Abstract
Staphylococcus aureus (S. aureus) is one of the most prevalent bacterial pathogens recovered from diabetic foot infections (DFIs). Most S. aureus isolates exhibit methicillin resistance, so treatment is recommended with antimicrobials active against methicillin-resistant S. aureus (MRSA) in patients who have risk factors associated with MRSA infections. The main goal of this study was to see if proteomics and molecular methods could be effective in identifying and distinguishing MRSA recovered from DFIs. Since MRSA is highly resistant to β-lactam antibiotics and usually does not respond to other antimicrobial drugs, we evaluated the resistance of MRSA isolates against different antibiotics. The standard procedures were followed for a culture of 250 skin swabs collected from diabetic foot patients. The phenotypic characteristics of 48 suspected S. aureus cultures were determined via microscopic examination, Gram staining, a coagulase test, a BBL™ Staphyloslide™ Latex test, a Staph ID 32 API system, and a Vitek 2 Compact system. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was used to examine the protein profile of all isolates, and real-time PCR was then used to identify mecA and PVL virulence genes. S aureus isolates were tested using the Vitek 2 Compact for antimicrobial susceptibility using Gram-positive cards (GP71). Among the 48 bacterial isolates tested, 45 (93.75%), 42 (87.5%), and 46 (95.83%) were positive in tube coagulase, the Staph ID 32 API system, and the Vitek 2 Compact system, respectively. We correctly identified all suspected S. aureus isolates (100%) via MALDI-TOF MS with a score value ≥2.00 and differentiated them into 22/48 MRSA (45.83%) and 26/48 MSSA (54.17%) isolates. A higher peak intensity at masses of 5530 Da, 6580 Da, 6710 Da, and 6820 Da was detected in MRSA, but not in MSSA. All MRSA isolates tested positive for the mecA gene, while all isolates tested negative for the PVL gene. The antibiotic susceptibility results showed that 22 (100%), 20 (90.91%), 19 (86.36%), 18 (81.82%), 17 (77.27%), 15 (68.18%), 13 (59.1%), and 12 (54.55%) MRSA strains were resistant to cefoxitin, daptomycin, erythromycin, benzylpenicillin, ciprofloxacin, oxacillin, and clindamycin, respectively. In contrast, all MRSA strains were extremely susceptible (100%) to linezolid, nitrofurantoin, quinupristin–dalfopristin, tigecycline, and vancomycin. Moreover, 20 (90.91%), 18 (81.82%), and 17 (77.27%) of the MRSA strains exhibited high sensitivity against rifampin, trimethoprim–sulfamethoxazole, and gentamicin, respectively. In DFIs, MALDI-TOF MS is a powerful and accurate method of identifying and distinguishing both MRSA and MSSA isolates. A high level of antimicrobial resistance was found in MRSA isolates, and antibiotic therapy based on antibiotic susceptibility patterns is essential for a successful outcome.
1. Introduction
Diabetes mellitus (DM) is a progressive and chronic endocrine disorder that is characterized by tenacious hyperglycemia. Diabetic foot infection (DFI) is considered one of the major severe complications of this disease [1,2] and is triggered by several bacteria [3]. Staphylococcus aureus (S. aureus) represents one of the main microorganisms responsible for the acute form of DFI. Nevertheless, numerous microorganisms are frequently detected in infected wounds [4]. It is already known that antibiotic resistance against different microorganisms recovered from diabetic foot patients is an emerging global problem. Previous studies conducted by Karmaker et al. [5] and Ornskov et al. [6] stated that DFI is caused by multiple antibiotic-resistant methicillin-resistant S. aureus (MRSA). A hospital-associated MRSA outbreak was first reported in the 1960s [6,7].
It has become increasingly common for MRSA to spread among foot ulcer patients over time, which often results in amputations [8,9]; 16.78% of DFIs are colonized by MRSA, though subsequent studies have shown higher prevalence rates in accordance with geographical variables [10,11].
In diabetic patients, it is difficult to accurately identify bacteria and diagnose DFI due to the polymicrobial nature of the infection and the confounding effects of neuropathy and ischemia [12]. There are several methods available for identifying and defining human microbiota. In clinical microbiology, diagnosis is mostly determined by phenotypic identification, which involves cultivating bacteria in order to isolate their colonies. Despite this, the traditional culture method cannot identify phenotypically similar bacteria [13].
