality control processing (where they were filtered, de-noised, and merged), 687,603 (90.84%) and 756,006 (87.57%) high-quality non-chimera reads were obtained and were clustered into OTUs. On average, each pre- and post-treated sample had 96 and 189 OTUs, respectively (Table S2) (Supplementary File S2) (Figure 6b). The rarefaction analysis suggested that the sequencing depth can recover most of the community’s diversity.
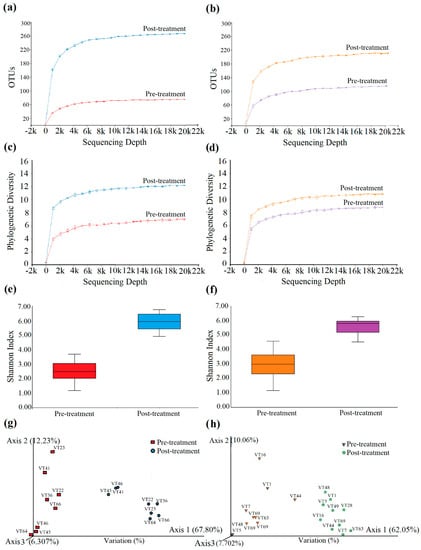
Figure 6.
Refraction analysis for the G1 and G2 samples. The rarefaction curves of observed OTUs and Pathway diversity were computed for G1 (a,c) and G2 (b,d). Shannon H-index alpha Diversity was measured and compared for the G1 (e) and G2 (f) samples. The inter and intragroup species differences were compared using PCoA for G1 (g) and G2 (h) samples.
3.4.3. Phylogenetic Diversity
The phylogenetic diversity was computed for the pre-and post-treated samples of G1 and G2. The pre-and post-treated G1 samples’ phylogenetic diversity index was 6.0 and 12.0 at maximum, respectively (Figure 6c). The pre-and post-treated G2 samples’ phylogenetic diversity index was 7.0 and 10.0 at maximum, respectively (Figure 6d). This shows that the diversity in the samples was high in pre-treated G2 samples. However, the diversity in the G2 samples was reduced significantly after being treated with DNMW.
3.4.4. Species Richness
The species richness in the pre-and post-treated G1 samples was found as quartile portions (Q1 = 3.06 and Q3 = 3.06, and Q1 = 5.43 and Q3 = 6.456, respectively) with the median of 2.48 and 5.93 and the whisker maximum of 3.68 and 6.76, respectively (Figure 6e). Likewise, the species richness in the pre-and post-treated G2 samples was found as quartile portions (Q1 = 2.32 and Q3 = 3.62, and Q1 = 5.19 and Q3 = 5.95, respectively) with the median of 2.99 and 5.82 and the whisker maximum of 4.58 and 6.28, respectively (Figure 6f).
This shows that the species evenness and richness were also increased in post-treated samples after being treated with mouthwashes. However, the overall species richness was higher in the pre-treated G2 samples. Additionally, the species richness and evenness were significantly reduced in post-treated G2 samples compared with the post-treated G1 samples. The species Stenotrophomonas maltophilia (7.601%), Serratia marcescens (7.304%), Morganella morganii (0.591%), and Lactococcus garvieae (0.296%) were uprooted completely in the G1 post-treated samples. Similarly, the species frequency in the genus Parvimonas was reduced from 0.208% to 0.198%, and the species Pseudomonas fragi (5.435%), Pseudomonas nitroreducens (5.320%), Serratia marcescens (3.552%), Lactobacillus zeae (0.127%), and Stenotrophomonas acidaminiphila (0.119%) were completely rooted out in the post-treated G2 samples.
3.4.5. Principal Coordinates Analysis
Differences in pre-and post-treated samples of G1 and G2 were represented in PCoA plots. The samples from the two groups significantly separated in the 3D space and scattered. The pre-and post-treated G1 samples were separated with 67.80% variation, resulting in significant differences at the species diversity level (Figure 6g). Likewise, the pre-and post-treated G2 samples were also disjointed, with 62.05% variation, which shows the notable differences at species divergence. However, the cluster formed between the samples indicates that some of the common species evolved (Figure 6h).
4. Discussion
Generally, the resulting oral microbial sequences were estimated using QIIME 2TM, which revealed the phyla Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, TM7, and Spirochaetes were observed predominantly in healthy subjects rather than Synergistetes, SR1, and Tenericutes. We previously reported that the use of DMW for weeks reduced the PI in healthy subjects without affecting the PBD, GI, and BOP values. The use of DMW and DNMW for four weeks reduced the PI in dental caries subjects [52]. The current study revealed the impact of DMW and DNMW (for 30 days) on the oral microbial community in healthy subjects.
