Multi-Angle Fast Neural Tangent Kernel Classifier
Abstract
1. Introduction
1.1. Our Contributions
- The traditional SVM is a single-kernel learning model, the SVM model with multi-kernel learning is introduced, using NTK, which is equivalent to an infinitely wide neural network, as the base kernel function for multi-kernel learning, and multi-kernel-NTK, RBF-Boolean, and POLY-Boolean models are selected for the classification task, where RBF- Boolean utilizes the multiscale multi-kernel idea, multi-kernel-NTK, POLY-Boolean utilizes the linear synthetic kernel idea.
- The HMPSO algorithm uses EDA to estimate and preserve the information of the historical optimal distribution of particles, which can avoid premature convergence of the model by competing among the information and maintaining fast convergence. Its performance is better than most of the PSO improvement algorithms. We use the HMPSO algorithm to select various parameters in multi-kernel-NTK and other multi-kernel models.
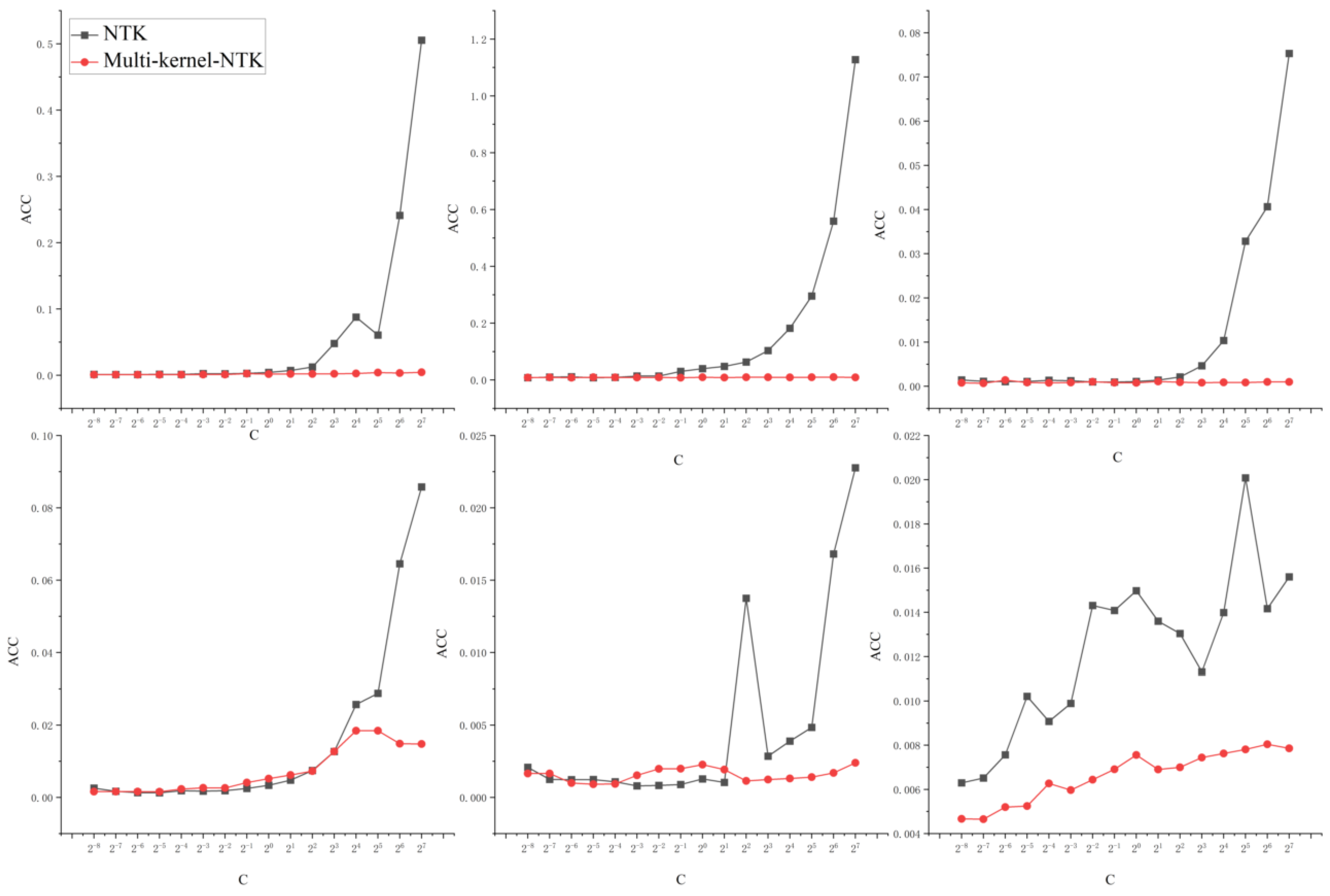
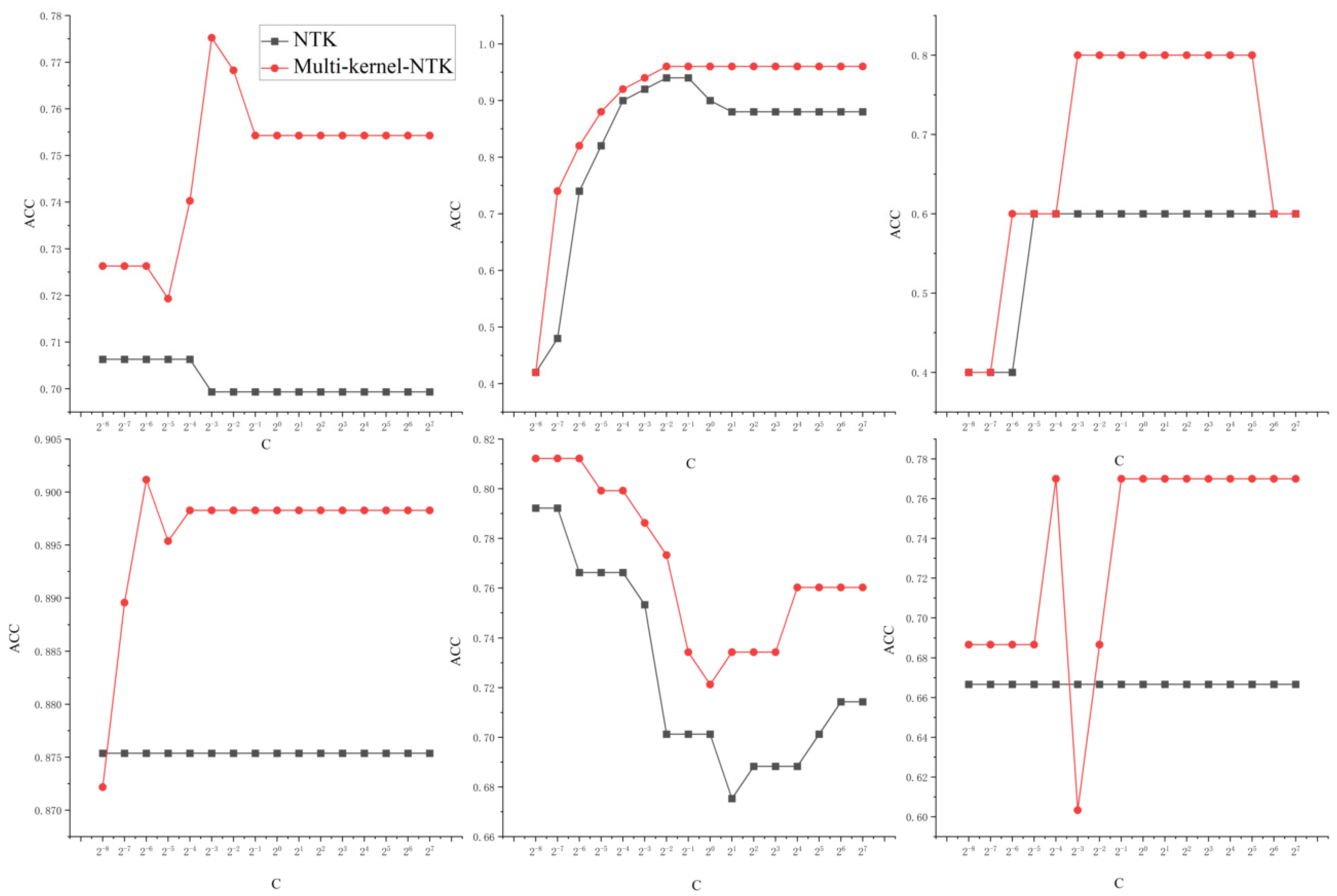
- The single kernel SVM model has a limited search space, and the diagnostic results of the model are more sensitive and less robust to the kernel parameters and penalty parameters. The sensitivity of multi-kernel-NTK and NTK-SVM models to the penalty parameter C and network depth L is studied visually and comparatively.