The identification of MRSA in DFI is based mainly on the Kirby–Bauer method and genetic analysis [2]. Cefoxitin and oxacillin disc diffusion are considered the most common phenotypic techniques applied in the identification of MRSA. Although the phenotypic approach is the gold standard, it is time-consuming, expensive, and seldom used to distinguish MRSA from methicillin-sensitive S. aureus (MSSA) [14]. A gene called mecA, which is responsible for determining penicillin-binding protein (PBP) production, is considered an indicator for the detection of MRSA. For that reason, detection of the mecA gene and PBPs via genetic approaches is still good practice for MRSA confirmation [7]. These PBPs usually have potent empathy for the β-lactam ring [15]. Nonetheless, in MRSA strains, alternative PBP2a has a very low ability for joining to β-lactam antibiotics, and this may lead to failure of methicillin antimicrobial drugs to damage the bacterial cell wall [16].
Molecular approaches, on the other hand, are nearly all time-consuming and costly. As a result, a quick, cost-effective, and reliable approach to identify the different bacteria in DFI is still needed. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a unique technique that gives efficient and fast characterization of diverse bacteria and is regarded as a significant step towards appropriate communicable and non-communicable disease control in clinical and public health diagnostics [17]. MALDI-TOF MS, as a novel method for the detection and classification of various microorganisms, is based on the protein complexes of the bacterial cells. Despite the fact that this technology has been used for a long time, only lately have investigations shown successful discovery of the species in a research facility [18]. During the last five to ten years, this methodology has been popular for the identification of species since it can be achieved within a short timeframe [19]. A key advantage of this approach is its speed and cost-effectiveness, provided that a library of spectral data that cover all pathogens is available.
Standard sample-organization techniques have arisen as a result of great efforts; hence, enhanced evaluation and management facilities have developed [20]. The treatment of MRSA is becoming a worldwide challenge. At the present time, MRSA creates resistance to various antimicrobial drugs [2,21]. The failure of the diabetic foot treatment process is due to several factors, the most important of which is the use of non-specific antibiotics isolated from the wound site [22,23]. Major antibiotic classes that are commonly used to treat MRSA infections are resistant to them [24,25], and MRSA infections are associated with longer hospital admissions and higher healthcare expenses that might reach EUR 44 million [9].
The majority of acute infections in patients who could not been cured with various antimicrobial drugs are commonly caused by a single type of bacterium (e.g., S. aureus). In contrast, chronic infections which have been treated with antibiotics are often caused by several types of bacteria (Gram-positive and/or Gram-negative bacteria [26]. In our region, the available information regarding DFI are scanty, and there is no satisfactory research or strategies to recommend suitable antibiotic selections according to our local information. Therefore, the main aim of the current investigation was to compare the PMFT with the biochemical and genotypic approaches for the detection and characterization of MRSA obtained from diabetic foot patients. In addition, the degree of susceptibility and resistance of MRSA against different types of antimicrobial drugs was studied in this investigation.
2. Materials and Methods
2.1. Ethical Statement
Human participants were approved by the general directorate of health affairs, Al-Qassim region, Ministry of Health, Kingdom of Saudi Arabia, with ethics approval no. 783211. All the clinical strains used in this study came from regular medical testing or strain collections, and only bacteria cultures obtained from these sources were used.
2.2. Sample Collection and Cultivation
This study was conducted at the King Fahd Specialist Hospital (Sheikh Fahad Al Owaidah Center for Diabetes Foot Care) in Buraydah city. The current investigation included approximately 250 patients with DM (type II) who were admitted with DFI to the Diabetic Center from March to October 2021. Additionally, the consent of all patients included in the study was obtained before starting the experiment. The swab samples (aspirates and/or pus) were collected from deep infected ulcers in sterile bottles under strict hygienic measures.
All samples were transported to the Microbiology Laboratory within two hours of collection or were left for 4 h in a refrigerator until we made the bacterial culture. After being cultivated for 24 h on blood agar, Baird-Parker agar, and MacConkey agar media (Sigma-Aldrich, Darmstadt, Germany) at 37 °C, all specimens were then subjected to the culture method via cultivation on blood agar complemented with 5% sheep blood. The pure cultures (3–5 pure colonies) were kept in CryoBank vials at −20 °C until further investigation (identification, qPCR, and Kirby–Bauer methods).
