Abstract
Antibiotic use causes dysbiosis; probiotic treatment may mitigate these effects by promoting the growth of beneficial bacteria and restoring a healthy gut microbiota. The effects of a probiotic formulation (MegaSporeBiotic™; Bacillus indicus [HU36], Bacillus subtilis [HU58], Bacillus coagulans [SC208], Bacillus licheniformis [SL307], and Bacillus clausii [SC109]) on the microbial community activity and composition of the baby gut microbiome were evaluated using the in vitro gut model, the mucosal simulator of the human intestinal microbial system (M-SHIME®), and fecal samples from four babies aged 6–24 months. Established Baby-Quad-M-SHIME® proximal colon vessels (control period) were treated with 75 mg amoxicillin:clavulanic acid (2:1) for five days (antibiotic period) and then with/without daily MegaSporeBiotic™ (2 weeks; follow-up period). Antibiotic-induced dysbiosis was confirmed by significantly decreased SCFA levels and changes in the microbial community composition in the antibiotic vs. the control periods. SCFA levels recovered for both untreated and treated chambers during the follow-up period; recovery was similar among donors. There were few differences in microbial community composition between untreated and treated chambers during the follow-up period. MegaSporeBiotic™ supplementation following antibiotic-induced dysbiosis had little effect on the recovery of the microbial community activity or composition of the baby gut microbiota. Future studies evaluating simultaneous antibiotic and probiotic treatment may be of interest.
1. Introduction
The gut microbiome influences both host metabolism and the immune system [1,2,3,4,5]. Some bacteria found in the gut are able to ferment indigestible dietary glycans into short-chain fatty acids (SCFAs) which play an important role in both gut health and overall health [6]. Acetate, propionate, and butyrate are the most predominant SCFAs involved in the maintenance of host health. Some important functions of SCFAs include that they are an energy source for intestinal epithelial cells, they help to strengthen the intestinal barrier, and they stimulate blood flow in the colon [7,8]. Prior to stabilization, which occurs at around 3 years of age, the composition of the gut microbiome is dynamic and may be influenced by a number of factors. One such factor is antibiotic use, which is relatively widespread among children. The microbiota of antibiotic-treated children is modestly less diverse than untreated children, though this is only apparent in those >1 year of age [9]. Short-term microbiota composition changes are observed after antibiotic treatment in those <3 years of age [9,10]. Decreased microbial diversity in early childhood may limit immune education, potentially leading to less robust microbial communities later in life [11].
Probiotics are defined by the World Health Organization as “live microorganisms which when administered in adequate amounts confer a health benefit to the host” [12]. Several benefits are reported for probiotic use, including improved gastrointestinal health and gut barrier function, and beneficial effects on both the mucosal and systemic immune systems [13]. Members of the spore-forming Bacillus spp. have been used as probiotics, having several advantages over non-spore-based probiotics. These advantages include resistance to desiccation and heat, and stability over a wide range of pHs, allowing them to survive the harsh acidic conditions of the stomach [14]. Bacillus-based probiotics have been shown to support increased SCFA production either directly or by promoting increased levels of SCFA-producing bacteria [15,16]. An in vitro study demonstrated that these probiotics aid in the recovery of the adult gut microbiome after antibiotic-induced dysbiosis, particularly for propionate-producing bacterial families [15].
The mucosal simulator of the human intestinal microbial ecosystem (M-SHIME®) is an in vitro modeling system that has been used to study the effects of probiotics, nutritional supplements, etc., on the activity and composition of the human gut microbiome [17,18]. This model has several advantages, including integration of the entire gastrointestinal tract, the ability to maintain microbiome stability over a long period of time (up to 7 weeks), and that the microbiota of different age groups can be explored (e.g., adult, toddler, baby) [18,19,20].
MegaSporeBiotic™ is a spore-based probiotic that contains five Bacillus strains (Bacillus indicus [HU36], Bacillus subtilis [HU58], Bacillus coagulans [SC208], Bacillus licheniformis [SL307], and Bacillus clausii [SC109]). This probiotic supplement was previously shown to support changes in gut microbiome metabolism, including increased propionate production, in an M-SHIME® model of the healthy adult gut microbiome [21]. In that study, changes to the microbial community composition with MegaSporeBiotic™ supplementation were also observed. Given that the gut microbiome of children <3 years is still developing and that antibiotics are known to affect the gut microbiome of young children, with potential impacts on microbial communities later in life, this study aimed to evaluate whether MegaSporeBiotic™ had any effects on the recovery of the activity and composition of the gut microbial community of 6–24-month-old babies following treatment with a broad-spectrum antibiotic using the Baby-M-SHIME® technology platform.
2. Materials and Methods
2.1. Test Products and Fecal Samples
All chemicals used in this study were obtained from Merck (Darmstadt, German), unless otherwise stated. MegaSporeBiotic™, which is a probiotic comprising spores of five different Bacillus strains (B. indicus [HU36], B. subtilis [HU58], B. coagulans [SC208], B. licheniformis [SL307], and B. clausii [SC109]) was provided by Microbiome Labs (Glenview, IL, USA).