Mouthwashes with chemicals are common among the general population due to their ease of use, reduced plaque biofilm, and breath-freshening impact. Although industrialized nations utilize various oral hygiene products to achieve oral hygiene, appropriate scientific proof must be available to back up such claims [53,54,55].
The minimal inhibitory concentration is also usually quoted as the primary indicator of the potential efficacy of plaque control agents. Oral care products often use the appropriate concentration, which can effectively help control plaque and gingivitis [56].
Dextran-degrading enzymes, commonly called dextranases, form a diverse group of different carbohydrates and transferases. They specifically hydrolyze (1→6)-α-glucosidic linkages in dextrans. Dextrans could diffuse into the biofilm, while most particles were excluded and inhibited the growth of S. mutans [57]. Nisin is an antimicrobial peptide that disturbs the bacterial cell membrane [58,59]. Nisin inhibited the growth of oral bacteria and the development of biofilms by preventing cell attachment and proliferation, as reported earlier [60].
In this study, dextranase showed the MIC of 4.5 unit/g against S. mutans. The nisin shows the inhibitory effect against all the tested microbes (P. gingivalis, A. actinomycetemcomitans, T. forsythia, and S. mutans) (Table 2). We selected S. mutans as the key microbial candidate to test the antibiofilm activity of the DMW (Formulation I) and DNMW (Formulation II), and it showed the inhibition against the biofilm formation (Figure 1). Formulation I and II were stable for three months at 4 °C, room temperature, and 45 °C (Table 3 and Table 4).
As per the results, the formation of plaque in the gingival sulcus and the development of periodontal pathogenesis might reduce significantly during the test period (Table 5). The DMW and DNMW have a closer effect on the oral health of healthy volunteers. However, DNMW reduces the PBD (Table 5). Moreover, on tested parameters, the impacts were insignificant (Table 6) among the mouthwash formulations, indicating that both DMW and DNMW exhibited a similar protective role.
Current methods for identifying and quantifying bacteria in dental biofilms, including genome sequencing, have shown a considerably more complicated ecology than previously thought [61,62]. The genera Catonella, Centipeda, Fretibacterium, Rhizobium, Ochrobactrum, Mogibacterium, Actinomyces, Streptococcus, Rothia, Selenomonas, and Veillonella were detected as major dental caries pathogens [63,64]. Atopobium, Bacteroides, Cryptobacterium, Lactobacillus, Pseudomonas, Mogibacterium, Ochrobactrum, Rhizobium, Centipeda, Campylobacter, Alloprevotella, Megasphaera, and Mycoplasma were the suspected pathogens of dental caries [65,66,67].
Bapat et al. reported that the common microorganisms implicated in oral disease are S. mutans and L. acidophilus [68]. Here, the results of our study showed that DMW completely eradicated the genera Stenotrophomonas, Serratia, Citrobacter, Enterococcus, Achromobacter Delftia, Chromobacterium, Klebsiella, Stenotrophomonas, and Psychrobacter in the post-treated samples of G1. However, the DMW effectively acts against the genera Citrobacter, Proteus, Enterococcus, Actinobacillus, and Psychrobacter, which are uniquely noted in Table S3 (Supplementary File S2). Similarly, the genera Pseudomonas, Veillonella, Stenotrophomonas, Achromobacter, Serratia, Brevundimonas, Klebsiella, Agrobacterium, Lactobacillus, and Treponema were completely rooted out in the post-treated samples of G2, as can be observed in Table S4 (Supplementary File S2).
Citrobacter, like other Enterobacteriaceae, may cause a variety of infections in humans, including urethritis, upper airways, sores, bone, peritoneal, endocardium, meninges, and bloodstream infections. The urinary system is the most frequent source of infection, preceded by the gut, skin/soft tissues (including surgical site infection), and influenza. Citrobacter spp. infection can be transmitted internally from the mother or laterally from carriers or other hospital sources [69,70,71,72]. Oral enterococcal colonies have the appropriate virulence characteristics, particularly antimicrobial resistance, to make them suitable candidates that might cause oral or systemic infections [57,73]. In addition, the genus Actinobacillus has been identified as a significant contender in the development of periodontitis [74,75,76]. Psychrobacter has been identified from blood, cerebrospinal fluid, and other internal organs in individuals and is thought to be an uncommon opportunistic infection [42,77]. However, Psychrobacter may be transmitted from periodontal blood, as it has existed in healthy subjects before using a mouthwash, which may enter the bloodstream and cause infectious disease in humans. Fortunately, the mouthwash formulation curtailed the growth of species from the genera Citrobacter, Actinobacillus, and Psychrobacter in G1 post-treated samples (Table S4) (Supplementary File S2).