2.3. Phenotypic Identification of S. aureus
2.3.1. Characteristics of Microscopic Examination and Gram Staining
The form of the colonies, the quantity of hemolysis, and the differential staining procedure were used to identify S. aureus strains at first. S. aureus was identified as a colony with a whipped cream or yellow appearance and positive catalase and coagulase results, along with full and partial degrees of hemolysis. All pure isolates stored in Cryobank vials were re-cultured for morphological determination of S. aureus strains utilizing a Gram-staining procedure [27].
2.3.2. BBL™ Staphyloslide™ Latex Test and Staph ID 32 API System
The BBL™ Staphyloslide™ Latex test kit (Thermo Fisher Scientific, Waltham, MA, USA) was used for separating staphylococci that had the properties of clumping factor and/or Protein A, commonly found in S. aureus, from those that did not, according to the protocol described by Kloos and Bannerman [28]. The Staph ID 32 API system was also utilized as a phenotypic method for S. aureus characterization [29]. The growth of tube coagulase on mannitol salt agar was tested with single colonies [30].
2.3.3. Kirby–Bauer Disc Diffusion Technique for Detection of MRSA
Based on criteria given by NCCLS, a revised Kirby–Bauer disc diffusion technique [31] was utilized to screen the isolates of S. aureus for methicillin resistance. In the current study, cefoxitin and oxacillin discs (1 µg), along with Muller–Hinton agar containing 4% NaCl, were used. After a 24 h incubation period at 37 °C, the inhibition zone was assessed. To estimate the occurrence of MRSA isolates found in all clinical specimens, the overall number of MRSA isolates was divided by the number of isolated S. aureus isolates.
2.3.4. Vitek 2 Compact System for Identification and Antimicrobial Resistance of MRSA
A Vitek 2 Compact device from bioMerieux, Craponne, France, which detects bacterial growth and drug resistance [32], was used to analyze all bacterial cultures that passed the coagulase and culturing tests. To prepare the isolate’s suspension, the manufacturer’s instructions instructed that it be corrected using McFarland standard between 0.5 and 0.63. Gram-positive identity cards for S. aureus were made using VITEK® 2 GP ID cards. These cards were injected as per the manufacturer’s instructions, and the isolated ID was supplied to the device in order it to pick the exact interpretative criteria. Throughout the experiment, S. aureus (ATCC® 29213TM) was used as the reference strain. All bacteria that showed an intermediate reaction to antimicrobial drugs were labeled as resistant.
2.4. Proteomic Screening for Identification of MSSA and MRSA
We used a MALDI Biotyper (MBT) obtained from Bruker Daltonics, Bremen, Germany, [13] for the identification and discrimination of MRSA from diabetic foot ulcers. All isolates were evaluated using FlexControl v1.3 and Compass software. All isolates were grown on blood agar obtained from Sigma-Aldrich, USA, and incubated for 24 h at 37 °C. According to Bruker Daltonics, an ethanol/formic acid extraction technique was utilized.
In a nutshell, two single colonies were placed in a clean microcentrifuge tube containing 300 µL of sterilized water and 900 µL of absolute ethanol (99.9%). Stirring at 13,000 rpm for two minutes thoroughly mixed the ingredients. After removing the excess, the particle was left to dry in the air for five min. Following two minutes of mixing at 13,000 rpm, 50 µL of 70% formic acid, and then, 70% acetonitrile were poured onto the air-dried pellet. Subsequently, 1 µL of supernatant from each isolate was put on an MBT target surface and dried at 23–25 °C. After that, 1 µL of cyano-4 hydroxy-cinnamic acid matrix liquid (matrix solution) was applied. The MBT equipment was used to identify its target and interpret the data. An Escherichia coli reference strain served as a bacterial test standard across the study. The score value of uncertain spectra was checked with the application’s spectral data. When the value is between 2 and 3, MBT’s capacity to accurately identify and discriminate distinct organisms increases. If this value is less than 1.69, however, misidentification occurs.
The Compass IVD software’s varied spectra were evaluated using m/z ranging from 2000 to 18,000 Da. The main spectrum profile (MSP) database, which contains over 7000 distinct strains of both bacteria and fungi, was used to generate hierarchical clustering. Furthermore, the MBT software’s single-peak intensity and principal component analysis (PCA) were utilized to investigate the divergence between MSSA and MRSA.