Fecal microbiota from healthy 6–24-month-old babies (4 individual donors) was used in this study. We chose to include four individual donors in our studies rather than a single donor given the variation in the gut microbiome among individuals. We hoped to identify effects that were common to all four donors. The fecal samples were processed as follows. An Oxoid™ AnaeroGen™ sachet (Thermo Fisher Scientific, MA, USA) was placed into the sample box immediately after fecal sample collection to remove all oxygen. This was followed by the addition of anaerobic phosphate-buffer and homogenization in a stomacher to prepare a fecal slurry. A brief centrifugation was utilized to remove large particles. The fecal slurry was then added to an equal volume of cryoprotectant solution in an anaerobic workstation. The slurry was then snap-frozen in liquid nitrogen and stored at –80 °C until used in the study. All donors were Belgian and were being bottle fed at the time the fecal sample was collected. Ages of the individual donors were as follows: donor A, 8 months; donor B, 11 months; donor C, 15 months; donor D, 12 months.
2.2. Baby-M-SHIME® Study
The SHIME® model was initially described by Molly et al. and was developed as an in vitro representation of the gastrointestinal tract of adult humans [17]. The model was later adapted to allow modeling of both the luminal and mucosal microbial communities (M-SHIME®) [22,23]. The Baby-M-SHIME® model was adapted from an adult model as described in Van den Abbeele et al. [24] to represent the gastrointestinal tract of weaned infants [25]. Adaptations included the use of medium to simulate both the intake of solid food and digested infant formula, and the use of optimized retention times and pH ranges to represent the infant gut (described in detail below). This study used the Baby-M-SHIME® model to assess the effects of the test product compared to an untreated control using the microbiota of four healthy 6–24-month-old babies. Therefore, two parallel Quad-Baby-M-SHIME® configurations were implemented, with each Quad-Baby-M-SHIME® assessing the effect of the test product versus the untreated control for two donors. Each arm of the Quad-Baby-M-SHIME® consisted of three reactors simulating the stomach/small intestine, proximal colon (PC), and distal colon (DC). The stomach/small intestine reactor operated using a fill-and-draw principle; peristaltic pumps added a defined amount of nutritional medium (140 mL) to the stomach (pH 3) for an incubation time of 1.5 h after which pancreatic and bile liquid (60 mL, 2.5 g/L NaHCO3, 0.9 g/L pancreatin, 4 g/L oxgall) were added for the small intestine simulation (pH 6; 1.5 h incubation). The intestinal suspension was then pumped to the PC (pH 5.8–6.0, volume 300 mL, retention time 12 h) and then the DC (pH 6.0–6.5, volume 500 mL, retention time 20 h); both were under constant stirring. A total retention time of 32 h was used in this study to mimic the short transit of the infant gastrointestinal tract. Each colonic reactor was inoculated with fecal microbiota (5% [v/v]) of 6–24-month-old babies (4 individual donors) and included a simulation of the mucosal environment by the inclusion of mucin-coated microcosms (Van den Abbeele et al. [22]). Medium simulating both the intake of solid food and digested infant formula was used and contained both carbohydrates and digested milk compounds (4 g/L mucin, 1 g/L yeast extract, 0.2 g/L cysteine, and approximately 10 g/L digested milk fragments (4.8 g/L lactose, 0.5 g/L casein, and 4.6 g/L lactalbumin)) as described by Van den Abbeele et al. [24]. Once initiated, the SHIME® cabinet and integrated software were run according to the manufacturer’s instructions (ProDigest, Ghent, Belgium).
The experimental timeline included a two-week stabilization period (d–14 to d0), allowing the fecal microbiota to differentiate into communities representative of each colon region; a subsequent two-week control period (d1 to d14), which provided reference values for the microbial community and composition in each of the colonic compartments; a 5-day antibiotic treatment period (d15 to d19); and a two-week follow-up period (d20 to d33). During the antibiotic treatment period, reactors were treated with 75 mg/day amoxicillin and clavulanic acid (2:1) administered in 3 doses of 25 mg each which corresponded with the reactor feeding schedule. During the follow-up period, the reactors were supplemented with 2 × 109 CFU MegaSporeBiotic™ each day.
2.3. Microbial Community Activity
Samples were collected three times per week from each colonic vessel and assayed for concentrations of SCFAs (acetate, propionate, butyrate) and lactate, a precursor of SCFA. SCFAs were measured as described by Ghyselinck et al. [26]. Briefly, 2-methyl hexanoic acid was added to the samples as an internal standard followed by SCFA extraction using diethyl ether. Extracts were injected (1 µL) into a GC-2014 gas chromatograph (Shimadzu, ‘s-Hertogenbosch, The Netherlands) for analysis (temperature profile: 110 to 160 °C; temperature increase: 6 °C min−1; injector temperature: 200 °C; detector temperature: 200 °C). Nitrogen was used as the carrier gas. The gas chromatograph was equipped with a GC SGE capillary column, 30 m × 0.32 mm ID-BP21x 0.25 μm (Achrom, Machelen, Belgium), a split injector, and a flame ionization detector. Total SCFA production was determined by summing the molar concentrations of acetate, propionate, and butyrate. Lactate was measured using a commercially available enzymatic assay kit (R-Biopharm, Darmstadt, Germany) according to the manufacturer’s instructions.