Pseudomonas spp. are significant bacteria linked to periodontal disorders in hospitalized and institutionalized patients. The mouth cavity might be a key reservoir for these infections, especially in the case of inadequate dental hygiene and periodontitis [78]. Veillonella species were found mainly in subjects who had poor oral hygiene compared to those with good or moderate oral hygiene. Additionally, Mashima et al. reported the distribution and high frequency of V. rogosae in tongue biofilms of Thai children, and the biofilms were resistant to penicillin and ampicillin [79,80]. Achromobacter species cause periodontal-related disease and impact immunocompromised individuals with high-risk malignancies followed by circulation and respiratory infections [81,82,83,84]. Brevundimonas diminuta was detected in the refractory periodontitis patients and plays a vital role in advancing periodontitis [85,86].
Similarly, B. diminuta was amplified and sequenced in patients with hematological malignancies, severely obstructed uropathy, and angiosarcoma of the heart [87]. Lactobacilli were the first microbes linked to the development of dental caries [88]. It is also depicted as an opportunistic bacterium that has adapted to a specialized niche produced by the instability of a healthy oral microbiome and has led to the development of dental caries and the formation of a significant reservoir in the gastrointestinal (GI) tract [26]. According to Zeng et al., Treponema species and their lineages are primarily found in the oral cavities of those suffering from periodontitis and gingivitis [89]. Likewise, the DNMW completely extirpated the growth of Veillonella, Achromobacter, Brevundimonas, Lactobacillus, and Treponema in G2 post-treated volunteers. In addition, the bacterial species B. diminuta was found in our healthy volunteers and was eradicated following the use of DNMW (Table S4, Supplementary File S2).
Prates et al., 2020 reported that S. maltophilia belongs to the genus Stenotrophomonas, an emerging multidrug-resistant opportunistic pathogen that leads to oral infections, malignancy, and respiratory tract infections [90]. Serratia marcescens is associated with urinary and respiratory infections, endocarditis, osteomyelitis, septicemia, wound infections, eye infections, meningitis, dental caries, and plaque [91]. Oral Klebsiella spp. was recently demonstrated to promote inflammatory response and dysbiosis in the gut upon aberrant colonization. It was postulated that the oral cavity serves as a storehouse for promising intestinal pathogens such as Klebsiella. However, the genera Serratia, Klebsiella, and Stenotrophomona were completely reduced by the mouthwash formulations. (Tables S3 and S4, Supplementary File S2).
Finally, the results showed that the DNMW reduces the growth of oral pathogens better than the DMW in healthy subjects, possibly due to the presence of nisin. The results exposed that the DMW and DNMW notably increased the growth of Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, and TM7 phylum (Figure 4).
Nonetheless, in the ongoing investigation, we were able to connect genome sequencing findings with other fundamental indices of oral health, allowing us to further examine the influence of DMW and DNMW on oral and systemic health. However, it is questionable whether these microbial alterations indicate a move toward a healthier oral environment or whether they may raise the risk of oral disorders, as both elevations and reductions in the bacteria linked with caries and periodontal disease have indeed been documented.
The current study also has some limitations. Both DMW and DNMW support the growth of some oral pathogenic species Veillonella dispar Veillonella parvula, Selenomonas noxia, Neisseria subflava, and Prevotella intermedia, shown in Tables S3 and S4 (Supplementary File S2). The PCoA plots explain that DMW supports the new species divergence (67.80%) more than DNMW (Figure 6g,h). The saccharolytic bacteria, including Streptococcus, Actinomyces, and some species in the genus Veillonella, metabolize carbohydrates through the Embden–Meyerhof–Parnas metabolic pathway, which yield acidic by-products, including lactate, acetate, ethanol, and formate, which cause the demineralization of dental enamel and the development of carious lesions with increased microbial diversity [92]. Furthermore, this study may be expanded to examine the effect of DNMW in individuals with oral health issues such as gingivitis and periodontitis.
5. Conclusions
The study has limitations. There was a limited number of participants, it was a short-term intervention, there was a difference in the group size, etc. Nonetheless, the results indicated that the DMW and DNMW reduced the PI without affecting the PBD, GI, and BOP scores in healthy subjects. Notably, the use of DNMW for thirty days could significantly reduce the oral pathogenic microbial load in healthy subjects. In detail, DNMW has a more significant impact on reducing oral pathogenic genus Pseudomonas, Veillonella, Stenotrophomonas, Achromobacter, Serratia, Brevundimonas, Klebsiella, Agrobacterium, Lactobacillus, and Treponema, which may cause oral diseases. Likewise, the DMW and DNMW showed a significant reduction in the biofilm of S. mutans. The current study proposed that the DMW and DNMW mouthwashes could be an adjuvant to treat and manage oral diseases. However, some of the pathogens’ loads also increased after the use of mouthwashes. Thus, the results need further confirmation by experimenting with more study subjects.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app12031650/s1, Supplementary File S1: Figure S1: Taxonomy and relative frequency of the pre-and post-treatment samples G1 (a; mouthwash formulation I) and G2 (b; mouthwash formulation II). Figure S2: The comparison of genus-level changes in pre-and post-treatment samples of G1 (mouthwash formulation I). Figure S3: The comparison of genus-level changes in pre-and post-treatment samples of G2 (mouthwash formulation II). Supplementary File S2: Table S1: The observed OTUs for group 1. Table S2: The observed OTUs for the group 2. Table S3: The effect of mouthwash formulation I on the genus and species level in post-treatment samples. Table S4: The effect of mouthwash formulation II on the genus and species level in post-treatment samples.