2.5. Detection of S. aureus and MRSA Genes Using qPCR
The mecA (methicillin resistance) and PVL (Panton–Valentine leucocidin) genes were designated in our study for the detection of MSSA and MRSA isolates recovered from DFIs. Based on the previous protocol carried out by Sastry and Bhat [33], we extracted DNA for both MRSA and MSSA isolates using artus MRSA/SA QS-RGQ Kit RUO (Qiagen, Hilden, Germany). As can be seen in Table 1, we used two sets of primers (forward and reverse) which were designed for both S. aureus and MRSA at the Applied Biosystem Company [27,34]. In brief, 100 pmol/µL of stock primers were prepared by adding 300 µL of each forward and reverse oligonucleotide primer in purified water according to the previous protocol [35].

Table 1.
Oligonucleotide primers and conditions of real-time PCR for running of mecA and PVL genes.
2.6. Antimicrobial Susceptibility of MRSA Using AST GP71 Cards
As instructed by the manufacturer, susceptibility testing was carried out using the Vitek 2 (bioMérieux, Inc., Durham, NC, USA) system and software v5.01 with AST-GP71 [36]. On the basis of the recommendations of the Clinical and Laboratory Standards Institute [37], MICs were classified as sensitive, moderate, or resistant.
3. Results
3.1. Preliminary Detection of S. aureus Isolates
Out of 250 samples, 48 isolates of S. aureus (19.2%) were isolated using a culture method. In order to examine the isolates further, a Bacterial Culture Freezing System (CryobankTM) was set at −20 °C with the initially detected isolates.
3.2. Morphological Characterization of S. aureus Strains
As shown in Table 2, 45 (93.75%) of the 48 bacterial isolates had positive tube coagulase test results and would have been identified as S. aureus. This test can occasionally produce false-negative results if the tube is shaken or agitated during preparation, which can cause the clot to disintegrate and never form again. Therefore, the remaining three strains were re-tested and eventually identified as S. aureus. According to the API system’s findings, 42 (87.5%) of the strains were successfully identified as S. aureus, while the remaining 6 (12.5%) were identified as coagulase-negative staphylococci (CNS), with three S. chromogens strains, two S. haemolyticus strains, and one S. epidermidis strain. In addition, the BBL™ Staphyloslide™ Latex test (Becton Dickinson, Franklin Lakes, NJ, USA) was conducted for the confirmation of S. aureus via detection of the clumping factor and/or Protein A, which is commonly not present in other staphylococci. According to our investigation, all tested isolates with the BBL™ Staphyloslide™ Latex were identified as S. aureus, including the six strains which were not identified using the Staph ID 32 API system. The Vitek 2 Compact was able to identify of 46 (95.83%) out of the total 48 isolates.

Table 2.
Comparison between different phenotypic techniques used for recognition of S. aureus recovered from clinical samples.
3.3. Routine Detection of MRSA Strains
According to the results obtained, out of 48 S. aureus isolates, 22 (45.83%) strains presented ≤21 mm and ≤10 mm cefoxitin and oxacillin inhibitory zones, respectively. They were categorized as MRSA according to the Clinical Laboratory Standard Institute’s (CLSI) protocols. Meanwhile, the rest of the isolates (54.17%) were identified as MSSA.