2.4. Microbial Community Composition
Microbial community composition was quantitatively (lumen) and relatively (mucus) evaluated using shallow shotgun sequencing. Samples were collected 3 times per week from the final week of the control period through the final week of the follow-up period from both the luminal and mucosal environments. For DNA library preparation, a modified protocol of the Illumina Nextera XT library preparation kit (Illumina, San Diego, CA, USA) was used; libraries were quantified using the Qubit (Thermo Fisher, Waltham, MA, USA). DNA libraries were sequenced on an Illumina HiSeq platform (2 × 150 bp). Unassembled sequencing reads were directly analyzed for multi-kingdom microbiome analysis and quantification of relative abundances as previously described [27,28,29,30]. Briefly, the analysis was conducted using curated genome databases and a high-performance data-mining algorithm that rapidly disambiguates hundreds of millions of metagenomic sequence reads to identify the specific microorganism for each identified sequence. For absolute abundance, the total number of bacterial cells was determined using BD FACSVerse Cell Analyzer (BD Biosciences, Franklin Lakes, NJ, USA) (high flow rate setting; threshold, 200 [SYTO channel]). The proportional values obtained using shallow shotgun sequencing were converted to absolute quantities by adjusting to the total bacteria quantification by flow cytometry (relative abundances of each population in a sample × the total cell count) as described by Vandeputte et al. [31].
2.5. Statistical Methods
A two-tailed homoscedastic t-test was used for both the comparison of the individual antibiotic and follow-up weeks to the control period (community activity), and for the comparison of the follow-up period to the control period (community activity and community composition). Statistical comparison of the untreated and treated arm was performed with a two-tailed paired t-test, since samples of the different conditions were taken at the same time points throughout the experiment. For microbial community composition, statistical tests were performed on the log-transformed data to accomplish a normal distribution. A p-value < 0.05 was considered statistically significant.
3. Results
3.1. Microbial Community Activity
SCFA levels for all donors combined are shown in Figure 1. There was a significant decrease from the control period to the antibiotic period for all three SCFAs. In the follow-up period, levels of acetate in the untreated chambers (i.e., no probiotic treatment) spontaneously recovered to levels significantly higher than the antibiotic period and near that observed during the control period, though the levels remained significantly different from the control period with the exception of the untreated PC samples (Figure 1a). This observation was similar for propionate; levels were significantly higher in the follow-up period versus the antibiotic period though the recovery levels remained significantly lower than those observed in the control period (Figure 1b). For butyrate, there was some recovery of the levels in the follow-up period, but to a lesser degree than observed with the other SCFAs (Figure 1c). In the PC, lactate levels were similar in the antibiotic period and significantly decreased in the follow-up period relative to the control period but not the antibiotic period (Figure 1d). In the DC, very little lactate was observed in the control period; the level significantly increased in the antibiotic period but returned to control period levels in the follow-up period. Patterns of SCFA levels were generally similar in the PC and DC, though the changes were more pronounced in the DC compared with the PC. There were no significant differences between untreated and treated for any of the SCFAs at any of the timepoints. In the antibiotic period, the lactate level in the treated chambers was significantly higher than that observed for the untreated chambers; there were no other significant differences between untreated and treated at any timepoint. Given the interindividual variability of the microbiome, we considered it likely that there would be some variation in the response to antibiotic treatment. Thus, SCFA and lactate levels for each individual donor are shown in Figure S1. The findings for individual donors were generally similar to those reported for the pooled data.
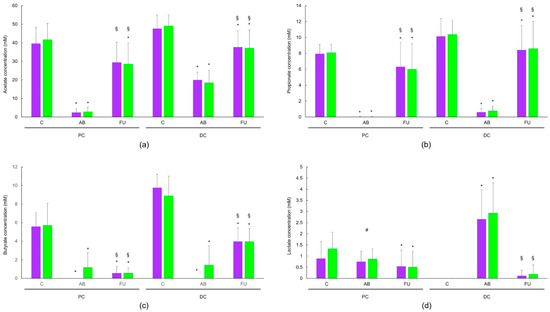
Figure 1.
Changes in acetate (a), propionate (b), butyrate (c), and lactate (d) over the control (n = 12), antibiotic (n = 12; final timepoint), and follow-up (n = 24) periods for pooled donors. Purple bars represent untreated chambers and green bars represent chambers treated with MegaSporeBiotic™. Levels in the AB and FU periods were compared to the control period and levels in the AB period were compared to the FU period using a two-tailed homoscedastic t-test; comparisons between the untreated versus treated groups were made using a two-tailed paired t-test. * indicates a significant (p < 0.05) difference versus the control period, § indicates a significant (p < 0.05) difference for the AB period versus the FU period, # indicates a significant (p < 0.05) difference for untreated versus treated. AB, antibiotic period; C, control period; DC = distal colon; FU, follow-up period; PC = proximal colon.
3.2. Microbial Community Composition
The reciprocal Simpson diversity index is a measure of the diversity and evenness of the microbial community. When all donors were averaged, there was a significant increase in the reciprocal Simpson diversity index between the control and antibiotic periods in the PC lumen for both the untreated and treated chambers; the index returned to similar to, or significantly higher levels than control in the follow-up period (Table 1). In the DC lumen and the PC and DC mucus, the reciprocal Simpson diversity index was significantly decreased during the antibiotic period when compared to the control period for both untreated and treated. In the follow-up period, diversity returned to levels similar to, or significantly higher than the control period; there was no difference between untreated and treated. Thus, probiotics had no effect on the recovery of diversity following antibiotic treatment.

