Deep Learning-Based Automatic Segmentation of Mandible and Maxilla in Multi-Center CT Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
2.2. Framework and Network Architectures
2.3. Performance Evaluation
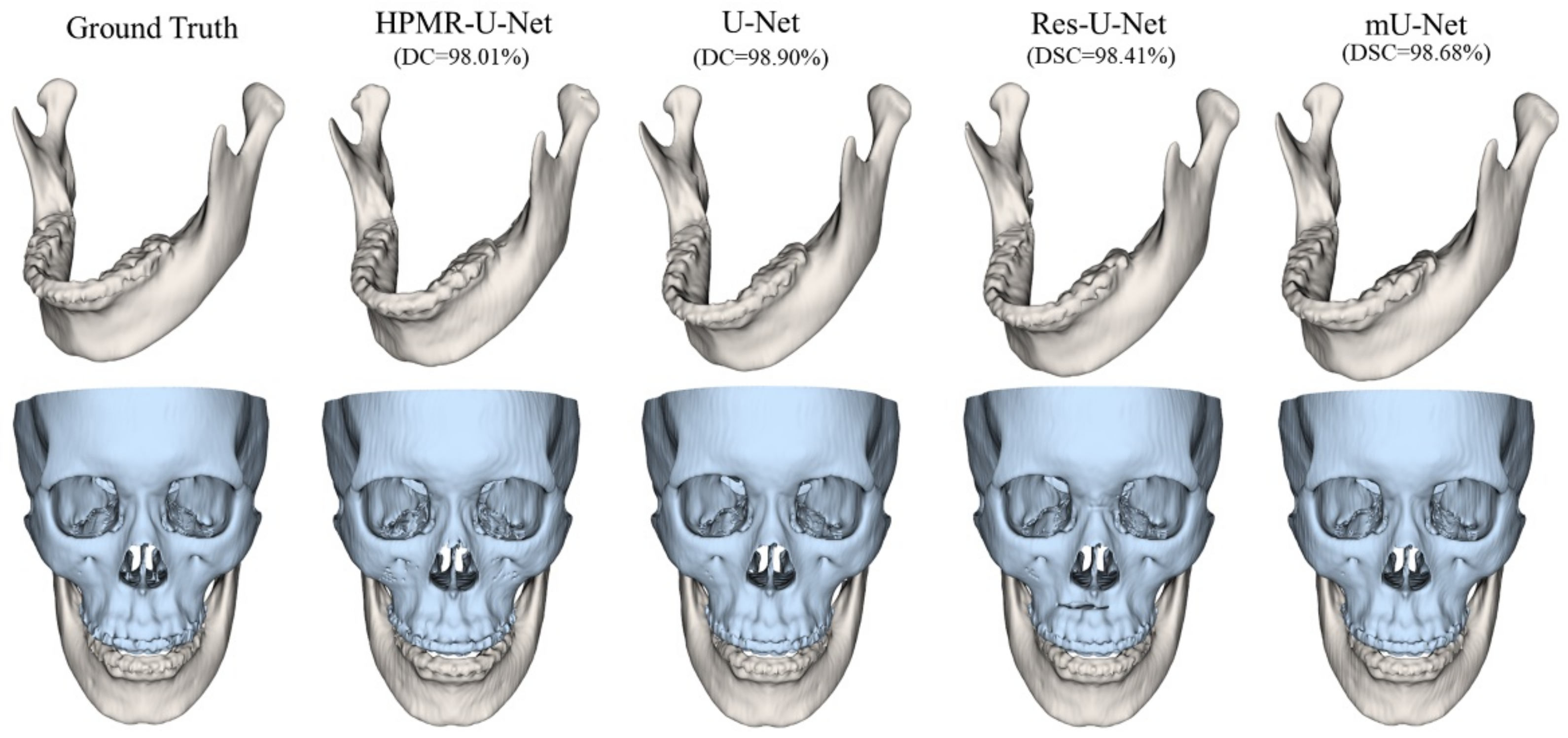
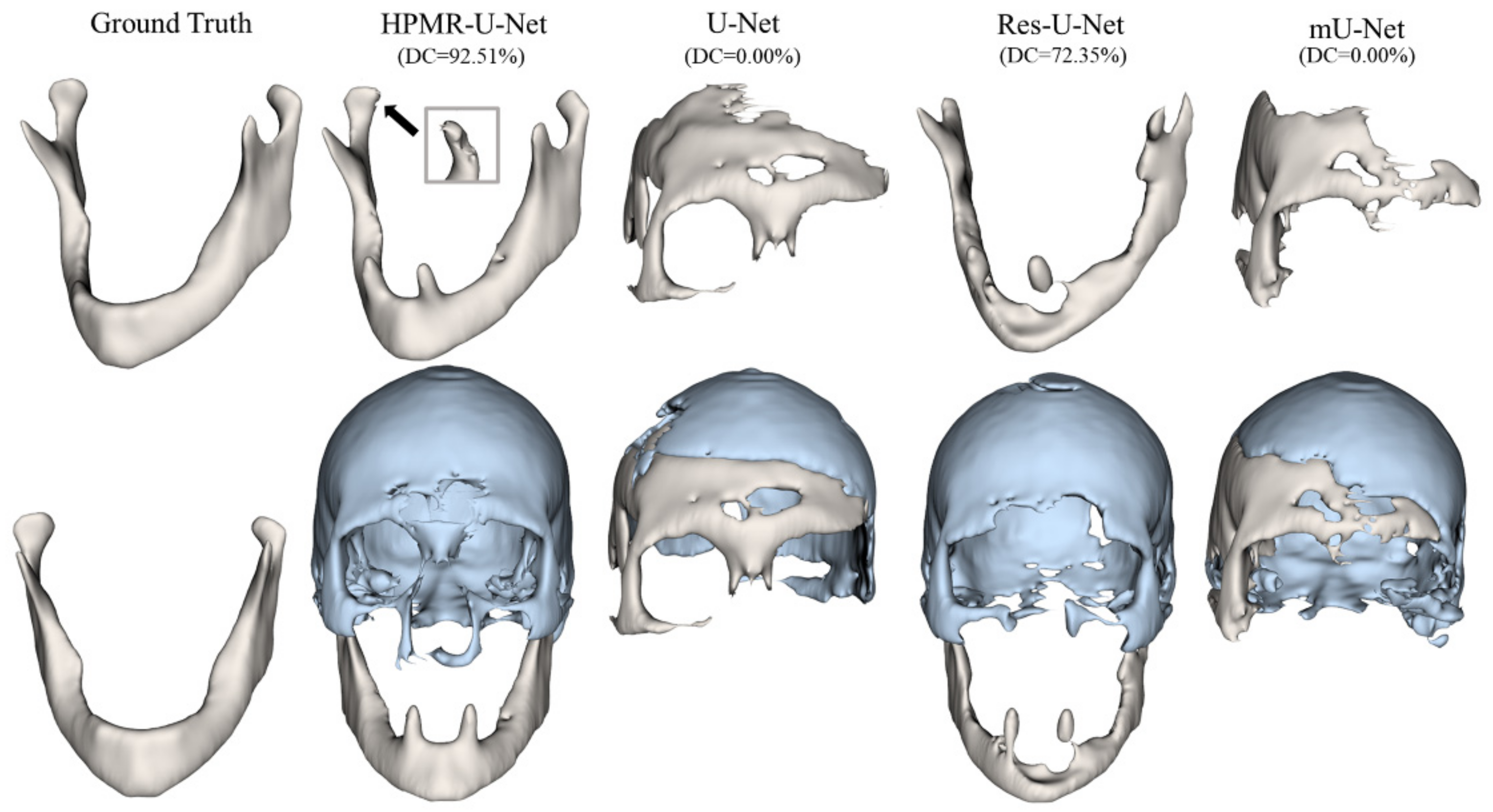
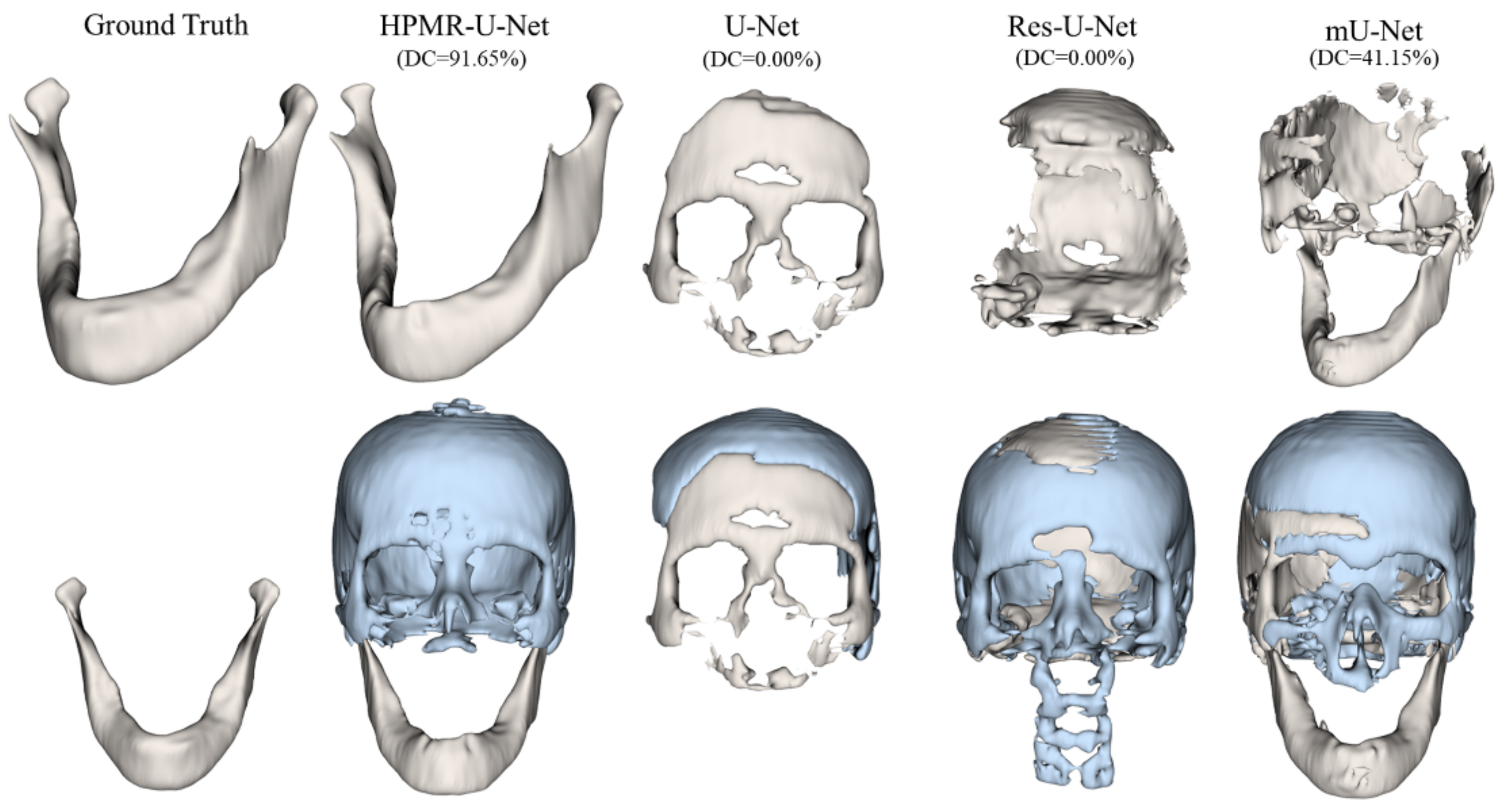
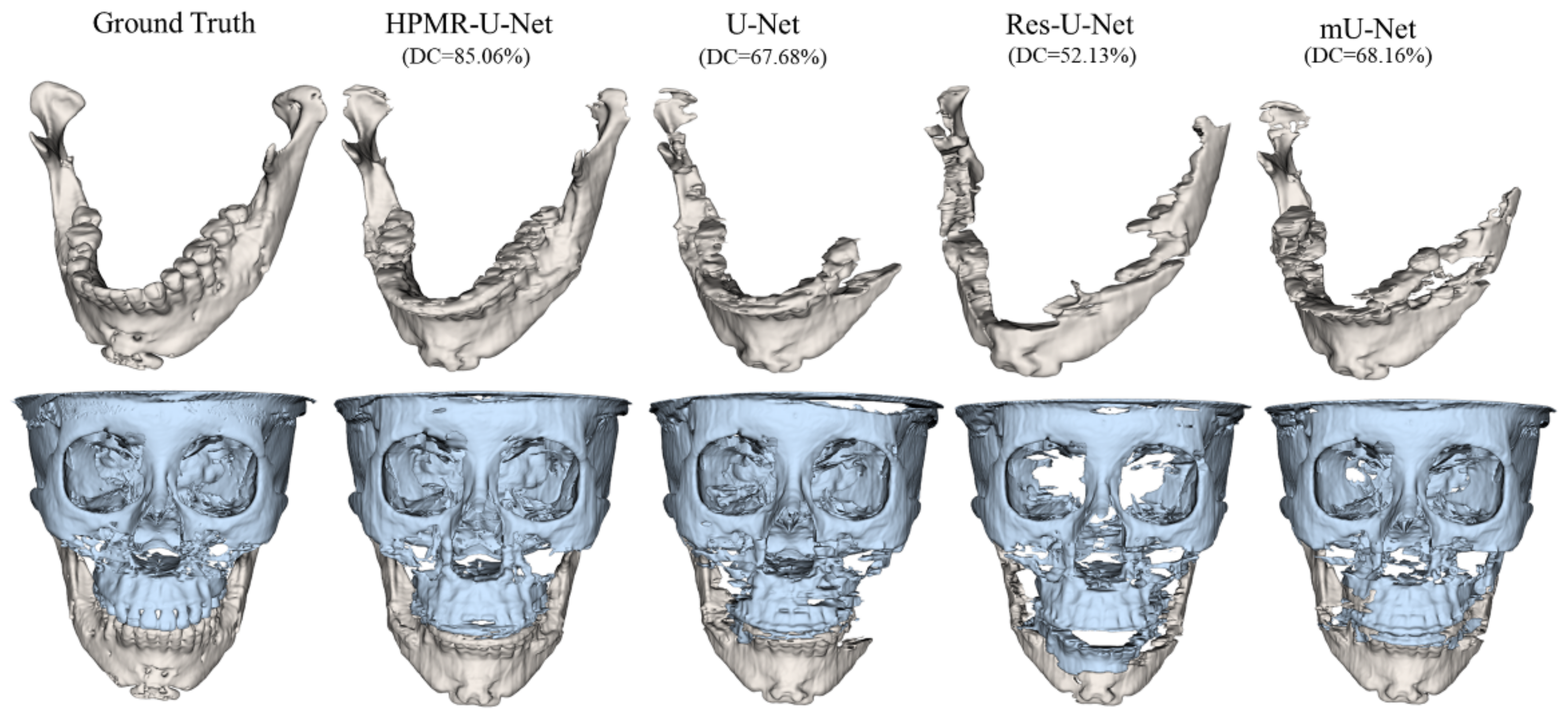
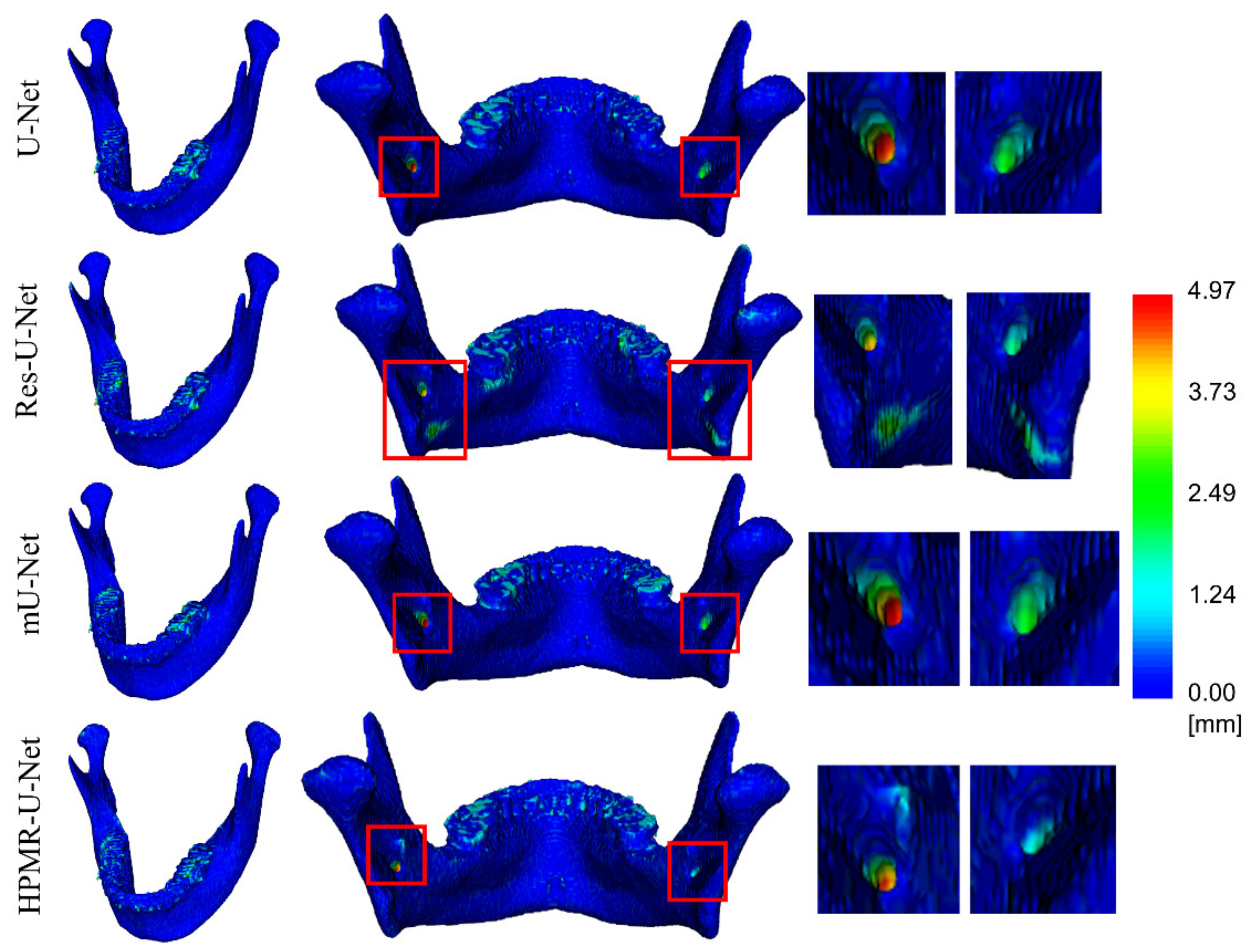
3. Results
4. Discussion & Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiu, B.; Guo, J.; Kraeima, J.; Glas, H.H.; Borra, R.J.; Witjes, M.J.; van Ooijen, P.M. Automatic Segmentation of the Mandible from Computed Tomography Scans for 3D Virtual Surgical Planning Using the Convolutional Neural Network. Phys. Med. Biol. 2019, 64, 175020. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, K.C.; Gao, Y.; Shi, F.; Liao, S.; Li, G.; Shen, S.G.; Yan, J.; Lee, P.K.; Chow, B. Automated Bone Segmentation from Dental CBCT Images Using Patch-based Sparse Representation and Convex Optimization. Med. Phys. 2014, 41, 043503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodym, O.; Španěl, M.; Herout, A. Segmentation of Head and Neck Organs at Risk Using Cnn with Batch Dice Loss. In Proceedings of the German Conference on Pattern Recognition; Springer: Stuttgart, Germany, 2018; pp. 105–114. [Google Scholar]
- Byrne, N.; Velasco Forte, M.; Tandon, A.; Valverde, I.; Hussain, T. A Systematic Review of Image Segmentation Methodology, Used in the Additive Manufacture of Patient-Specific 3D Printed Models of the Cardiovascular System. JRSM Cardiovasc. Dis. 2016, 5, 2048004016645467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huff, T.J.; Ludwig, P.E.; Zuniga, J.M. The Potential for Machine Learning Algorithms to Improve and Reduce the Cost of 3-Dimensional Printing for Surgical Planning. Expert Rev. Med. Devices 2018, 15, 349–356. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, L.; Wang, L.; Gao, Y.; Chen, W.; Shen, D. Hierarchical Vertex Regression-Based Segmentation of Head and Neck CT Images for Radiotherapy Planning. IEEE Trans. Image Process. 2017, 27, 923–937. [Google Scholar] [CrossRef]
- Qiu, B.; Guo, J.; Kraeima, J.; Glas, H.H.; Borra, R.J.; Witjes, M.J.; Ooijen, P.M.V. Recurrent Convolutional Neural Networks for Mandible Segmentation from Computed Tomography. arXiv 2020, arXiv:2003.06486. [Google Scholar]
- Raudaschl, P.F.; Zaffino, P.; Sharp, G.C.; Spadea, M.F.; Chen, A.; Dawant, B.M.; Albrecht, T.; Gass, T.; Langguth, C.; Lüthi, M. Evaluation of Segmentation Methods on Head and Neck CT: Auto-segmentation Challenge 2015. Med. Phys. 2017, 44, 2020–2036. [Google Scholar] [CrossRef]
- Chen, A.; Dawant, B. A Multi-Atlas Approach for the Automatic Segmentation of Multiple Structures in Head and Neck CT Images. MIDAS J. 2015. [Google Scholar] [CrossRef]
- Mannion-Haworth, R.; Bowes, M.; Ashman, A.; Guillard, G.; Brett, A.; Vincent, G. Fully Automatic Segmentation of Head and Neck Organs Using Active Appearance Models. MIDAS J. 2015. [Google Scholar] [CrossRef]
- Han, X.; Hoogeman, M.S.; Levendag, P.C.; Hibbard, L.S.; Teguh, D.N.; Voet, P.; Cowen, A.C.; Wolf, T.K. Atlas-Based Auto-Segmentation of Head and Neck CT Images; Springer: Berlin/Heidelberg, Germany, 2008; pp. 434–441. [Google Scholar]
- Linares, O.C.; Bianchi, J.; Raveli, D.; Neto, J.B.; Hamann, B. Mandible and Skull Segmentation in Cone Beam Computed Tomography Using Super-Voxels and Graph Clustering. Vis. Comput. 2019, 35, 1461–1474. [Google Scholar]