- The NTK kernel has a multi-layer structure, which consumes a lot of time in its computation. Because of its special function construction (four-dimensional GRAM matrix), matrix decomposition and chunking acceleration techniques are difficult to be applied to the NTK kernel function. We extract the features belonging to the Boolean type from the original dataset and use the Boolean kernel (Monotone Disjunctive Kernel) to encode them into a binary square array for training, which improves the model accuracy and compresses the model training time at the same time.
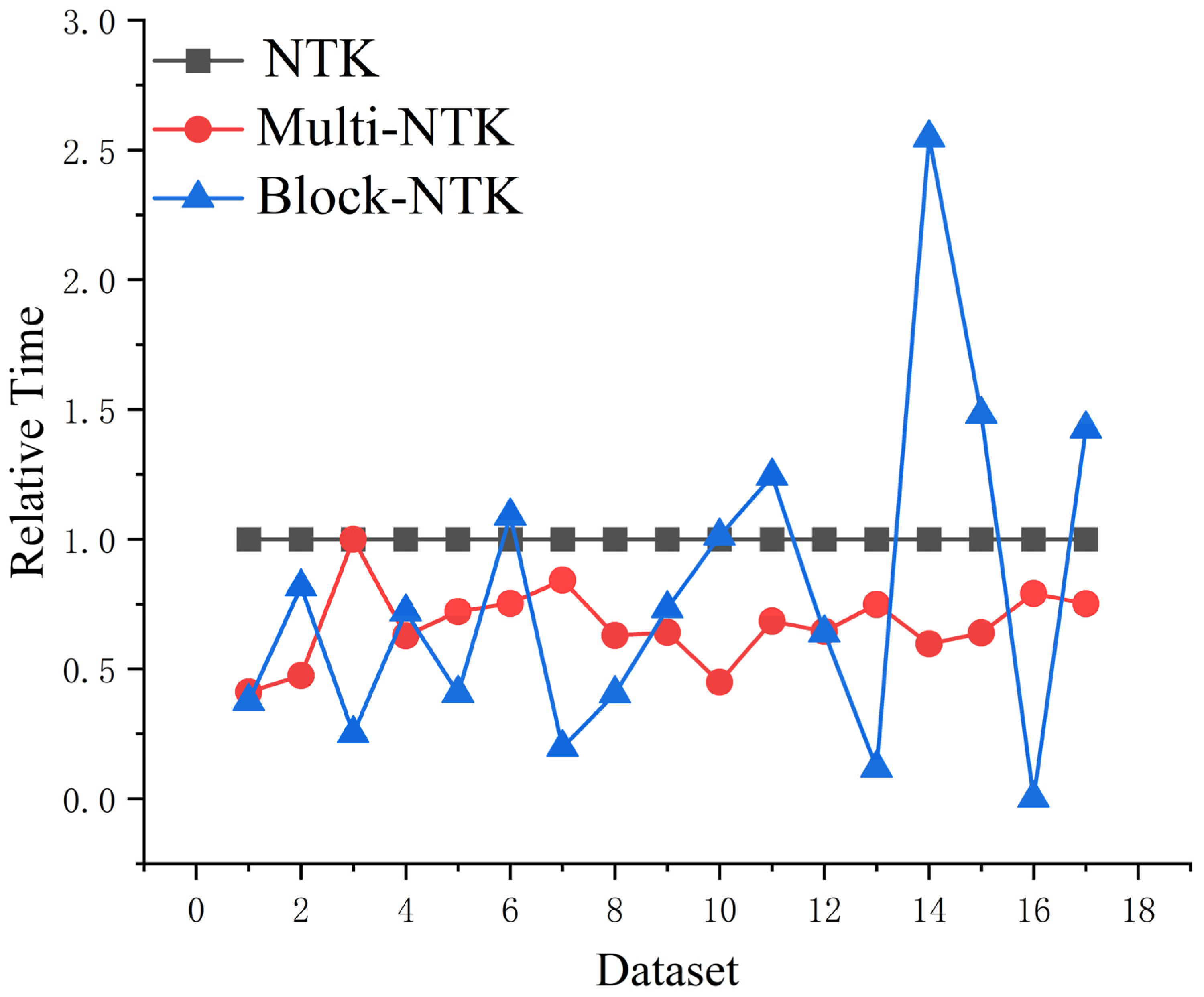
- The Multi-NTK model is accelerated regarding the number of features and the multi-kernel model. We also used the data-cutting technique to construct the Block-NTK model to accelerate the model regarding the number of samples.