Table 1.
Average reciprocal Simpson diversity index.
Antibiotic treatment resulted in changes at both the phylum level (Figure 2, Table S1 (individual donors, lumen), Table S2 (individual donors, mucus)) and family level (Figure 3, Table S3 (individual donors, lumen), Table S4 (individual donors, mucus)) as expected. Antibiotic treatment resulted in a significant decrease in all bacterial families in both the PC and DC of the lumen versus the control period (Figure 2a–e). In the mucus layer of the PC and DC, changes in the proportional abundance varied among phyla (Figure 2f). The abundance of Bifidobacteriaceae changed notably with antibiotic treatment, accounting for the bulk of the changes in Actinobacteria levels (Figure 3b). In the follow-up period, the levels of Actinobacteria recovered and changes at the family level were similar for treated and untreated (Figure 3a). Overall, the results indicate a similar recovery in the untreated and treated arms.
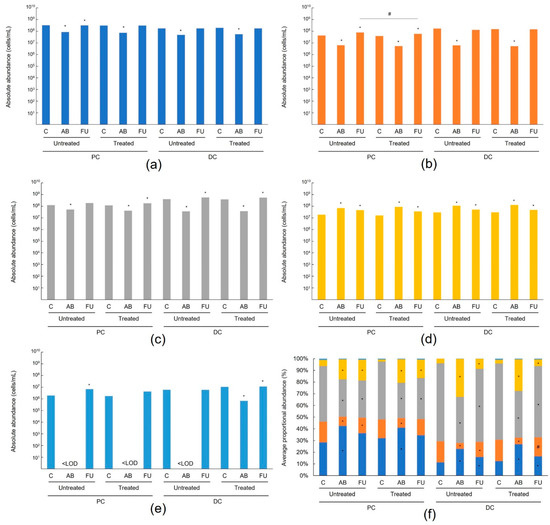
Figure 2.
Average absolute abundance (cells/mL) of different phyla in the lumen. Actinobacteria (a), Bacteroidetes (b), Firmicutes (c), Proteobacteria (d), Verrucomicrobia (e), and average proportional abundance (%) of different phyla in the mucus (f) of the untreated and treated PC and DC compartment. The average values (n = 12) of different phyla for data pooled from four different 6–24-month-old baby fecal donors (n = 3 for each donor). Samples were collected at the end of the control period, at the end of the antibiotic period, and at the end of the follow-up period. Dark blue bars represent Actinobacteria, orange bars represent Bacteroidetes, gray bars represent Firmicutes, yellow bars represent Proteobacteria, and light blue bars represent Verrucomicrobia. Levels in the antibiotic and follow-up periods were compared to the control period using a two-tailed homoscedastic t-test; comparisons between the untreated and treated groups were made using a two-tailed paired t-test. * indicates a significant (p < 0.05) difference versus the control period, # indicates a significant (p < 0.05) difference for untreated versus treated. AB = antibiotic; C = control period; DC = distal colon; FU = follow-up period; PC = proximal colon.
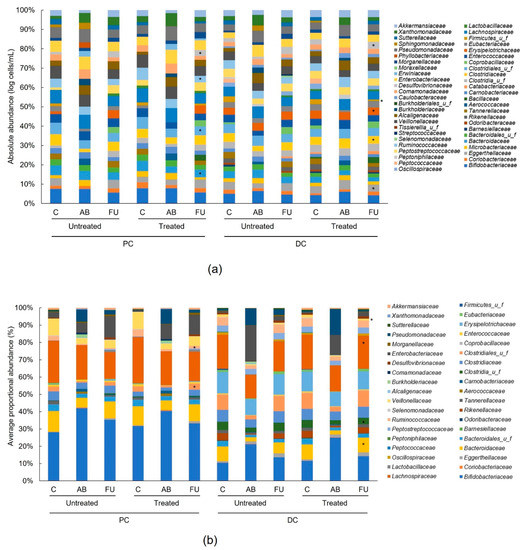
Figure 3.
Average proportional abundance (%) of different families in the lumen (a) and in the mucus (b) of the untreated and treated PC and DC compartment. The average values (n = 12) of different families for data pooled from four different 6–24-month-old baby fecal donors (n = 3 for each donor). Samples were collected at the end of the control period, at the end of the antibiotic period, and at the end of the follow-up period. Levels in the antibiotic and follow-up periods were compared to the control period using a two-tailed homoscedastic t-test; comparisons between the untreated and treated groups were made using a two-tailed paired t-test. * indicates a significant (p < 0.05) difference versus the control period. AB = antibiotic; C = control period; DC = distal colon; FU = follow-up period; PC = proximal colon; u f = unidentified family.
Bacteroidetes levels were significantly reduced in all colon regions in the antibiotic versus control periods (Figure 2a,b), which was mainly due to decreases in the following families: Bacteroidaceae and Tannerellaceae in all colon regions and Rikenellaceae in the DC (Figure 3a,b). These levels recovered during the follow-up period and recovery was similar for the untreated and treated chambers. In the mucosal DC, the Bacteroidaceae levels recovered to a higher extent in the treated arm (p < 0.05 versus the control period) than the untreated arm (Figure 3b).