Author Contributions
Conceptualization, C.C., S.S. and P.S.; methodology, C.C., S.S., N.J., P.T. and R.W.-S.; software, P.P., P.K. and M.B.; validation, C.C., S.S., B.S.S., M.B. and P.S.; formal analysis, N.J., P.T., B.S.S., P.P., P.K. and M.B.; investigation, N.J. and P.T.; resources, S.P.; data curation, N.J., P.T., W.O., P.P., P.K. and M.B.; writing—original draft preparation, C.C., N.J., B.S.S., M.B. and P.S.; writing—review and editing, C.C., S.S., B.S.S., P.P., P.K., M.B. and P.S.; visualization, S.S., P.T., R.W.-S. and P.S.; supervision, C.C., S.S., R.W.-S. and P.S.; project administration, S.P.; funding acquisition, C.C. and S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Supporting Research Funds for Industries, SuRF (RDG58I0002).
Institutional Review Board Statement
The study was approved (No. 145/2562) by the Research Ethics Committee, Mae Fah Luang University, Thailand.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All the related data has been provided in the manuscript.
Acknowledgments
This study was partially supported by Chiang Mai University, Thailand and Neuropsychological Research Laboratory, Department of Anti-Aging and Regenerative Science, School of Anti-Aging and Regenerative Medicine, Mae Fah Luang University, Thailand.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Patrick, D.L.; Lee, R.S.; Nucci, M.; Grembowski, D.; Jolles, C.Z.; Milgrom, P. Reducing oral health disparities: A focus on social and cultural determinants. BMC Oral Health 2006, 15, S4. [Google Scholar] [CrossRef] [PubMed]
- Northridge, M.E.; Kumar, A.; Kaur, R. Disparities in access to oral health care. Annu. Rev. Public Health 2020, 41, 513–535. [Google Scholar] [CrossRef]
- Kamieńska, P.T.; Walkowiak, T.N.; Lewicka, B.M. The relationship between oral hygiene level and gingivitis in children. Adv. Clin. Exp. Med. 2018, 27, 1397–1401. [Google Scholar] [CrossRef] [PubMed]
- Heng, C. Tooth decay is the most prevalent disease. Fed. Pract. 2016, 33, 31–33. [Google Scholar]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Borgnakke, W.S. IDF diabetes atlas: Diabetes and oral health—A two-way relationship of clinical importance. Diabetes Res. Clin. Pract. 2019, 157, 107839. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jha, P.C.; Negi, B.S.; Haidry, N.; Kulkarni, P.; Gulati, S.; Muttu, J. Oral health status and treatment need in geriatric patients with different degrees of cognitive impairment and dementia: A cross-sectional study. J. Family Med. Prim. Care. 2021, 10, 2171–2176. [Google Scholar]
- Khor, B.; Snow, M.; Herrman, E.; Ray, N.; Mansukhani, K.; Patel, K.A.; Said-Al-Naief, N.; Maier, T.; Machida, C.A. Interconnections between the oral and gut microbiomes: Reversal of microbial dysbiosis and the balance between systemic health and disease. Microorganisms 2021, 9, 496. [Google Scholar] [CrossRef]
- Kamieńska, P.T.; Lewicka, B.M.; Gabryel, B.H. Salivary Biomarkers and oral microbial load in relation to the dental status of adults with cystic fibrosis. Microorganisms 2019, 7, 692. [Google Scholar] [CrossRef]
- Cwik, J.; Gonzalez, L.A.; Shi, X.; Spirgel, C.C.; Yankell, S. Plaque reduction and tensile strength evaluations of three dental floss products. Am. J. Dent. 2021, 34, 123–126. [Google Scholar]
- Naik, R.; Mujib, A.B.R.; Telagi, N.; Anil, B.S.; Spoorthi, B.R. Contaminated tooth brushes-potential threat to oral and general health. J. Family Med. Prim. Care 2015, 4, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Worthington, H.V.; MacDonald, L.; Pericic, P.T.; Sambunjak, D.; Johnson, T.M.; Imai, P.; Clarkson, J.E. Home use of interdental cleaning devices, in addition to toothbrushing, for preventing and controlling periodontal diseases and dental caries. Cochrane Database Syst. Rev. 2019, 4, CD012018. [Google Scholar] [CrossRef] [PubMed]
- Zinn, M.K.; Schages, L.; Bockmühl, D. The toothbrush microbiome: Impact of user age, period of use and bristle material on the microbial communities of toothbrushes. Microorganisms 2020, 8, 1379. [Google Scholar] [CrossRef]