3.4. Identification of S. aureus via Mass Peptide Analysis
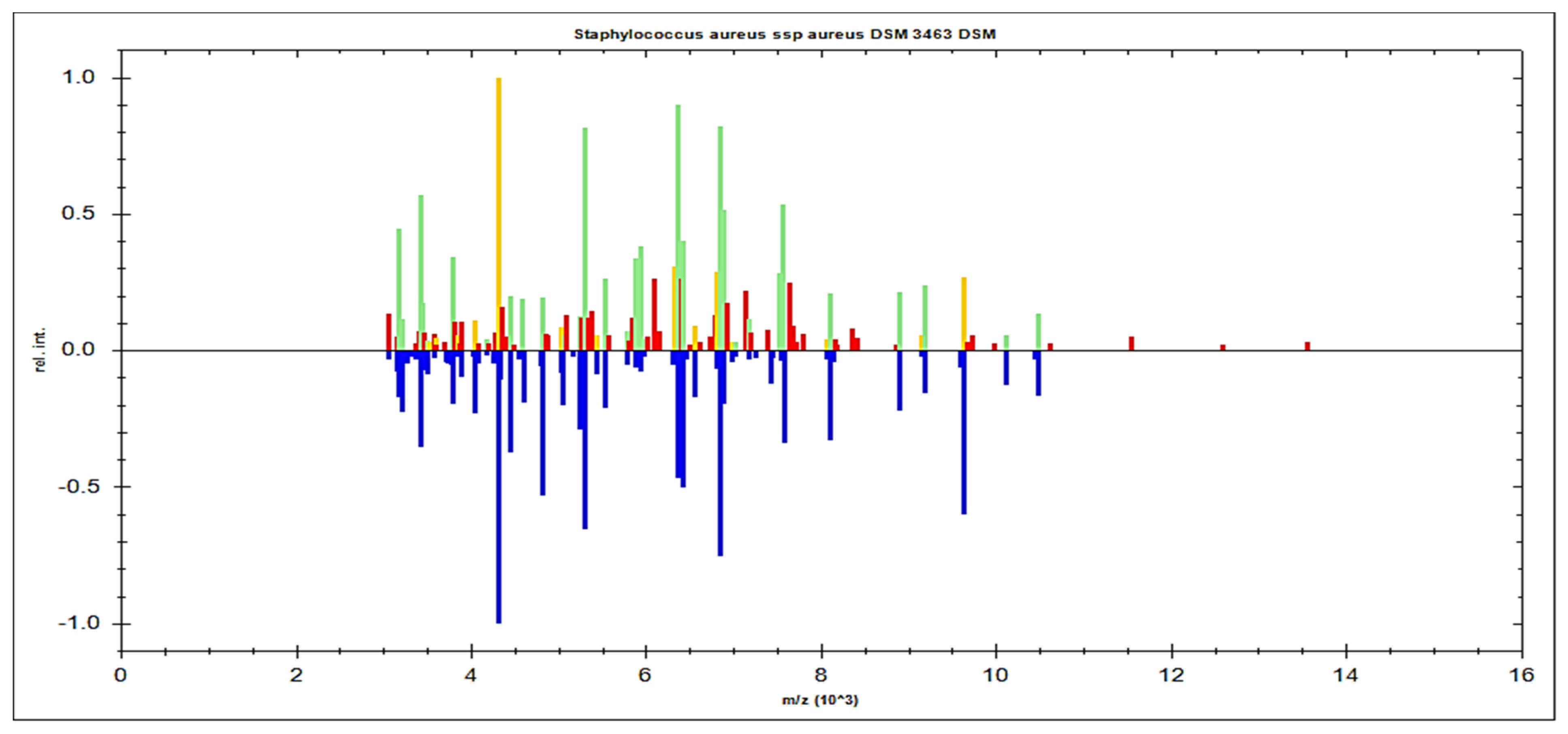
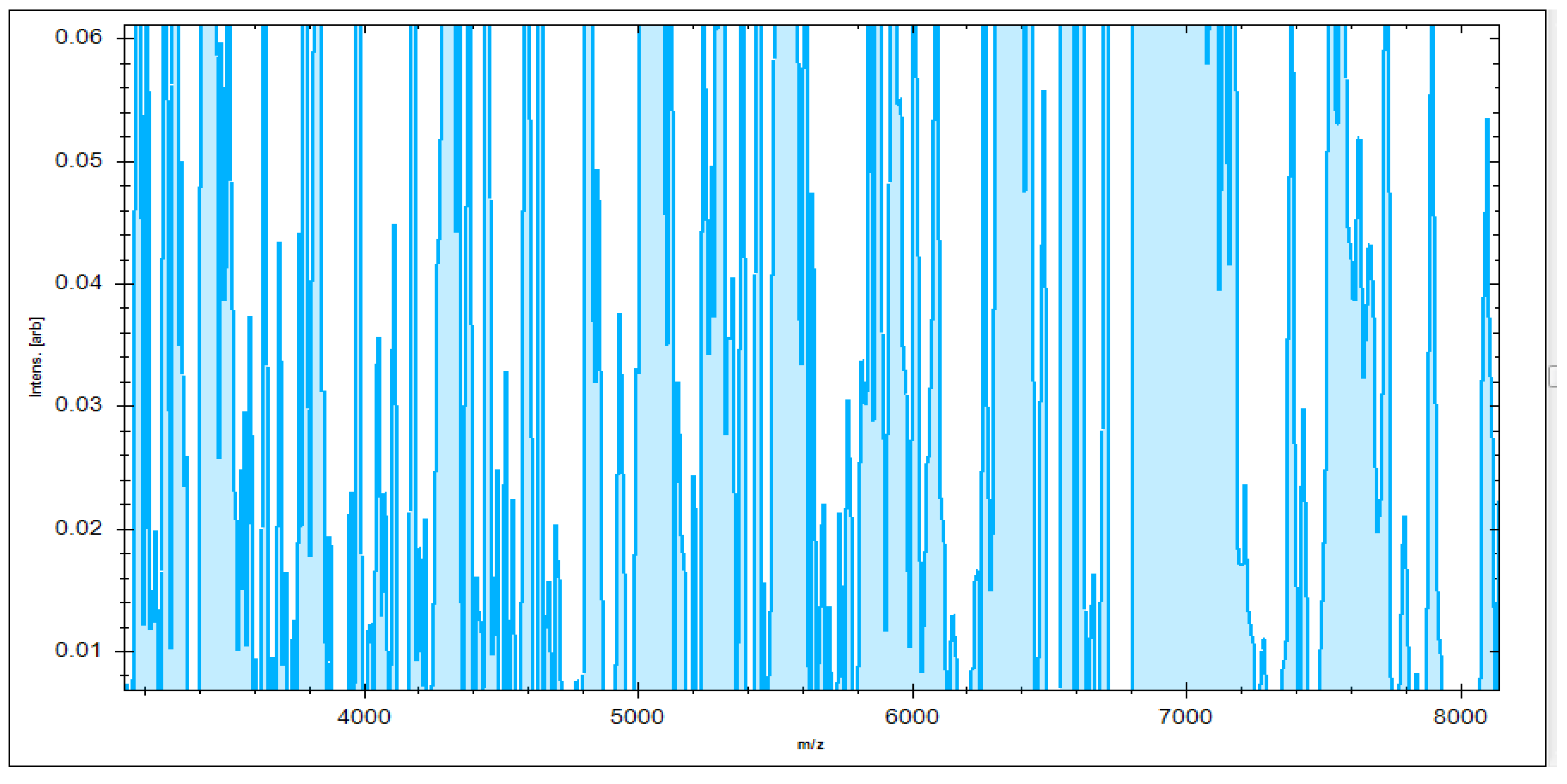
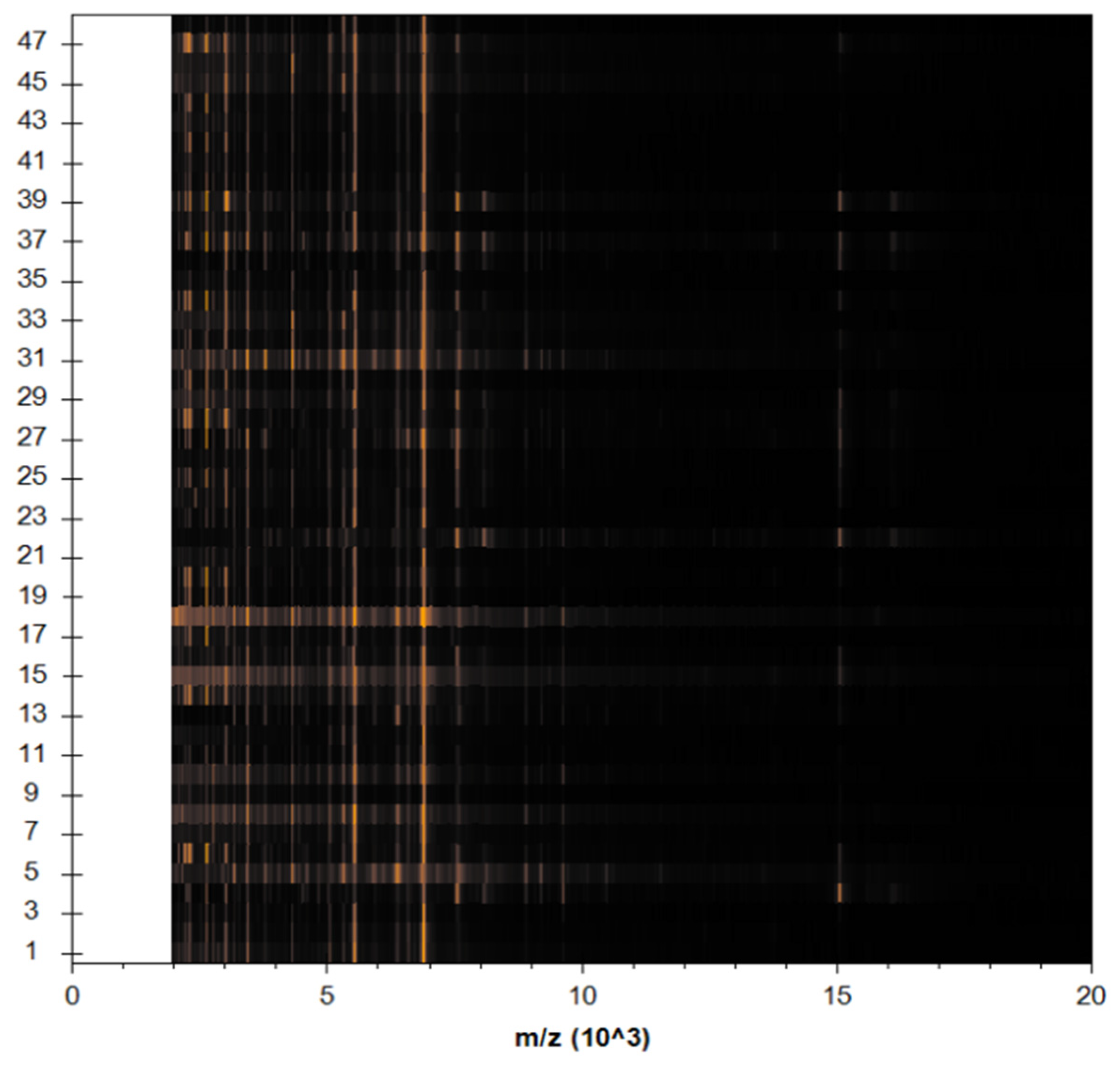
MBT’s Compass software was used to analyze the recovered microorganisms in the ongoing experiment, and the acquired spectra were evaluated by comparing them to those in the MBT database. MBT’s Compass software displayed a standard evaluation of many S. aureus strains acquired from inpatient hospital specimens. According to our findings, up to 20 distended ion peaks