Firmicutes levels were also significantly reduced between the control period and the antibiotic period in all colonic regions (Figure 2a,b), indicating a dysbiosed microbial community. This was mainly linked to a reduction in the butyrate-producing Lachnospiraceae, Clostridiaceae, and Ruminococcaceae families, and the propionate-producing Veillonellaceae family, and to a lesser extent, the lactate-producing Enterococcaceae family (Figure 3a,b). Generally, Firmicutes levels recovered during the follow-up period and differences between the untreated and treated arms were limited (Figure 2). Bacillaceae levels were significantly increased in the luminal PC and DC with treatment, which can be linked with the presence of bacterial species of this family in the MegaSporeBiotic™ probiotic (Figure 3a). Ruminococcaceae levels recovered to a significantly higher extent with treatment than without in both the luminal and mucosal PC; levels recovered to those observed in the control period with treatment but remained significantly decreased versus the control period without treatment (p < 0.05) (Figure 3a,b). In the luminal DC the levels of Clostridiaceae were significantly enhanced with treatment versus untreated and reached higher levels with treatment than were observed in the control period (Figure 3a). In the mucosal DC, a similar observation was made regarding the recovery of the butyrate-producing Lachnospiraceae and the propionate-producing Veillonellaceae families, and the opposite for unclassified Clostridia (Figure 3b).
The Proteobacteria were increased with antibiotic treatment; increases were significant in most colon regions (Figure 2a,b) and were mainly linked with increases in Enterobacteriaceae, Pseudomonadaceae, and Xanthomonadaceae (Figure 3a,b). Proteobacteria levels gradually decreased in the follow-up period, though they remained significantly elevated compared with the control period in all colonic regions (Figure 2a,b). There were limited differences between untreated and treated. Enterobacteriaceae levels tended to decrease to a greater extent in the treatment arm versus untreated in the lumen (Figure 3a).
For Akkermansia muciniphila, the only representative of the Verrucomicrobia phylum, levels were significantly reduced after antibiotic treatment, most notably in the lumen (Figure 3a). While there was recovery of Verrucomicrobia abundance in the follow-up period, there were no differences between untreated and treated.
4. Discussion
The effect of daily supplementation of MegaSporeBiotic™ spore-based probiotic formulation on the recovery of the microbial community activity and microbial community composition after antibiotic-induced dysbiosis was assessed using a Baby-Quad-M-SHIME® model evaluating the microbiota from four individual baby donors. Recovery with probiotic treatment was compared to that of untreated (i.e., spontaneous recovery).
SCFA levels decreased significantly from the control period upon antibiotic treatment and recovered similarly in both the untreated and treated groups. In the follow-up period, levels of acetate and propionate had recovered to levels similar to the control period for most donors while levels of butyrate were increased over the antibiotic period but remained significantly lower than in the control period, indicating that a longer recovery time was necessary. The recovery of lactate levels in the PC varied somewhat between donors, making the results difficult to interpret, but there were no major differences in recovery for untreated versus treated. In the DC, lactate levels were very low in the control period and increased significantly in the antibiotic period. Given that lactate is a substrate for SCFA production [32,33,34,35] and the fact that SCFA production was significantly decreased during the antibiotic period, this may indicate a loss of lactate-utilizing bacteria after antibiotic treatment, as lactate accumulation is associated with dysbiosis [36,37,38]. Lactate levels were reduced in the follow-up period, potentially reflecting the observed microbiota recovery. Overall, our findings showed a similar recovery of microbial metabolic activity with or without treatment with MegaSporeBiotic™, suggesting that probiotic intervention did not aid in recovery from antibiotic-induced dysbiosis in the Baby-SHIME® model. Treatment appeared safe in that there were neither increases in microbial pathogens nor signals of dysbiosis with treatment. These findings are important given that there is a paucity of data for the use of probiotics in infants following antibiotic treatment.
Regarding the microbial community composition, antibiotic treatment reduced the abundance of Actinobacteria, Bacteroidetes, Firmicutes, and Verrucomicrobia and increased the levels of Proteobacteria, indicating a dysbiosed microbial community. When antibiotics were stopped, a consistent spontaneous recovery of the microbial community composition was observed for all four donors. There were minimal differences between untreated and treated, indicating an overall similar microbial recovery for both test conditions. Probiotic supplementation to infants throughout the first 6 months of life revealed minimal changes in the infant gut microbiome, with the only difference from untreated being an increase in the abundance of Bifidobacterium in the first week of treatment [39]. Therefore, it may not be surprising that probiotic supplementation had little effect on the infant gut microbiome. However, it must be noted that the participants in that study were administered a variety of probiotics (parent’s choice), so the findings are very generalized and do not relate to specific probiotic strains. There is also some evidence that breastfeeding versus formula feeding affects the response of the infant microbiome to probiotic treatment. A strong impact on the gut microbiota composition was observed for infants who received antibiotics and probiotics and were breastfed while the same was not true for those who were formula fed [40]. The feed used in our study was based on formula feed rather than breastmilk, which may have affected our results.