- Zhu, W.; Huang, Y.; Zeng, L.; Chen, X.; Liu, Y.; Qian, Z.; Du, N.; Fan, W.; Xie, X. AnatomyNet: Deep Learning for Fast and Fully Automated Whole-volume Segmentation of Head and Neck Anatomy. Med. Phys. 2019, 46, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cootes, T.F.; Edwards, G.J.; Taylor, C.J. Active Appearance Models. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 681–685. [Google Scholar] [CrossRef] [Green Version]
- Fritscher, K.D.; Peroni, M.; Zaffino, P.; Spadea, M.F.; Schubert, R.; Sharp, G. Automatic Segmentation of Head and Neck CT Images for Radiotherapy Treatment Planning Using Multiple Atlases, Statistical Appearance Models, and Geodesic Active Contours. Med. Phys. 2014, 41, 051910. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T.; Gass, T.; Langguth, C.; Lüthi, M. Multi Atlas Segmentation with Active Shape Model Refinement for Multi-Organ Segmentation in Head and Neck Cancer Radiotherapy Planning. MIDAS J. 2015. [Google Scholar] [CrossRef]
- Aghdasi, N.; Li, Y.; Berens, A.; Moe, K.; Hannaford, B. Automatic Mandible Segmentation on CT Images Using Prior Anatomical Knowledge. MIDAS J. 2016. [Google Scholar] [CrossRef]
- Chuang, Y.J.; Doherty, B.M.; Adluru, N.; Chung, M.K.; Vorperian, H.K. A Novel Registration-Based Semi-Automatic Mandible Segmentation Pipeline Using Computed Tomography Images to Study Mandibular Development. J. Comput. Assist. Tomogr. 2018, 42, 306. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [Green Version]
- Ibragimov, B.; Xing, L. Segmentation of Organs-at-risks in Head and Neck CT Images Using Convolutional Neural Networks. Med. Phys. 2017, 44, 547–557. [Google Scholar] [CrossRef] [Green Version]
- Tong, N.; Gou, S.; Yang, S.; Ruan, D.; Sheng, K. Fully Automatic Multi-organ Segmentation for Head and Neck Cancer Radiotherapy Using Shape Representation Model Constrained Fully Convolutional Neural Networks. Med. Phys. 2018, 45, 4558–4567. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Liu, L.; Cheng, J.; Quan, Q.; Wu, F.-X.; Wang, Y.