1.2. Article Structure
2. Related Works
3. Background Techniques
3.1. Particle Swarm Algorithms
3.2. Multi-Kernel Learning
3.3. Kernel Functions
3.3.1. Boolean Kernel
3.3.2. Neural Tangent Kernel
4. Experimental Setup
4.1. Data Setup and Parameter Selection
4.2. Multi-Kernel-NTK and Other Multi-Kernel Models
4.3. Multi-Kernel-NTK Model Robustness Test
4.4. Multi-Kernel-NTK Model Robustness Test
5. Analysis of Experimental Results
5.1. Comparison of Multi-Model Results
5.2. Multi-Kernel-NTK Model Acceleration
5.3. Multi-Kernel-NTK Robustness Testing
5.3.1. Network Parameters and Multi-Kernel-NTK
5.3.2. Network Parameters and Multi-Kernel-NTK
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Q.; Liu, Y.; Chen, T. Federated machine learning: Concept and applications. ACM Trans. Intell. Syst. Technol. 2019, 10, 1–19. [Google Scholar] [CrossRef]
- McMahan, B.; Moore, E.; Ramage, D.; Hampson, S.; Arcas, B.A. Communication-efficient learning of deep networks from decentralized data. Artif. Intell. Stat. 2017, 1, 1273–1282. [Google Scholar]
- Wei, K.; Li, J.; Ma, C.; Ding, M.; Wei, S.; Wu, F.; Ranbaduge, T. Vertical Federated Learning: Challenges, Methodologies and Experiments. arXiv 2022, arXiv:2202.04309. [Google Scholar]
- Gupta, O.; Ramesh, R. Distributed learning of deep neural network over multiple agents. J. Netw. Comput. Appl. 2018, 116, 1–8. [Google Scholar] [CrossRef]
- Thapa, C.; Arachchige, P.C.M.; Camtepe, S.; Sun, L. Splitfed: When federated learning meets split learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Pomona, CA, USA, 24–28 October 2022. [Google Scholar]
- Iñaki, E.A.; Abdellatif, Z. Distributed information bottleneck method for discrete and Gaussian sources. In Proceedings of the International Zurich Seminar on Information and Communication, Zurich, Switzerland, 21–23 February 2018; ETH Zurich: Zurich, Switzerland, 2018. [Google Scholar]
- Moldoveanu, M.; Abdellatif, Z. On in-network learning. A comparative study with federated and split learning. In Proceedings of the IEEE 22nd International Workshop on Signal Processing Advances in Wireless Communications, Lucca, Italy, 27–30 September 2021; IEEE: Piscataway, NJ, USA, 2021. [Google Scholar]
- Lanckriet, G.; Crisetianini, N.; Bartlett, P. Learning the kernel matrix with semidefinite programming. J. Mach. Learn. Res. 2002, 5, 27–72. [Google Scholar]
- Sefidgaran, M.; Chor, R.; Zaidi, A. Rate-distortion bounds on the generalization error of distributed learning. In Proceedings of the Thirty-Sixth Annual Conference on Neural Information Processing Systems, New Orleans, LA, USA, 20 October 2022. [Google Scholar]
- Siewert, C.E.; Williams, M.M.R. The effect of anisotropic scattering on the critical slab problem in neutron transport theory using a synthetic kernel. J. Phys. D Appl. Phys. 1977, 10, 2031. [Google Scholar] [CrossRef]
- Lewis, D.P.; Tony, J.; William, S.N. Support vector machine learning from heterogeneous data: An empirical analysis using protein sequence and structure. Bioinformatics 2006, 22, 2753–2760. [Google Scholar] [CrossRef]