It is of interest to evaluate whether probiotic strains engraft with treatment. We observed a significant increase in the Bacillaceae family during the follow-up period in both the PC and DC of the treated chambers, indicating that there was at least partial engraftment of the probiotic strains. Although this observation of potential engraftment is encouraging, the authors note that we were not able to evaluate long-term engraftment in this study. Studies are mixed as to whether orally administered probiotics can stably or transiently engraft [41,42]; while stable engraftment appears possible for some probiotics, this may be dependent on the gut microbiome of the host as well as other host-related factors [41,43,44]. This was demonstrated by a study of Bifidobacterium longum AH1206 which reported long-term persistence (> 6 months) in only 30% of study participants [41], as well as mouse studies that suggest the presence of an exclusive metabolic niche enables engraftment in the gut microbiota [43]. It has been suggested that prebiotics improve engraftment [45]; therefore, it would be interesting to test for engraftment of the MSB strains with and without prebiotics and to determine whether any specific prebiotics facilitate improved engraftment of these strains.
MegaSporeBiotic™ supplementation has been shown to result in changes in community activity and composition, such as increased propionate levels, decreased lactate levels, and increased Akkermansia muciniphila, Bifidobacteria spp., and Firmicutes in an M-SHIME® model using the fecal microbiota of adult donors under healthy conditions [21]. Because of this, we hypothesized that MegaSporeBiotic™ treatment may enhance recovery following antibiotic-induced dysbiosis in an M-SHIME® model using fecal microbiota of baby donors. While the chambers treated with MegaSporeBiotic™ did recover both microbial community activity and composition following antibiotic-induced dysbiosis, the recovery was not enhanced relative to untreated (i.e., spontaneous recovery). A study in premature infants found that supplementing probiotics containing B. longum, Lactobacillus acidophilus, and Enterococcus faecalis within two days of receiving cefotaxime antibiotics resulted in increased beneficial bacteria relative to the infants who received antibiotics only [46], indicating that there may be a role for probiotics in antibiotic-induced dysbiosis recovery. Another study evaluated probiotic supplementation (Bifidobacterium breve, Propionibacterium freundenreichii, Lactobacillus rhamnosus Lc705, and L. rhamnosus GG) to mothers during pregnancy and their infants after birth for 3 months [40]. Using fecal samples collected from the infants at 3 months, the study reported that there was a strong effect of antibiotic use and birth mode (i.e., vaginal birth versus caesarean section) in the group that did not receive probiotics, which was eliminated in the group that did, indicating that consistent probiotic use could mitigate the damaging effects of antibiotic use on the fecal microbiota. Together, these studies may indicate the importance of earlier probiotic treatment (i.e., within two days of antibiotic treatment or concurrently with antibiotic treatment) for an improved effect of probiotics in babies treated with antibiotics.
Our study had several limitations. This was an in vitro study. As such, the results cannot directly translate to biological responses, meaning that studies in vivo are needed to confirm these findings. Probiotic treatment was initiated following five days of antibiotic treatment; thus, we were not able to evaluate the potential benefits of earlier or simultaneous probiotic supplementation. Additionally, the study was not designed to evaluate whether the probiotic strains established themselves within the gut microbiota; therefore, it is unclear whether the lack of effect was due to a lack of transient persistence or a true lack of effect. Finally, in the real world, antibiotics are given to treat an infection; however, in our study, the donors were healthy and did not have a known infection.
In general, recovery from antibiotic-induced dysbiosis was similar for untreated and treated chambers, indicating that intervention with MegaSporeBiotic™ did not result in a faster or different recovery in microbial community activity or composition relative to spontaneous recovery. We speculate that the timing of probiotic intervention may be important and that earlier intervention (i.e., co-administration of antibiotics and probiotics) may provide benefit. Additional studies evaluating different timings of MegaSporeBiotic™ treatment may be of interest.
Supplementary Materials
The following supporting information are available online at https://www.mdpi.com/article/10.3390/app122312302/s1, Figure S1: Changes in acetate (a), propionate (b), butyrate (c), and lactate (d), Table S1: Absolute abundance (log cells/mL) for different phyla in the lumen for each individual donor, Table S2: Proportional abundance (%) for different phyla in the mucus for each individual donor, Table S3: Absolute abundance (log cells/mL) for different bacterial families in the lumen for each individual donor, Table S4: Proportional abundance (%) for different bacterial families in the mucus for each individual donor.
Author Contributions
Conceptualization, M.M., T.B., K.K., M.G.; methodology, M.M., T.B., K.K., M.G.; formal analysis, M.M.; data curation, M.M.; writing—original draft preparation, S.B.; writing—review and editing, M.M., T.B., K.K., M.G.; visualization, S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study and the article processing charges were funded by Microbiome Labs.
Institutional Review Board Statement
Ethical approval of the University Hospital Ghent (reference number B670201836585) was obtained for the collection of the fecal samples.
Informed Consent Statement
Informed consent of donors or their legal representatives was obtained.
Data Availability Statement
The data presented in this study are available on reasonable request from the corresponding author.
Conflicts of Interest
T.B., K.K., and M.G. are employees of Microbiome Labs. S.B. received payment from Microbiome Labs. The funders were involved in the design of the study, in the writing of the manuscript, and in the decision to publish the results.