-P.; Wang, J. A Survey on U-Shaped Networks in Medical Image Segmentations. Neurocomputing 2020, 409, 244–258. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, L.; Wang, M.; Song, Z. Organ at Risk Segmentation in Head and Neck Ct Images Using a Two-Stage Segmentation Framework Based on 3D U-Net. IEEE Access 2019, 7, 144591–144602. [Google Scholar] [CrossRef]
- Rundo, L.; Han, C.; Nagano, Y.; Zhang, J.; Hataya, R.; Militello, C.; Tangherloni, A.; Nobile, M.S.; Ferretti, C.; Besozzi, D.; et al. USE-Net: Incorporating Squeeze-and-Excitation Blocks into U-Net for Prostate Zonal Segmentation of Multi-Institutional MRI Datasets. Neurocomputing 2019, 365, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention Gated Networks: Learning to Leverage Salient Regions in Medical Images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.; Sala, E.; Schönlieb, C.-B.; Rundo, L. Focus U-Net: A Novel Dual Attention-Gated CNN for Polyp Segmentation during Colonoscopy. Comput. Biol. Med. 2021, 137, 104815. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Nguyen, D.; Jiang, S.B. Generalizability Issues with Deep Learning Models in Medicine and Their Potential Solutions: Illustrated with Cone-Beam Computed Tomography (CBCT) to Computed Tomography (CT) Image Conversion. Mach. Learn. Sci. Technol. 2020, 2, 015007. [Google Scholar] [CrossRef]
- Qiu, B.; van der Wel, H.; Kraeima, J.; Glas, H.H.; Guo, J.; Borra, R.J.H.; Witjes, M.J.H.; van Ooijen, P.M.A. Automatic Segmentation of Mandible from Conventional Methods to Deep Learning—A Review. J. Pers. Med. 2021, 11, 629. [Google Scholar] [CrossRef]
- Hesse, L.S.; Kuling, G.; Veta, M.; Martel, A.L. Intensity Augmentation to Improve Generalizability of Breast Segmentation Across Different MRI Scan Protocols. IEEE Trans. Biomed. Eng. 2021, 68, 759–770. [Google Scholar] [CrossRef]