were found in the existing bands from the region, ranging from three hundred to eleven thousand Daltons (Da) (Figure 1). Strong peaks were found between 3000 and 8000 Da (Figure 2). MRSA isolates from the field were matched with two MRSA benchmark strains, including S. aureus DSM 3463 and S. aureus DSM 20232, whereas MSSA isolates were matched with five MSSA reference strains, including S. aureus ATCC 29213, S. aureus ATCC 25923, S. aureus DSM 20231, S. aureus DSM 346, and S. aureus DSM 799.
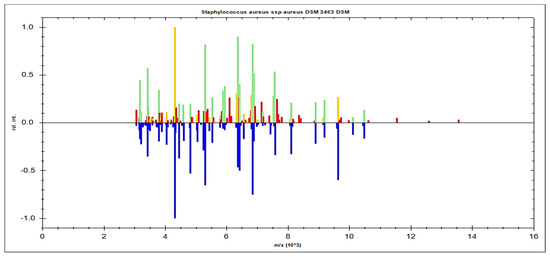
Figure 1.
S. aureus spp. aureus DSM 3463 DSM was used as a reference strain to compare spectral protein profiles obtained from clinical specimens. The blue color in the bottom section of the spectrum represents the deposited peaks that were utilized to fit the sequence; the green color in the top section of the spectrum represents extremely well-matched peaks; and the red and yellow colors represent misaligned and transitional peaks, in both.
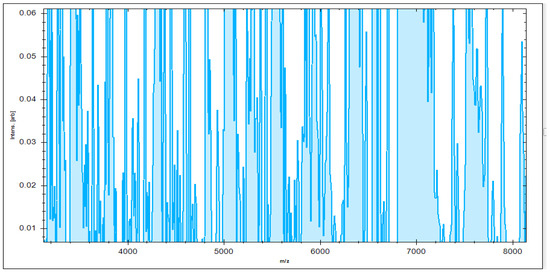
Figure 2.
Higher peak intensity was concentrated between 3000 and 8000 Da.