- Liu, R.; Wang, F.; Yang, B.; Qin, S.J. Multiscale kernel based residual convolutional neural network for motor fault diagnosis under nonstationary conditions. IEEE Trans. Ind. Inform. 2020, 16, 3797–3806. [Google Scholar] [CrossRef]
- Opfer, R. Multiscale kernels. Comput. Math. 2006, 25, 357–380. [Google Scholar] [CrossRef]
- Mansouri, M.; Mohamed, N.N.; Hazem, N.N. Multiscale kernel pls-based exponentially weighted-glrt and its application to fault detection. IEEE Trans. Emerg. Top. Comput. Intell. 2017, 3, 49–58. [Google Scholar] [CrossRef]
- Özöğür, A.S.; Weber, G.W. Infinite kernel learning via infinite and semi-infinite programming. Optim. Methods Softw. 2010, 25, 937–970. [Google Scholar] [CrossRef]
- Özöğür, A.S.; Weber, G.W. On numerical optimization theory of infinite kernel learning. J. Glob. Optim. 2010, 48, 215–239. [Google Scholar] [CrossRef]
- Liu, Y.; Liao, S.; Lin, H.; Yue, Y.; Wang, W. Infinite kernel learning: Generalization bounds and algorithms. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Huang, C.R.; Chen, Y.T.; Chen, W.Y. Gastroesophageal reflux disease diagnosis using hierarchical heterogeneous descriptor fusion support vector machine. IEEE Trans. Biomed. Eng. 2016, 63, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.L.; Hu, Q.H.; Chen, Y.L. Improved support vector machine algorithm for heterogeneous data. Pattern Recognit. 2015, 48, 2072–2083. [Google Scholar] [CrossRef]
- Shi, Y.; Eberhart, R.C. A modified particle swarms optimizer. In Proceedings of the IEEE Congress on Evolutionary Computation, Anchorage, AK, USA, 4–9 May 1998; IEEE: Piscataway, NJ, USA, 1998; pp. 69–73. [Google Scholar]
- Dey, A.; Sharma, K.D.; Sanyal, T.; Bhattacharjee, P.; Bhattacharjee, P., Sr. Identification of Biomarkers for Arsenicosis Employing Multiple Kernel Learning Embedded Multi-objective Swarm Intelligence. IEEE Trans. Nanobiosci. 2022; online ahead of print. [Google Scholar]
- Chien, C.; Seiler, M.; Eitel, F.; Schmitz-Hübsch, T.; Paul, F.; Ritter, K. Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity. Mult. Scler. J. Exp. Transl. Clin. 2022, 8, 20552173221109770. [Google Scholar] [CrossRef]
- Jiang, H.; Shen, D.; Ching, W.K.; Qiu, Y. A high-order norm-product regularized multiple kernel learning framework for kernel optimization. Inf. Sci. 2022, 606, 72–91. [Google Scholar] [CrossRef]
- Price, S.R.; Anderson, D.T.; Havens, T.C.; Price, S.R. Kernel Matrix-Based Heuristic Multiple Kernel Learning. Mathematics 2022, 10, 2026. [Google Scholar] [CrossRef]
- Alavi, F.; Hashemi, S. A bi-level formulation for multiple kernel learning via self-paced training. Pattern Recognit. 2022, 129, 108770. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Z.; Li, Y.; Zhou, Y.; Li, D.; Du, W. Gravitation balanced multiple kernel learning for imbalanced classification. Neural Comput. Appl. 2022, 34, 13807–13823. [Google Scholar] [CrossRef]
- Archibald, R.; Bao, F. Kernel learning backward SDE filter for data assimilation. J. Comput. Phys. 2022, 455, 111009. [Google Scholar] [CrossRef]
- Tian, Y.; Fu, S.; Tang, J. Incomplete-view oriented kernel learning method with generalization error bound. Inf. Sci. 2021, 581, 951–977. [Google Scholar] [CrossRef]