- Kamal, D.; Hassanein, H.; Akah, M.; Abdelkawy, M.A.; Hamza, H. Caries preventive and antibacterial effects of two natural mouthwashes vs. chlorhexidine in high caries-risk patients: A randomized clinical trial. J. Contemp. Dent. Pract. 2020, 21, 1316–1324. [Google Scholar] [CrossRef]
- Son, E.J.; Kim, Y.J.; Kim, D.M.; Chung, H.J. The effect of dextranase-containing mouthwash in human experimental gingivitis. J. Korean Acad. Periodontol. 2001, 31, 401–420. [Google Scholar] [CrossRef]
- Lai, X.; Liu, X.; Liu, X.; Deng, T.; Feng, Y.; Tian, X.; Lyu, M.; Wang, A.S. The marine Catenovulum agarivorans MNH15 and dextranase: Removing dental plaque. Mar. Drugs 2019, 17, 592. [Google Scholar] [CrossRef]
- Malvania, E.A.; Sharma, A.S.; Sheth, S.A.; Rathod, S.; Chovatia, N.R.; Kachwala, M.S. In vitro analysis of licorice (glycyrrhiza glabra) root extract activity on Streptococcus mutans in comparison to chlorhexidine and fluoride mouthwash. J. Contemp. Dent. Pract. 2019, 20, 1389–1394. [Google Scholar] [CrossRef]
- Padiyar, B.; Marwah, N.; Gupta, S.; Padiyar, N. Comparative evaluation of effects of Triphala, garlic extracts, and chlorhexidine mouthwashes on salivary Streptococcus mutans counts and oral hygiene status. Int. J. Clin. Pediatr. Dent. 2018, 11, 299–306. [Google Scholar]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [PubMed]
- Leung, R.; Covasa, M. Do gut microbes taste? Nutrients 2021, 13, 2581. [Google Scholar] [CrossRef]
- Offenbacher, S.; Divaris, K.; Barros, S.P.; Moss, K.L.; Marchesan, J.T.; Morelli, T.; Zhang, S.; Kim, S.; Sun, L.; Beck, J.D.; et al. Genome-wide association study of biologically informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum. Mol. Genet. 2016, 25, 2113–2129. [Google Scholar] [CrossRef]
- Marchesan, J.T.; Girnary, M.S.; Moss, K.; Monaghan, E.T.; Egnatz, G.J.; Jiao, Y.; Zhang, S.; Beck, J.; Swanson, K.V. Role of inflammasomes in the pathogenesis of periodontal disease and therapeutics. Periodontology 2000 2020, 82, 93–114. [Google Scholar] [CrossRef] [PubMed]
- Herrera, H.A.; Ospina, F.L.; Fang, L.; Caballero, D.A. Susceptibility of Porphyromonas gingivalis and Streptococcus mutans to antibacterial effect from Mammea americana. Adv. Pharmacol. Sci. 2014, 2014, 384815. [Google Scholar]
- Seneviratne, C.J.; Zhang, C.F.; Samaranayake, L.P. Dental plaque biofilm in oral health and disease. Chin. J. Dent. Res. 2001, 14, 87–94. [Google Scholar]
- Heimisdottir, L.H.; Lin, B.M.; Cho, H.; Orlenko, A.; Ribeiro, A.A.; Simon-Soro, A.; Roach, J.; Shungin, D.; Ginnis, J.; Simancas-Pallares, M.A.; et al. Metabolomics insights in early childhood caries. J. Dent. Res. 2021, 100, 615–622. [Google Scholar] [CrossRef]
- Caufield, P.W.; Schön, C.N.; Saraithong, P.; Li, Y.; Argimón, S. Oral lactobacilli and dental caries: A model for niche adaptation in humans. J. Dent. Res. 2015, 94, 110S–118S. [Google Scholar] [CrossRef]
- Kobayashi, R.; Ogawa, Y.; Hashizume-Takizawa, T.; Kurita-Ochiai, T. Oral bacteria affect the gut microbiome and intestinal immunity. Pathog. Dis. 2020, 78, ftaa024. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Olsen, I.; Yamazaki, K. Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 2019, 11, 1586422. [Google Scholar] [CrossRef]
- Bui, F.Q.; Almeida-da-Silva, C.L.C.; Huynh, B.; Trinh, A.; Liu, J.; Woodward, J.; Asadi, H.; Ojcius, D.M. Association between periodontal pathogens and systemic disease. Biomed. J. 2019, 42, 27–35. [Google Scholar] [CrossRef]
- Abed, J.; Maalouf, N.; Manson, A.L.; Earl, A.M.; Parhi, L.; Emgård, J.E.M.; Klutstein, M.; Tayeb, S.; Almogy, G.; Atlan, K.A.; et al. Colon cancer-associated Fusobacterium nucleatum may originate from the oral cavity and reach colon tumors via the circulatory system. Front. Cell Infect. Microbiol. 2020, 10, 400. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Diaz, P.I. Porphyromonas gingivalis: Immune subversion activities and role in periodontal dysbiosis. Curr. Oral Health Rep. 2020, 7, 12–21. [Google Scholar] [CrossRef]