- Bosniak, M.A. The Current Radiological Approach to Renal Cysts. Radiology 1986, 158, 1–10. [Google Scholar] [CrossRef]
- Bluemke, D.A.; Moy, L.; Bredella, M.A.; Ertl-Wagner, B.B.; Fowler, K.J.; Goh, V.J.; Halpern, E.F.; Hess, C.P.; Schiebler, M.L.; Weiss, C.R. Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers—From the Radiology Editorial Board. Radiology 2020, 294, 487–489. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Shim, E.; Park, J.; Kim, Y.-J.; Lee, U.; Kim, Y. Web-Based Fully Automated Cephalometric Analysis by Deep Learning. Comput. Methods Programs Biomed. 2020, 194, 105513. [Google Scholar] [CrossRef]
- Tao, Q.; Yan, W.; Wang, Y.; Paiman, E.H.M.; Shamonin, D.P.; Garg, P.; Plein, S.; Huang, L.; Xia, L.; Sramko, M.; et al. Deep Learning–Based Method for Fully Automatic Quantification of Left Ventricle Function from Cine MR Images: A Multivendor, Multicenter Study. Radiology 2019, 290, 81–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- B, S.; R, N. Transfer Learning Based Automatic Human Identification Using Dental Traits- An Aid to Forensic Odontology. J. Forensic Leg. Med. 2020, 76, 102066. [Google Scholar] [CrossRef] [PubMed]
- Ghafoorian, M.; Mehrtash, A.; Kapur, T.; Karssemeijer, N.; Marchiori, E.; Pesteie, M.; Guttmann, C.R.G.; de Leeuw, F.-E.; Tempany, C.M.; van Ginneken, B.; et al. Transfer Learning for Domain Adaptation in MRI: Application in Brain Lesion Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2017; pp. 516–524. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.-S.; Jung, S.-K.; Ryu, J.-J.; Shin, S.-W.; Choi, J. Evaluation of Transfer Learning with Deep Convolutional Neural Networks for Screening Osteoporosis in Dental Panoramic Radiographs. J. Clin. Med. 2020, 9, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A Survey of Transfer Learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Frei, S.; Cao, Y.; Gu, Q. Algorithm-Dependent Generalization Bounds for Overparameterized Deep Residual Networks. arXiv 2019, arXiv:1910.02934. [Google Scholar]