- Saeedi, S.; Panahi, A.; Arodz, T. Quantum semi-supervised kernel learning. Quantum Mach. Intell. 2021, 3, 24. [Google Scholar] [CrossRef]
- Guo, W.; Wang, Z.; Ma, M.; Chen, L.; Yang, H.; Li, D.; Du, W. Semi-supervised multiple empirical kernel learning with pseudo empirical loss and similarity regularization. Int. J. Intell. Syst. 2021, 37, 1674–1696. [Google Scholar] [CrossRef]
- Hengyue, S.; Dong, W.; Peng, W.; Yi, C.; Yuehui, C. Deep Multiple Kernel Learning for Prediction of MicroRNA Precursors. Sci. Program. 2021, 2021, 1–9. [Google Scholar]
- Shifei, D.; Yuting, S.; Yuexuan, A.; Weikuan, J. Multiple birth support vector machine based on dynamic quantum particle swarm optimization algorithm. Neurocomputing 2022, 480, 146–156. [Google Scholar]
- Han, B.; Ji, K.; Singh, B.P.M.; Qiu, J.; Zhang, P. An Optimization Method for Mix Proportion of Wet-Mix Shotcrete: Combining Artificial Neural Network with Particle Swarm Optimization. Appl. Sci. 2022, 12, 1698. [Google Scholar] [CrossRef]
- Guo, Q.; Chen, Z.; Yan, Y.; Xiong, W.; Jiang, D.; Shi, Y. Model Identification and Human-robot Coupling Control of Lower Limb Exoskeleton with Biogeography-based Learning Particle Swarm Optimization. Int. J. Control. Autom. Syst. 2022, 20, 589–600. [Google Scholar] [CrossRef]
- Polato, M.; Lauriola, I.; Aiolli, F. A Novel Boolean Kernels Family for Categorical Data. Entropy 2018, 20, 444. [Google Scholar] [CrossRef]
- Mirko, P.; Fabio, A. Boolean kernels for rule based interpretation of support vector machines. Neurocomputing 2019, 342, 113–124. [Google Scholar]
- Alfaro, C.; Gomez, J.; Moguerza, J.M.; Castillo, J.; Martinez, J.I. Toward Accelerated Training of Parallel Support Vector Machines Based on Voronoi Diagrams. Entropy 2021, 23, 1605. [Google Scholar] [CrossRef]
- Yunsheng, S.; Jiye, L.; Feng, W. An accelerator for support vector machines based on the local geometrical information and data partition. Int. J. Mach. Learn. Cybern. 2019, 10, 2389–2400. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, Australia, 27 November–1 December 1995; IEEE: Piscataway, NJ, USA, 1995; pp. 1942–1948. [Google Scholar]
- Li, J.; Zhang, J.Q.; Jiang, C.J.; Zhou, M. Composite Particle Swarm Optimizer with Historical Memory for Function Optimization. IEEE Trans. Cybern. 2017, 45, 2350–2363. [Google Scholar] [CrossRef] [PubMed]
- Jacot, A.; Gabriel, F.; Clément, H. Neural Tangent Kernel: Convergence and Generalization in Neural Networks. In Proceedings of the Neural Information Processing Systems, Montréal, QC, Canada, 3–8 December 2018; pp. 8571–8580. [Google Scholar]
- Neal, R.M. Priors for infinite networks. In Bayesian Learning for Neural Networks; Springer: New York, NY, USA, 1996; pp. 29–53. [Google Scholar]
- Matthews, A.G.D.G.; Rowland, M.; Hron, J.; Turner, R.E.; Ghahramani, Z. Gaussian Process Behaviour in Wide Deep Neural Networks. In Proceedings of the 7th International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Arora, S.; Du, S.S.; Li, Z.; Salakhutdinov, R.; Wang, R.; Yu, D. Harnessing the Power of Infinitely Wide Deep Nets on Small-data Tasks. In Proceedings of the 8th International Conference on Learning Representations, Addis Ababa, Ethiopia, 26–30 April 2020. [Google Scholar]