- Nakayama, M.; Inoue, T.; Naito, M.; Nakayama, K.; Ohara, N. Attenuation of the phosphatidylinositol 3-kinase/Akt signaling pathway by Porphyromonas gingivalis gingipains RgpA, RgpB, and Kgp. J. Biol. Chem. 2015, 290, 5190–5202. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Xiong, C.; The, S.W.; Lim, J.C.W.; Kumar, S.; Thilakavathy, K. Mechanisms of oral bacterial virulence factors in pancreatic cancer. Front Cell Infect. Microbiol. 2019, 9, 412. [Google Scholar] [CrossRef] [PubMed]
- Lassalle, F.; Spagnoletti, M.; Fumagalli, M.; Shaw, L.; Dyble, M.; Walker, C.; Thomas, M.G.; Migliano, B.A.; Balloux, F. Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet. Mol. Ecol. 2018, 27, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.R.; Gabaldón, T. The human oral microbiome in health and disease: From sequences to ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef]
- Huang, Y.Z.; Jin, Z.; Wang, Z.M.; Qi, L.B.; Song, S.; Zhu, B.W.; Dong, X.P. Marine bioactive compounds as nutraceutical and functional food ingredients for potential oral health. Front. Nutr. 2021, 8, 686663. [Google Scholar] [CrossRef]
- Nallabelli, N.; Patil, P.P.; Pal, V.K.; Singh, N.; Jain, A.; Patil, P.B.; Grover, V.; Korpole, S. Biochemical and genome sequence analyses of Megasphaera sp. strain DISK18 from dental plaque of a healthy individual reveals commensal lifestyle. Sci. Rep. 2016, 6, 33665. [Google Scholar] [CrossRef][Green Version]
- Juntarachot, N.; Sivamaruthi, B.S.; Sirilun, S.; Tongpong, P.; Sittiprapaporn, P.; Kantachote, D.; Chaiyasut, C. An introductory report on the effect of use of dextranase-containing mouthwash on oral health status of human volunteers. Asian J. Med. Sci. 2020, 11, 22–25. [Google Scholar] [CrossRef]
- McBirney, S.E.; Trinh, K.; Wong-Beringer, A.; Armani, A.M. Wavelength-normalized spectroscopic analysis of Staphylococcus aureus and Pseudomonas aeruginosa growth rates. Biomed. Opt. Express 2016, 7, 4034–4042. [Google Scholar] [CrossRef]
- Sato, Y.; Unno, Y.; Ubagai, T.; Ono, Y. Sub-minimum inhibitory concentrations of colistin and polymyxin B promote Acinetobacter baumannii biofilm formation. PLoS ONE 2018, 13, e0194556. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qi, J.; Zhao, H.; He, S.; Zhang, Y.; Wei, S.; Zhao, F. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci. Rep. 2013, 3, 1843. [Google Scholar] [CrossRef] [PubMed]
- Feier, I.; Onisei, D.; Onisei, D. The plurivalence of the interpretation of correlation between plaque score and bleeding score. J. Romanian Med. Dent. 2009, 13, 45–48. [Google Scholar]
- Checchi, L.; Montevecchi, M.; Checchi, V.; Zappulla, F. The relationship between bleeding on probing and subgingival deposits. An endoscopical evaluation. Open Dent. J. 2009, 3, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Rath, S.K.; Lohra, P. Comparative evaluation of accuracy of periodontal probing depth and attachment levels using a Florida probe versus traditional probes. Med. J. Armed. Forces India 2015, 71, 352–358. [Google Scholar] [CrossRef]
- Gopinath, V.K.; Rahman, B.; Awad, M.A. Assessment of gingival health among school children in Sharjah, United Arab Emirates. Eur. J. Dent. 2015, 9, 36–40. [Google Scholar] [CrossRef]
- Loe, H.; Silness, J. Periodontal disease in pregnancy. I. prevalence and severity. Acta Odontol. Scand. 1963, 21, 533–551. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Tongpong, P.; Chaiyavat, C.; Sasithorn, S.; Phakkharawat, S. The relationship between dextranase enzyme in mouthwash and dental caries. Asian J. Med. Sci. 2020, 11, 80–84. [Google Scholar] [CrossRef]