- Huang, K.; Tao, M.; Wang, Y.; Zhao, T. Why Do Deep Residual Networks Generalize Better than Deep Feedforward Networks? — A Neural Tangent Kernel Perspective. 2020, 12. arXiv 2020, arXiv:2002.06262. [Google Scholar]
- Ang, K.K.; Zhang, Q.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Sherman, E.J.; Weber, R.S.; Galvin, J.M.; Bonner, J.A.; Harris, J.; El-Naggar, A.K. Randomized Phase III Trial of Concurrent Accelerated Radiation plus Cisplatin with or without Cetuximab for Stage III to IV Head and Neck Carcinoma: RTOG 0522. J. Clin. Oncol. 2014, 32, 2940. [Google Scholar] [CrossRef]
- Nikolov, S.; Blackwell, S.; Zverovitch, A.; Mendes, R.; Livne, M.; De Fauw, J.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B. Deep Learning to Achieve Clinically Applicable Segmentation of Head and Neck Anatomy for Radiotherapy. arXiv 2018, arXiv:1809.04430. [Google Scholar]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- Bosch, W.R.; Straube, W.L.; Matthews, J.W.; Purdy, J.A. Data from Head-Neck_cetuximab. Cancer Imaging Arch. 2015, 10, K9. [Google Scholar]
- Han, X. Automatic Liver Lesion Segmentation Using A Deep Convolutional Neural Network Method. Med. Phys. 2017, 44, 1408–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulat, A.; Tzimiropoulos, G. Binarized Convolutional Landmark Localizers for Human Pose Estimation and Face Alignment with Limited Resources. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 3706–3714. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Seo, H.; Huang, C.; Bassenne, M.; Xiao, R.; Xing, L. Modified U-Net (MU-Net) with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images. IEEE Trans. Med. Imaging 2019, 39, 1316–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Gupta, R.; Ojha, M.; Singh, K.P. A Comparative Analysis of Various Regularization Techniques to Solve Overfitting Problem in Artificial Neural Network. In Proceedings of the Data Science and Analytics; Panda, B., Sharma, S., Roy, N.R., Eds.; Springer: Singapore, 2018; pp. 363–371. [Google Scholar]
- He, F.; Liu, T.; Tao, D. Why ResNet Works? Residuals Generalize. arXiv 2019, arXiv:1904.01367. [Google Scholar]
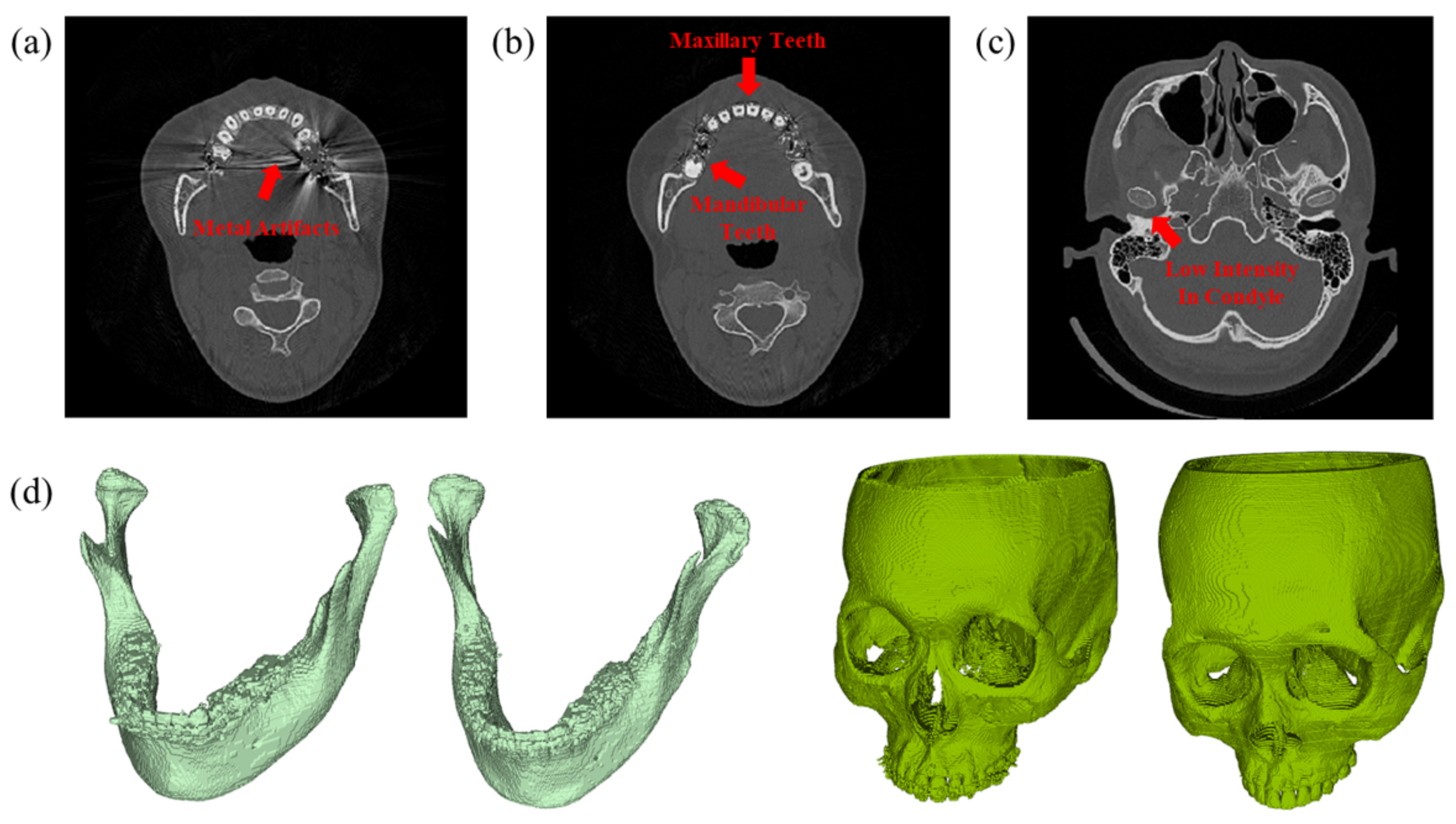
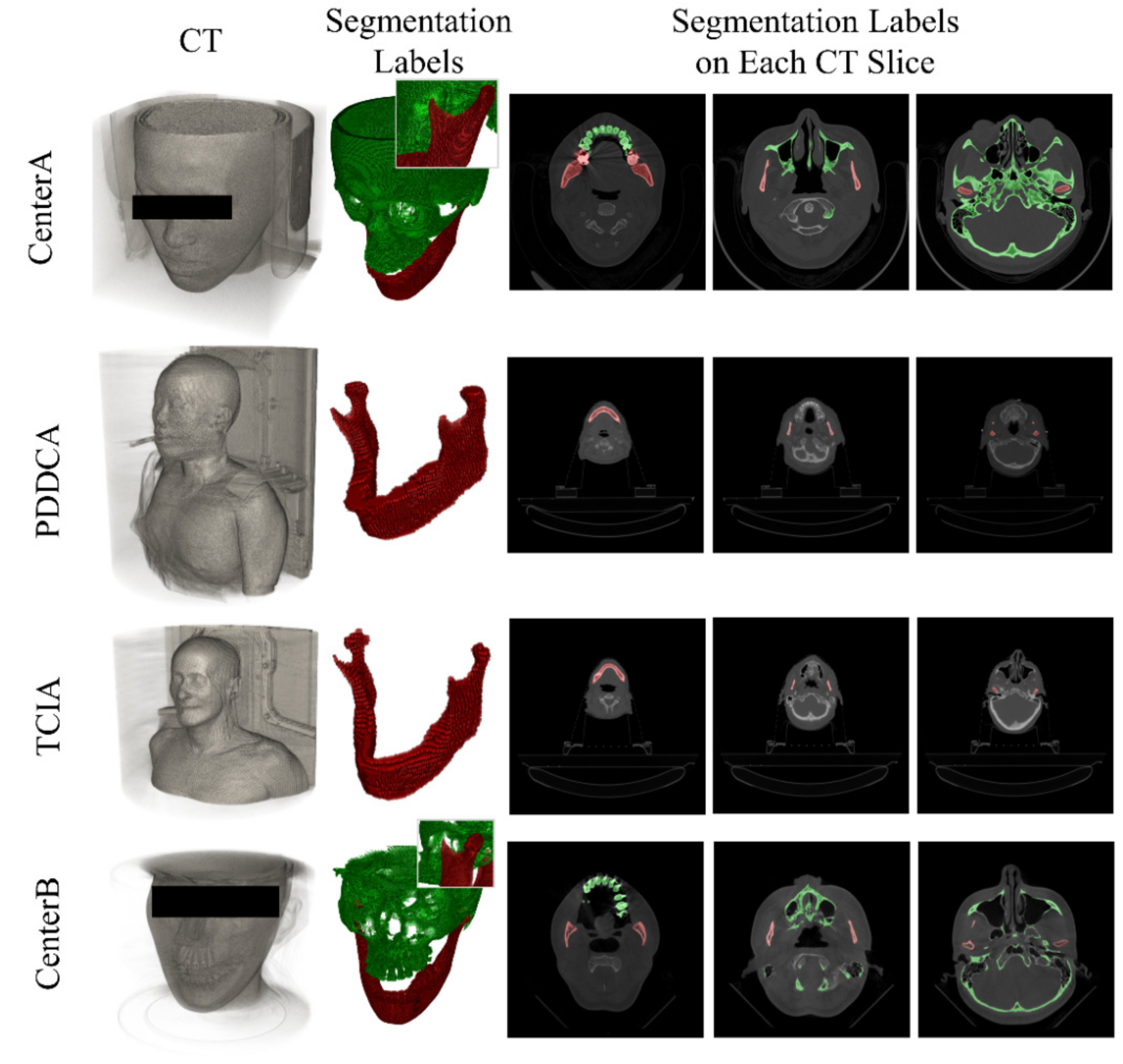
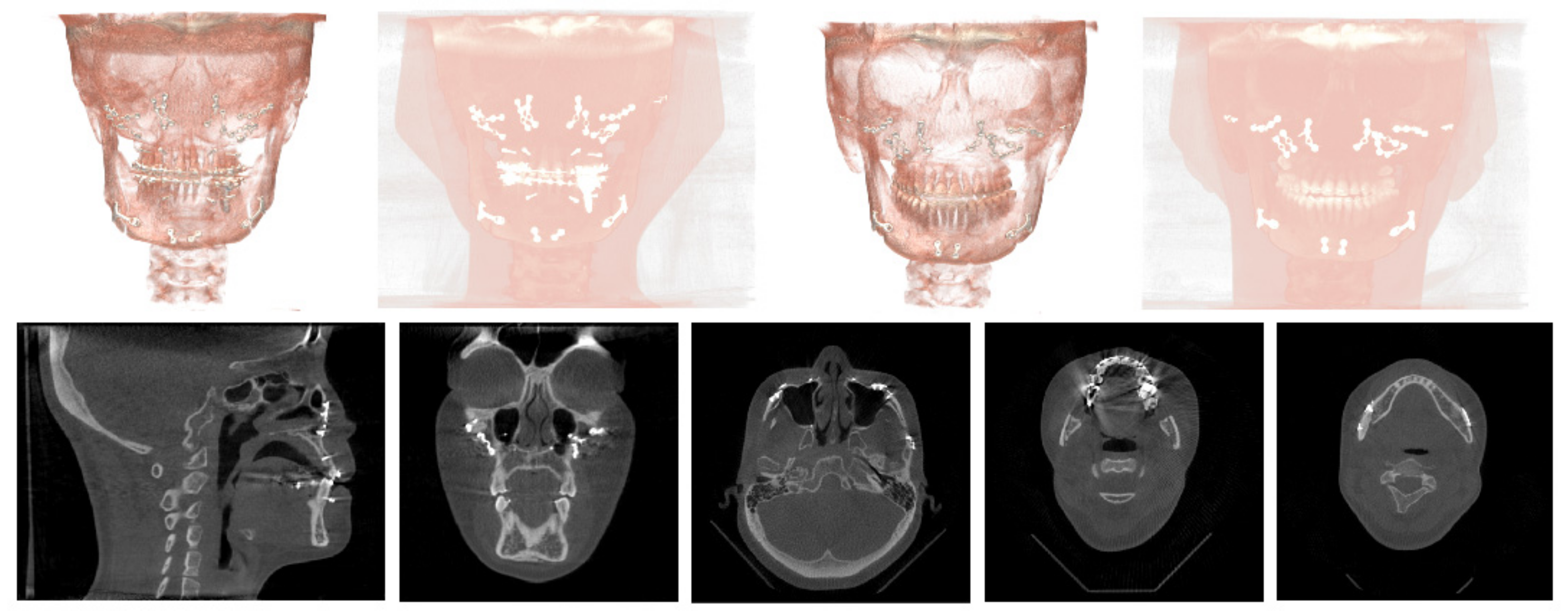
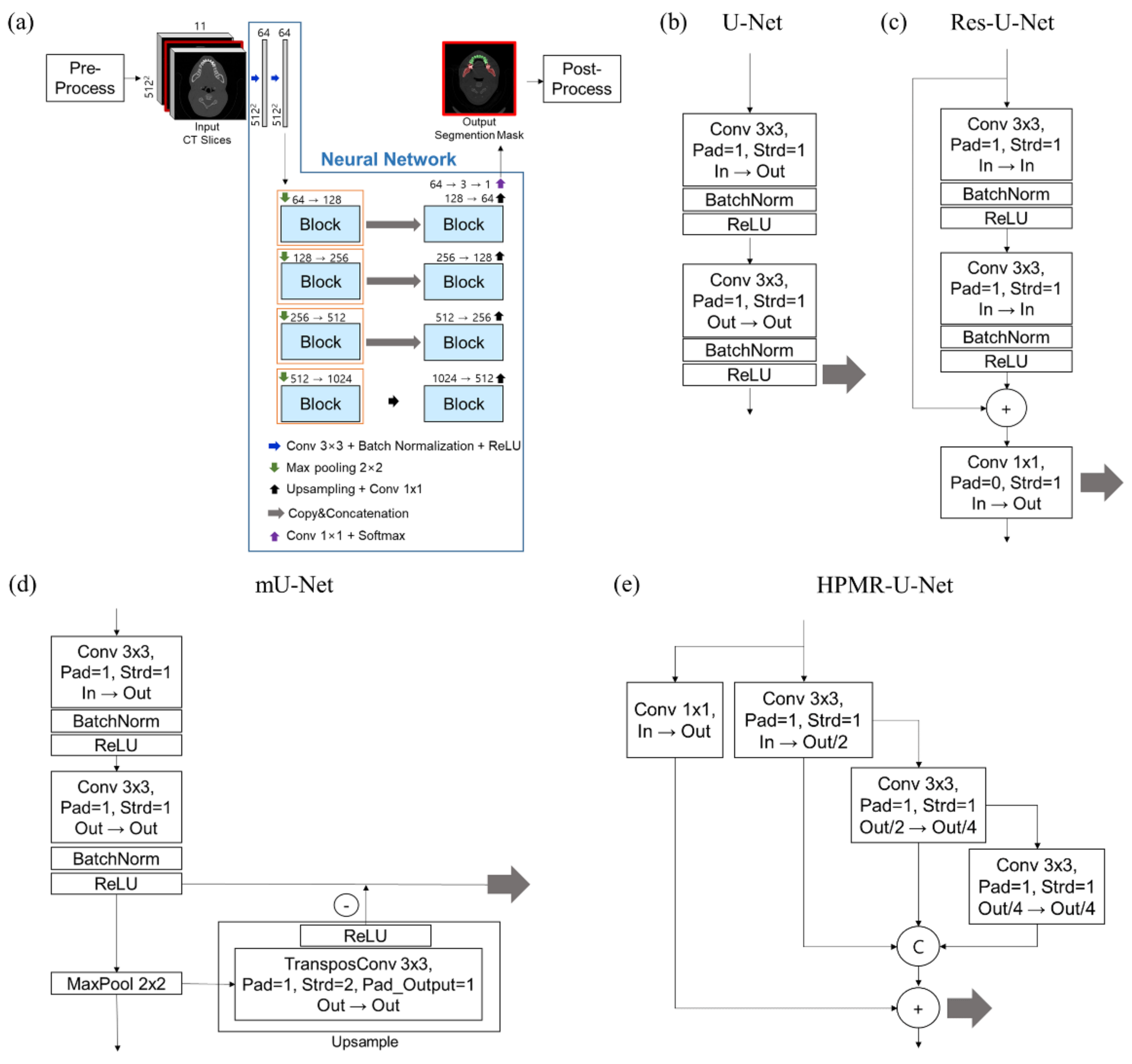
| Dataset | CenterA | PDDCA † | TCIA † | CenterB |
|---|---|---|---|---|
| No. of sets | 171 (Train: 146, Validation: 10, Test: 15) | 15 | 28 | 15 |
| Acquisition type | MDCT | MDCT | MDCT | CBCT |
| Target structure | Mandible & Maxilla | OARs | OARs | Mandible & Maxilla |
| No. of slices | 166–450, 208 ± 32 | 109–263, 154 ± 36 | 61–110, 93 ± 12 | 432 |
| Slice size [pixel] | 512 | 576 | 512 | 512 |
| Pixel spacing [mm] | 0.36–0.49, 0.44 ± 0.03 | 0.98–1.27, 1.11 ± 0.10 | 0.94–1.27, 1.04 ± 0.10 | 0.40 |
| Slice thickness [mm] | 0.50–1.04, 0.99 ± 0.07 | 2.0–3.0, 2.73 ± 0.31 | 2.50 | 0.40 |
| In-House Test | External Test | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CenterA | PDDCA | TCIA | CenterB | |||||||||
| DC [%] | 95HD [mm] | ASD [mm] | DC [%] | 95HD [mm] | ASD [mm] | DC [%] | 95HD [mm] | ASD [mm] | DC [%] | 95HD [mm] | ASD [mm] | |
| U-Net | 98.3 ± 0.4 | 0.4 ± 0.1 | 0.0 ± 0.0 | 63.4 ± 20.2 | 7.3 ± 5.8 | 1.8 ± 3.5 | 62.8 ± 26.3 | 9.6 ± 11.8 | 3.2 ± 7.9 | 61.2 ± 17.9 | 33.7 ± 26.1 | 4.1 ± 4.0 |
| Res-U-Net | 98.2 ± 0.4 | 0.4 ± 0.1 | 0.1 ± 0.0 | 51.3 ± 20.1 | 13.5 ± 12.3 | 2.0 ± 1.8 | 46.3 ± 25.5 | 18.0 ± 19.8 | 6.5 ± 13.5 | 48.5 ± 13.1 | 28.8 ± 20.6 | 4.1 ± 3.3 |