- Du, S.S.; Hou, K.; Salakhutdinov, R.R.; Poczos, B.; Wang, R.; Xu, K. Graph Neural Tangent Kernel: Fusing Graph Neural Networks with Graph Kernels. In Proceedings of the Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Novak, R.; Xiao, L.; Hron, J.; Lee, J.; Alemi, A.A.; Sohl-Dickstein, J.; Schoenholz, S.S. Neural Tangents: Fast and Easy Infinite Neural Networks in Python. In Proceedings of the 9th International Conference on Learning Representations, Virtual Event, Austria, 3–7 May 2021. [Google Scholar]


| Kernel | Para.1 | Range | Para.2 | Range | Para.3 | Range |
|---|---|---|---|---|---|---|
| Poly | C | 0.001, …, 1024 | Coef0 | 0.02, …, 2 | degree | 1, 2, 3 |
| Rbf | C | 0.001, …, 1024 | gamma | 0.1, …, 50 | ||
| MDK | C | 0.004, …, 1024 | d | 0.003, …, 32 | ||
| NTK | C | 0.004, …, 128 | dep | 1, …, 13 | fix | 0, …, 12 |
| Datasets | POLY | M-POLY | RBF | M-RBF | NTK | M-NTK | B-NTK |
|---|---|---|---|---|---|---|---|
| breast-cancer | 0.808 | 0.857 | 0.788 | 0.808 | 0.818 | 0.858 | 0.831 |
| credit-approval | 0.884 | 0.892 | 0.884 | 0.872 | 0.883 | 0.901 | 0.825 |
| echocardiogram | 0.892 | 0.881 | 0.862 | 0.896 | 0.854 | 0.898 | 0.942 |
| fertility | 0.9 | 0.9 | 0.9 | 0.91 | 0.9 | 0.92 | 0.875 |
| hepatitis | 0.805 | 0.799 | 0.844 | 0.822 | 0.818 | 0.882 | 0.856 |
| libras | 0.772 | 0.77 | 0.689 | 0.702 | 0.817 | 0.846 | 0.775 |
| Parkinsons | 0.918 | 0.873 | 0.938 | 0.883 | 0.938 | 0.968 | 0.877 |
| bridges-MATERIAL | 0.868 | 0.888 | 0.887 | 0.898 | 0.887 | 0.887 | 0.928 |
| bridges-REL-L | 0.784 | 0.845 | 0.824 | 0.875 | 0.765 | 0.875 | 0.755 |
| bridges-SPAN | 0.696 | 0.685 | 0.674 | 0.695 | 0.739 | 0.803 | 0.799 |
| bridges-T-OR-D | 0.902 | 0.943 | 0.902 | 0.943 | 0.887 | 0.907 | 0.926 |
| bridges-TYPE | 0.654 | 0.697 | 0.654 | 0.728 | 0.716 | 0.728 | 0.738 |
| german-credit | 0.76 | 0.772 | 0.75 | 0.798 | 0.786 | 0.798 | 0.770 |
| trains | 0.6 | 0.6 | 0.6 | 0.6 | 0.6 | 0.8 | 0.6 |
| wine | 0.989 | 0.953 | 0.978 | 0.908 | 0.978 | 0.989 | 0.870 |
| zoo | 0.96 | 0.96 | 0.96 | 0.9 | 0.9 | 0.96 | 0.8 |
| Friedman test | |||||||
| Kendall test | |||||||
| Dataset | Boolean(%) | M-Time Saving (%) | M-Acc Improv. (%) | B-Time Saving(%) | B-Acc Improv.(%) |
|---|---|---|---|---|---|
| breast-cancer 1 | 22.2 | 58.9 | 2.47 | 62.5 | 8.01 |
| breast-cancer 2 | 6.1 | 52.5 | 3.65 | 62.5 | 1.57 |
| credit-approval | 26.7 | 30.0 | 2.04 | 18.3 | −7.89 |
| echocardiogram | 30.0 | 37.2 | 5.15 | 74.9 | 10.30 |
| heart-cleveland | 23.1 | 27.9 | 8.46 | 28.0 | 15.15 |
| hepatitis | 63.2 | 24.7 | 7.83 | 59.3 | 4.62 |
| libras | 23.3 | 15.7 | 3.55 | −9.0 | −5.14 |
| Parkinsons | 13.6 | 37.1 | 3.20 | 80.2 | −5.96 |
| bridges-REL-L | 28.6 | 35.9 | 14.38 | 59.5 | −1.31 |
| bridges-T-OR-D | 28.6 | 55.1 | 2.25 | 26.6 | −9.92 |
| bridges-SPAN | 28.6 | 31.6 | 8.66 | −1.3 | 8.53 |
| bridges-TYPE | 28.6 | 35.5 | 22.15 | −24.4 | 3.07 |
| Australian-credit | 28.6 | 25.2 | 3.36 | 36.0 | 13.04 |
| zoo | 62.5 | 40.3 | 6.67 | 88.1 | −11.11 |