- Singh, A.; Purohit, B. Tooth brushing, oil pulling and tissue regeneration: A review of holistic approaches to oral health. J. Ayurveda Integr. Med. 2011, 2, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Kishore, M.; Panat, S.R.; Aggarwal, A.; Agarwal, N.; Upadhyay, N.; Alok, A. Evidence based dental care: Integrating clinical expertise with systematic research. J. Clin. Diagn. Res. 2014, 8, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Shetty, H. Use of natural products for oral hygiene maintenance: Revisiting traditional medicine. J. Complement. Integr. Med. 2018, 15, 20150103. [Google Scholar] [CrossRef] [PubMed]
- Vyas, T.; Bhatt, G.; Gaur, A.; Sharma, C.; Sharma, A.; Nagi, R. Chemical plaque control—A brief review. J. Family Med. Prim. Care 2021, 10, 1562–1568. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Z.; Hou, B. Diverse bacterial profile in extraradicular biofilms and periradicular lesions associated with persistent apical periodontitis. Int. Endod. J. 2021, 54, 1425–1433. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical applications of nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef]
- Najmi, Z.; Kumar, A.; Scalia, A.C.; Cochis, A.; Obradovic, B.; Grassi, F.A.; Leigheb, M.; Lamghari, M.; Loinaz, I.; Gracia, R.; et al. Evaluation of nisin and LL-37 antimicrobial peptides as tool to preserve articular cartilage healing in a septic environment. Front. Bioeng. Biotechnol. 2020, 8, 561. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Hadjirin, N.F.; Miller, E.L.; Murray, G.G.R.; Yen, P.L.K.; Phuc, H.D.; Wileman, T.M.; Hernandez-Garcia, J.; Williamson, S.M.; Parkhill, J.; Maskell, D.J.; et al. Large-scale genomic analysis of antimicrobial resistance in the zoonotic pathogen Streptococcus suis. BMC Biol. 2021, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jiang, W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 2014, 5, 508. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, J.; Wang, X.; Shi, J. Oral microbial diversity analysis among atrophic glossitis patients and healthy individuals. J. Oral Microbiol. 2021, 13, 1984063. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.; Mathney, J.M.; Kent, R.L.; Chalmers, N.I.; Hughes, C.V.; Loo, C.Y.; Pradhan, N.; Kanasi, E.; Hwang, J.; Dahlan, M.A.; et al. Cultivable anaerobic microbiota of severe early childhood caries. J. Clin. Microbiol. 2011, 49, 1464–1474. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Ran, S.; Huang, Z.; Liang, J. Bacterial diversity and community structure of supragingival plaques in adults with dental health or caries revealed by 16S pyrosequencing. Front. Microbiol. 2016, 7, 1145. [Google Scholar] [CrossRef]
- Jiang, W.; Ling, Z.; Lin, X.; Chen, Y.; Zhang, J.; Yu, J.; Xiang, C.; Chen, H. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb. Ecol. 2014, 67, 962–969. [Google Scholar] [CrossRef]
- Obata, J.; Takeshita, T.; Shibata, Y.; Yamanaka, W.; Unemori, M.; Akamine, A.; Yamashita, Y. Identification of the microbiota in carious dentin lesions using 16S rRNA gene sequencing. PLoS ONE 2014, 9, e103712. [Google Scholar] [CrossRef]
- Bapat, S.; Nagarajappa, R.; Ramesh, G.; Bapat, K. Effect of propolis mouth rinse on oral microorganisms—A randomized controlled trial. Clin. Oral Investig. 2021, 25, 6139–6146. [Google Scholar] [CrossRef]
- Samonis, G.; Anaissie, E.; Elting, L.; Bodey, G.P. Review of Citrobacter bacteremia in cancer patients over a sixteen-year period. Eur. J. Clin. Microbiol. Infect. Dis. 1991, 10, 479–485. [Google Scholar] [CrossRef]
- Shih, C.C.; Chen, Y.C.; Chang, S.C.; Luh, K.T.; Hsieh, W.C. Bacteremia due to Citrobacter species: Significance of primary intraabdominal infection. Clin. Infect. Dis. 1996, 23, 543–549. [Google Scholar] [CrossRef]
- Samonis, G.; Karageorgopoulos, D.E.; Kofteridis, D.P.; Matthaiou, D.K.; Sidiropoulou, V.; Maraki, S.; Falagas, M.E. Citrobacter infections in a general hospital: Characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 61–68. [Google Scholar] [CrossRef]
- Ranjan, K.P.; Ranjan, N. Citrobacter: An emerging health care associated urinary pathogen. Urol. Ann. 2013, 5, 313–314. [Google Scholar] [PubMed]