References
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.M.; Sitarik, A.R.; Havstad, S.L.; Fujimura, K.E.; Wegienka, G.; Cassidy-Bushrow, A.E.; Kim, H.; Zoratti, E.M.; Lukacs, N.W.; Boushey, H.A.; et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci. Rep. 2016, 6, 31775. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hamalainen, A.M.; et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Keeney, K.M.; Yurist-Doutsch, S.; Arrieta, M.C.; Finlay, B.B. Effects of antibiotics on human microbiota and subsequent disease. Annu. Rev. Microbiol. 2014, 68, 217–235. [Google Scholar] [CrossRef] [PubMed]
- Koropatkin, N.M.; Cameron, E.A.; Martens, E.C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012, 10, 323–335. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Y.; Wang, P.; Huang, Y.; Wang, F. Short-chain fatty acids manifest stimulative and protective effects on intestinal barrier function through the inhibition of NLRP3 inflammasome and autophagy. Cell Physiol. Biochem. 2018, 49, 190–205. [Google Scholar] [CrossRef]
- Topping, D.L.; Clifton, P.M. Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001, 81, 1031–1064. [Google Scholar] [CrossRef]
- Yassour, M.; Vatanen, T.; Siljander, H.; Hamalainen, A.M.; Harkonen, T.; Ryhanen, S.J.; Franzosa, E.A.; Vlamakis, H.; Huttenhower, C.; Gevers, D.; et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016, 8, 343ra81. [Google Scholar] [CrossRef]
- Wei, S.; Mortensen, M.S.; Stokholm, J.; Brejnrod, A.D.; Thorsen, J.; Rasmussen, M.A.; Trivedi, U.; Bisgaard, H.; Sorensen, S.J. Short- and long-term impacts of azithromycin treatment on the gut microbiota in children: A double-blind, randomized, placebo-controlled trial. EBioMedicine 2018, 38, 265–272. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization and World Health Organization Expert Consultation. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. 2006. Available online: https://www.fao.org/3/a0512e/a0512e.pdf (accessed on 28 September 2022).
- Pyne, D.B.; West, N.P.; Cox, A.J.; Cripps, A.W. Probiotics supplementation for athletes—Clinical and physiological effects. Eur. J. Sport Sci. 2015, 15, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Cutting, S.M. Bacillus probiotics. Food Microbiol. 2011, 28, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Marzorati, M.; Abbeele, P.V.D.; Bubeck, S.S.; Bayne, T.; Krishnan, K.; Young, A.; Mehta, D.; DeSouza, A. Bacillus subtilis HU58 and Bacillus coagulans SC208 probiotics reduced the effects of antibiotic-induced gut microbiome dysbiosis in an M-SHIME((R)) model. Microorganisms 2020, 8, 1028. [Google Scholar] [CrossRef]
- Sasaki, K.; Sasaki, D.; Inoue, J.; Hoshi, N.; Maeda, T.; Yamada, R.; Kondo, A. Bacillus coagulans SANK 70258 suppresses Enterobacteriaceae in the microbiota of ulcerative colitis in vitro and enhances butyrogenesis in healthy microbiota. Appl. Microbiol. Biotechnol. 2020, 104, 3859–3867. [Google Scholar] [CrossRef]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME((R))). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models. Cham (CH); Verhoeckx, K., Cotter, P., Lopez-Exposito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wicher, H., Eds.; Springer: New York, NY, USA, 2015; pp. 305–317. [Google Scholar]
- Van den Abbeele, P.; Sprenger, N.; Ghyselinck, J.; Marsaux, B.; Marzorati, M.; Rochat, F. A comparison of the in vitro effects of 2′fucosyllactose and lactose on the composition and activity of gut microbiota from infants and toddlers. Nutrients 2021, 13, 726. [Google Scholar] [CrossRef]
- Bondue, P.; Lebrun, S.; Taminiau, B.; Everaert, N.; LaPointe, G.; Crevecoeur, S.; Daube, G.; Delcenserie, V. A toddler SHIME(R) model to study microbiota of young children. FEMS Microbiol. Lett. 2020, 367, 726. [Google Scholar] [CrossRef]
- Marzorati, M.; Van den Abbeele, P.; Bubeck, S.; Bayne, T.; Krishnan, K.; Young, A. Treatment with a spore-based probiotic containing five strains of Bacillus induced changes in the metabolic activity and community composition of the gut microbiota in a SHIME(R) model of the human gastrointestinal system. Food Res. Int. 2021, 149, 110676. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.M.; Van de Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Roos, S.; Eeckhaut, V.; MacKenzie, D.A.; Derde, M.; Verstraete, W.; Marzorati, M.; Possemiers, S.; Vanhoecke, B.; Van Immerseel, F.; et al. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 2012, 5, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Duysburgh, C.; Vazquez, E.; Chow, J.; Buck, R.; Marzorati, M. 2′-Fucosyllactose alters the composition and activity of gut microbiota from formula-fed infants receiving complementary feeding in a validated intestinal model. J. Funct. Foods 2019, 61, 103484. [Google Scholar] [CrossRef]