| lenses | 75.0 | 36.1 | 25.00 | 14.0 | 4.34 |
| waveform | 28.6 | 20.9 | 4.81 | −48.6 | 5.96 |
| wine | 30.8 | 24.9 | 1.12 | 99.8 | −0.80 |
| Datasets | RF | ADABOOST | KNN | Decision Trees | M-RBF | M-NTK | B-NTK |
|---|---|---|---|---|---|---|---|
| breast-cancer | 0.798 | 0.677 | 0.768 | 0.727 | 0.808 | 0.858 | 0.831 |
| credit-approval | 0.870 | 0.817 | 0.846 | 0.803 | 0.872 | 0.901 | 0.825 |
| echocardiogram | 0.846 | 0.831 | 0.815 | 0.8 | 0.896 | 0.898 | 0.942 |
| fertility | 0.9 | 0.86 | 0.880 | 0.9 | 0.91 | 0.92 | 0.875 |
| hepatitis | 0.818 | 0.792 | 0.831 | 0.701 | 0.822 | 0.882 | 0.856 |
| libras | 0.678 | 0.767 | 0.633 | 0.572 | 0.702 | 0.846 | 0.775 |
| Parkinsons | 0.876 | 0.856 | 0.876 | 0.845 | 0.883 | 0.968 | 0.877 |
| bridges-MATERIAL | 0.849 | 0.868 | 0.868 | 0.830 | 0.898 | 0.887 | 0.928 |
| bridges-REL-L | 0.765 | 0.706 | 0.667 | 0.706 | 0.875 | 0.875 | 0.755 |
| bridges-SPAN | 0.674 | 0.717 | 0.696 | 0.587 | 0.695 | 0.803 | 0.799 |
| bridges-T-OR-D | 0.843 | 0.882 | 0.902 | 0.804 | 0.943 | 0.907 | 0.926 |
| bridges-TYPE | 0.635 | 0.596 | 0.558 | 0.538 | 0.728 | 0.728 | 0.738 |
| German-credit | 0.75 | 0.73 | 0.724 | 0.696 | 0.798 | 0.798 | 0.770 |
| trains | 0.6 | 0.6 | 0.4 | 0.6 | 0.6 | 0.8 | 0.6 |
| wine | 0.966 | 0.955 | 0.966 | 0.876 | 0.908 | 0.989 | 0.870 |
| zoo | 0.96 | 0.8 | 0.92 | 0.94 | 0.9 | 0.96 | 0.8 |
| Datasets | RF | ADABOOST | KNN | Decision Trees | M-NTK | B-NTK |
|---|---|---|---|---|---|---|
| breast-cancer | 124.19 | 16.05 | 7.41 | 1.63 | 1.13 | 1.00 |
| credit-approval | 112.83 | 14.69 | 9.74 | 1.00 | 1.32 | 16.32 |
| echocardiogram | 236.59 | 28.55 | 7.58 | 1.00 | 10.64 | 1.72 |
| fertility | 323.30 | 43.64 | 5.39 | 1.00 | 5.42 | 5.19 |
| hepatitis | 479.56 | 33.72 | 5.51 | 1.00 | 2.19 | 1.52 |
| libras | 279.17 | 46.18 | 9.43 | 7.13 | 1.00 | 3.97 |
| Parkinsons | 186.28 | 24.77 | 4.87 | 1.00 | 1.29 | 1.34 |
| bridges-MATERIAL | 302.00 | 39.42 | 5.05 | 1.00 | 1.38 | 1.36 |
| bridges-REL-L | 235.11 | 30.93 | 3.54 | 1.00 | 1.21 | 1.29 |
| bridges-SPAN | 221.42 | 31.12 | 3.79 | 1.00 | 2.19 | 1.39 |
| bridges-T-OR-D | 305.00 | 39.12 | 4.53 | 1.00 | 1.90 | 1.25 |
| bridges-TYPE | 306.06 | 38.88 | 4.70 | 1.00 | 3.29 | 2.02 |
| German-credit | 163.34 | 24.71 | 13.00 | 1.98 | 1.00 | 12.49 |
| trains | 122.63 | 19.45 | 17.42 | 1.53 | 1.12 | 1.00 |
| wine | 358.86 | 5.72 | 93.48 | 1.00 | 6.40 | 4.05 |
| zoo | 247.87 | 32.61 | 7.16 | 1.00 | 2.07 | 1.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, Y.; Li, Z.; Liu, H. Multi-Angle Fast Neural Tangent Kernel Classifier. Appl. Sci. 2022, 12, 10876. https://doi.org/10.3390/app122110876
Zhai Y, Li Z, Liu H. Multi-Angle Fast Neural Tangent Kernel Classifier. Applied Sciences. 2022; 12(21):10876. https://doi.org/10.3390/app122110876
Chicago/Turabian StyleZhai, Yuejing, Zhouzheng Li, and Haizhong Liu. 2022. "Multi-Angle Fast Neural Tangent Kernel Classifier" Applied Sciences 12, no. 21: 10876. https://doi.org/10.3390/app122110876
APA StyleZhai, Y., Li, Z., & Liu, H. (2022). Multi-Angle Fast Neural Tangent Kernel Classifier. Applied Sciences, 12(21), 10876. https://doi.org/10.3390/app122110876