- Komiyama, E.Y.; Lepesqueur, L.S.; Yassuda, C.G.; Samaranayake, L.P.; Parahitiyawa, N.B.; Balducci, I.; Koga-Ito, C.Y. Enterococcus species in the oral cavity: Prevalence, virulence factors and antimicrobial susceptibility. PLoS ONE 2016, 11, e0163001. [Google Scholar] [CrossRef] [PubMed]
- Slots, J.; Listgarten, M.A. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J. Clin. Periodontol. 1988, 15, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Buhlin, K.; Sorsa, T.; Pussinen, P.J. Systemic Aggregatibacter actinomycetemcomitans leukotoxin-neutralizing antibodies in periodontitis. J. Periodontol. 2017, 88, 122–129. [Google Scholar] [CrossRef]
- Zhu, Z.; He, Z.; Xie, G.; Fan, Y.; Shao, T. Altered oral microbiota composition associated with recurrent aphthous stomatitis in young females. Medicine 2021, 100, e24742. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. The Genus psychrobacter. In The Prokaryotes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 920–930. [Google Scholar]
- Souto, R.; Silva-Boghossian, C.M.; Colombo, A.P. Prevalence of Pseudomonas aeruginosa and Acinetobacter spp. in subgingival biofilm and saliva of subjects with chronic periodontal infection. Braz. J. Microbiol. 2014, 45, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Ready, D.; Bedi, R.; Mullany, P.; Wilson, M. Penicillin and amoxicillin resistance in oral Veillonella spp. Int. J. Antimicrob. Agents 2012, 40, 188–189. [Google Scholar] [CrossRef]
- Mashima, I.; Theodorea, C.F.; Thaweboon, B.; Thaweboon, S.; Nakazawa, F. Identification of Veillonella species in the tongue biofilm by using a novel one-step polymerase chain reaction method. PLoS ONE 2016, 11, e0157516. [Google Scholar] [CrossRef]
- Ramos, J.M.; Fernandez-Roblas, R.; Garcia-Ruiz, P.; Soriano, F. Meningitis caused by Alcaligenes (Achromobacter) xylosoxidans associated with epidural catheter. Infection 1995, 23, 395–396. [Google Scholar] [CrossRef]
- Aisenberg, G.; Rolston, K.V.; Safdar, A. Bacteremia caused by Achromobacter and Alcaligenes species in 46 patients with cancer (1989–2003). Cancer 2004, 101, 2134–2140. [Google Scholar] [CrossRef] [PubMed]
- Gelfand, M.S.; Cleveland, K.O. Successful treatment with doripenem of ventriculitis due to Achromobacter xylosoxidans. QJM 2014, 107, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Isler, B.; Kidd, T.J.; Stewart, A.G.; Harris, P.; Paterson, D.L. Achromobacter infections and treatment options. Antimicrob. Agents Chemother. 2020, 64, e01025-20. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.P.; Boches, S.K.; Cotton, S.L.; Goodson, J.M.; Kent, R.; Haffajee, A.D.; Socransky, S.S.; Hasturk, H.; Van Dyke, T.E.; Dewhirst, F.; et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 2009, 80, 1421–1432. [Google Scholar] [CrossRef]
- Krishnan, K.; Chen, T.; Paster, B.J. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017, 23, 276–286. [Google Scholar] [CrossRef]
- Han, X.Y.; Andrade, R.A. Brevundimonas diminuta infections and its resistance to fluoroquinolones. J. Antimicrob. Chemother. 2005, 55, 853–859. [Google Scholar] [CrossRef]
- Owen, O.W. A study of bacterial counts (lactobacilli) in saliva related to orthodontic appliances. Am. J. Orthodont. 1949, 35, 672–678. [Google Scholar] [CrossRef]
- Zeng, H.; Chan, Y.; Gao, W.; Leung, W.K.; Watt, R.M. Diversity of Treponema denticola and other oral Treponeme lineages in subjects with periodontitis and gingivitis. Microbiol. Spectr. 2021, 9, e0070121. [Google Scholar] [CrossRef]
- Prates, M.; Fernandes, F.; Proença, F.; Mussá, Y.; Tavares, A.; Pereira, A. Oral infection caused by Stenotrophomonas maltophilia: A rare presentation of an emerging opportunistic pathogen. Case Rep. Infect. Dis. 2020, 2020, 6346120. [Google Scholar]
- Buckle, J. Clinical Aromatherapy, 3rd ed.; Elsevier Health Sciences: London, UK, 2015; pp. 130–167. [Google Scholar]
- Sedghi, L.; DiMassa, V.; Harrington, A.; Lynch, S.V.; Kapila, Y.L. The oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontology 2000 2021, 87, 107–131. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).