| mU-Net | 98.4 ± 0.3 | 0.4 ± 0.0 | 0.0 ± 0.0 | 72.3 ± 21.6 | 5.6 ± 6.4 | 1.5 ± 3.5 | 71.4 ± 27.8 | 8.4 ± 12.9 | 2.5 ± 5.0 | 63.6 ± 14.7 | 22.5 ± 18.9 | 2.6 ± 2.2 |
| HPMR-U-Net | 97.4 ± 0.4 | 0.4 ± 0.1 | 0.1 ± 0.0 | 86.5 ± 3.9 | 1.8 ± 1.3 | 0.2 ± 0.1 | 86.4 ± 6.2 | 2.8 ± 7.7 | 0.3 ± 0.7 | 77.7 ± 4.1 | 3.4 ± 0.6 | 0.7 ± 0.2 |
| In-House Test | External Test | |||||
|---|---|---|---|---|---|---|
| CenterA | CenterB | |||||
| DC [%] | 95HD [mm] | ASD [mm] | DC [%] | 95HD [mm] | ASD [mm] | |
| U-Net | 96.5 ± 0.8 | 0.4 ± 0.1 | 0.1 ± 0.0 | 75.0 ± 5.7 | 9.0 ± 8.8 | 1.1 ± 0.8 |
| Res-U-Net | 96.2 ± 0.8 | 0.4 ± 0.1 | 0.1 ± 0.0 | 67.6 ± 12.3 | 17.1 ± 18.1 | 2.5 ± 3.4 |
| mU-Net | 96.5 ± 0.7 | 0.4 ± 0.0 | 0.1 ± 0.0 | 75.9 ± 5.1 | 8.6 ± 7.9 | 1.0 ± 0.7 |
| HPMR-U-Net | 90.2 ± 19.5 | 0.5 ± 0.1 | 0.1 ± 0.0 | 82.8 ± 3.2 | 2.7 ± 1.6 | 0.4 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Kim, H.; Shim, E.; Hwang, B.-Y.; Kim, Y.; Lee, J.-W.; Seo, H. Deep Learning-Based Automatic Segmentation of Mandible and Maxilla in Multi-Center CT Images. Appl. Sci. 2022, 12, 1358. https://doi.org/10.3390/app12031358
Park S, Kim H, Shim E, Hwang B-Y, Kim Y, Lee J-W, Seo H. Deep Learning-Based Automatic Segmentation of Mandible and Maxilla in Multi-Center CT Images. Applied Sciences. 2022; 12(3):1358. https://doi.org/10.3390/app12031358
Chicago/Turabian StylePark, Seungbin, Hannah Kim, Eungjune Shim, Bo-Yeon Hwang, Youngjun Kim, Jung-Woo Lee, and Hyunseok Seo. 2022. "Deep Learning-Based Automatic Segmentation of Mandible and Maxilla in Multi-Center CT Images" Applied Sciences 12, no. 3: 1358. https://doi.org/10.3390/app12031358
APA StylePark, S., Kim, H., Shim, E., Hwang, B.-Y., Kim, Y., Lee, J.-W., & Seo, H. (2022). Deep Learning-Based Automatic Segmentation of Mandible and Maxilla in Multi-Center CT Images. Applied Sciences, 12(3), 1358. https://doi.org/10.3390/app12031358