- De Boever, P.; Wouters, R.; Vermeirssen, V.; Boon, N.; Verstraete, W. Development of a six-stage culture system for simulating the gastrointestinal microbiota of weaned infants. Microb. Ecol. Health Dis. 2001, 13, 111–123. [Google Scholar] [CrossRef]
- Ghyselinck, J.; Verstrepen, L.; Moens, F.; Van Den Abbeele, P.; Bruggeman, A.; Said, J.; Smith, B.; Barker, L.A.; Jordan, C.; Leta, V.; et al. Influence of probiotic bacteria on gut microbiota composition and gut wall function in an in-vitro model in patients with Parkinson’s disease. Int. J. Pharm. X 2021, 3, 100087. [Google Scholar] [CrossRef] [PubMed]
- Ottesen, A.; Ramachandran, P.; Reed, E.; White, J.R.; Hasan, N.; Subramanian, P.; Ryan, G.; Jarvis, K.; Grim, C.; Daquiqan, N.; et al. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 2016, 16, 275. [Google Scholar] [CrossRef]
- Ponnusamy, D.; Kozlova, E.V.; Sha, J.; Erova, T.E.; Azar, S.R.; Fitts, E.C.; Kirtley, M.L.; Tiner, B.L.; Andersson, J.A.; Grim, C.J.; et al. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci. USA 2016, 113, 722–727. [Google Scholar] [CrossRef]
- Hasan, N.A.; Young, B.A.; Minard-Smith, A.T.; Saeed, K.; Li, H.; Heizer, E.M.; McMillan, N.J.; Isom, R.; Abdullah, A.S.; Bornman, D.M.; et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS. ONE 2014, 9, e97699. [Google Scholar] [CrossRef]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef]
- Vandeputte, D.; Kathagen, G.; D’Hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef]
- Bourriaud, C.; Robins, R.J.; Martin, L.; Kozlowski, F.; Tenailleau, E.; Cherbut, C.; Michel, C. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J. Appl. Microbiol. 2005, 99, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Belenguer, A.; Duncan, S.H.; Calder, A.G.; Holtrop, G.; Louis, P.; Lobley, G.E.; Flint, H.J. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 2006, 72, 3593–3599. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Mackay, W.G.; Edwards, C.A.; Preston, T.; Dodson, B.; Weaver, L.T. Butyrate production from oligofructose fermentation by the human faecal flora: What is the contribution of extracellular acetate and lactate? Br. J. Nutr. 2006, 96, 570–577. [Google Scholar] [PubMed]
- Hove, H.; Nordgaard-Andersen, I.; Mortensen, P.B. Faecal DL-lactate concentration in 100 gastrointestinal patients. Scand. J. Gastroenterol. 1994, 29, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Belenguer, A.; Duncan, S.H.; Holtrop, G.; Anderson, S.E.; Lobley, G.E.; Flint, H.J. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl. Environ. Microbiol. 2007, 73, 6526–6533. [Google Scholar] [CrossRef]
- Louis, P.; Duncan, S.H.; Sheridan, P.O.; Walker, A.W.; Flint, H.J. Microbial lactate utilisation and the stability of the gut microbiome. Gut. Microbiome. 2022, 3, e3. [Google Scholar] [CrossRef]
- Quin, C.; Estaki, M.; Vollman, D.M.; Barnett, J.A.; Gill, S.K.; Gibson, D.L. Probiotic supplementation and associated infant gut microbiome and health: A cautionary retrospective clinical comparison. Sci. Rep. 2018, 8, 8283. [Google Scholar] [CrossRef]
- Korpela, K.; Salonen, A.; Vepsäläinen, O.; Suomalainen, M.; Kolmeder, C.; Varjosalo, M.; Miettinen, S.; Kukkonen, K.; Savilahti, E.; Kuitunen, M.; et al. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome 2018, 6, 182. [Google Scholar] [CrossRef]
- Maldonado-Gómez, M.X.; Martínez, I.; Bottacini, F.; O’Callaghan, A.; Ventura, M.; van Sinderen, D.; Hillmann, B.; Vangay, P.; Knights, D.; Hutkins, R.W.; et al. Stable Engraftment of Bifidobacterium. longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe 2016, 20, 515–526. [Google Scholar] [CrossRef]
- Jacobsen, C.N.; Rosenfeldt Nielsen, V.; Hayford, A.E.; Møller, P.L.; Michaelsen, K.F.; Paerregaard, A.; Sandström, B.; Tvede, M.; Jakobsen, M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 1999, 65, 4949–4956. [Google Scholar] [CrossRef]
- Frese, S.A.; Hutkins, R.W.; Walter, J. Comparison of the colonization ability of autochthonous and allochthonous strains of Lactobacilli in the human gastrointestinal tract. Adv. Microbiol. 2012, 2, 399–409. [Google Scholar] [CrossRef]
- Shepherd, E.S.; DeLoache, W.C.; Pruss, K.M.; Whitaker, W.R.; Sonnenburg, J.L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 2018, 557, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.P.; Rao, K.; Young, V.B. Probiotics for prevention of Clostridium difficile infection. Curr. Opin. Gastroenterol. 2018, 34, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Yang, L.; Liu, K.; Shen, S.; Zhang, Q.; Li, H.; Cheng, Y. Effects of antibiotic treatment and probiotics on the gut microbiome of 40 infants delivered before term by cesarean section analysed by using 16S rRNA quantitative polymerase chain reaction sequencing. Med. Sci. Monit. 2021, 27, e928467. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).