In the present investigation, 26/26 (100%) MSSA isolates and 22/22 (100%) MRSA isolates were successfully identified, with a logarithmic score ranging between 2.300 and 3.000 for 15 MSSA isolates and 10 MRSA isolates, and a logarithmic value ranging from 2.00 to 2.29 for 10 MSSA isolates and 12 MRSA isolates. Interestingly, one MSSA was found to have a score ranging from 1.7 to 1.99. MSSA and MRSA isolates were identified by comparing their profiles to those in the MBT device dataset, which includes over 290 strains from 16 species from the ATCC and the DSMZ reference strains.
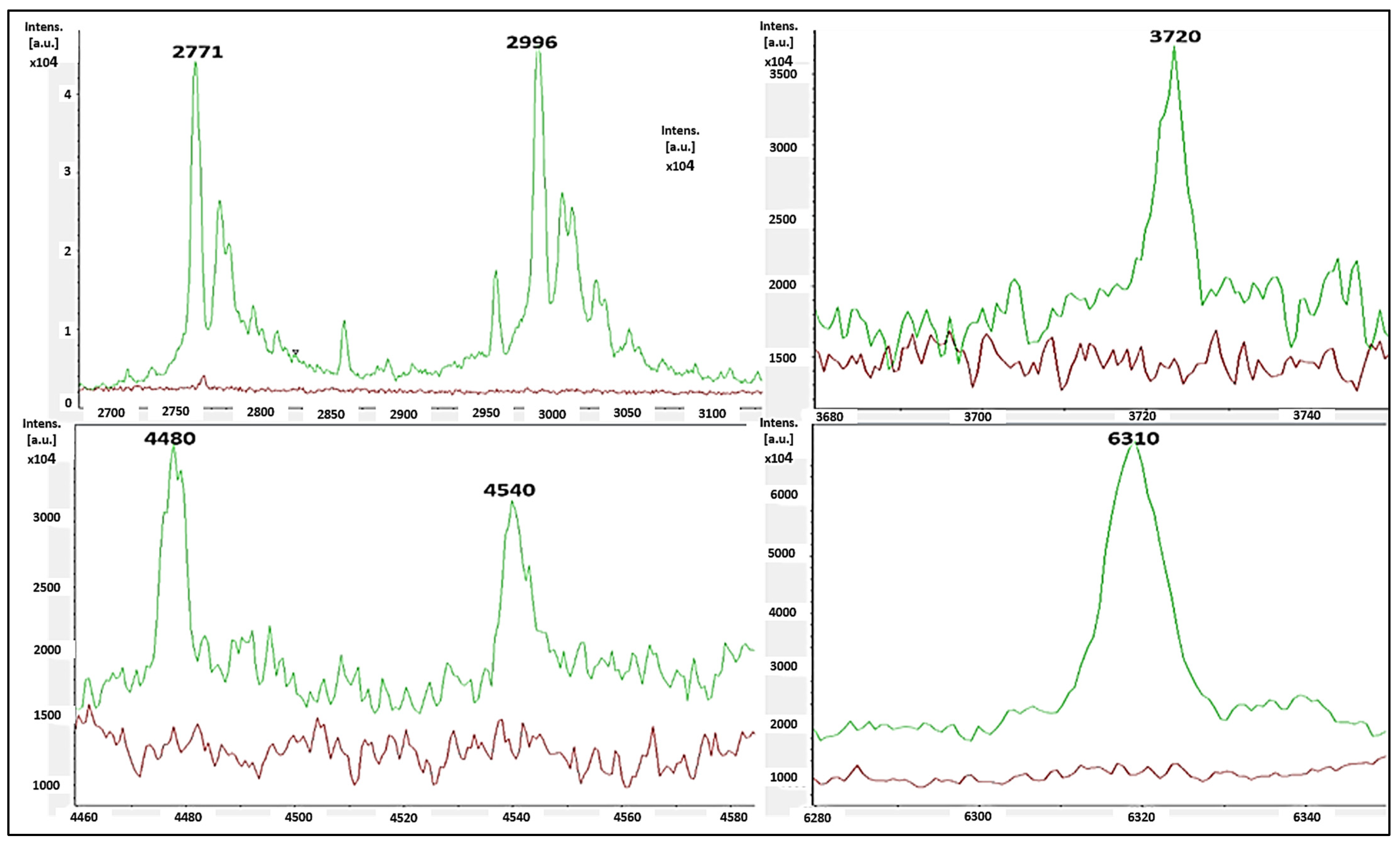
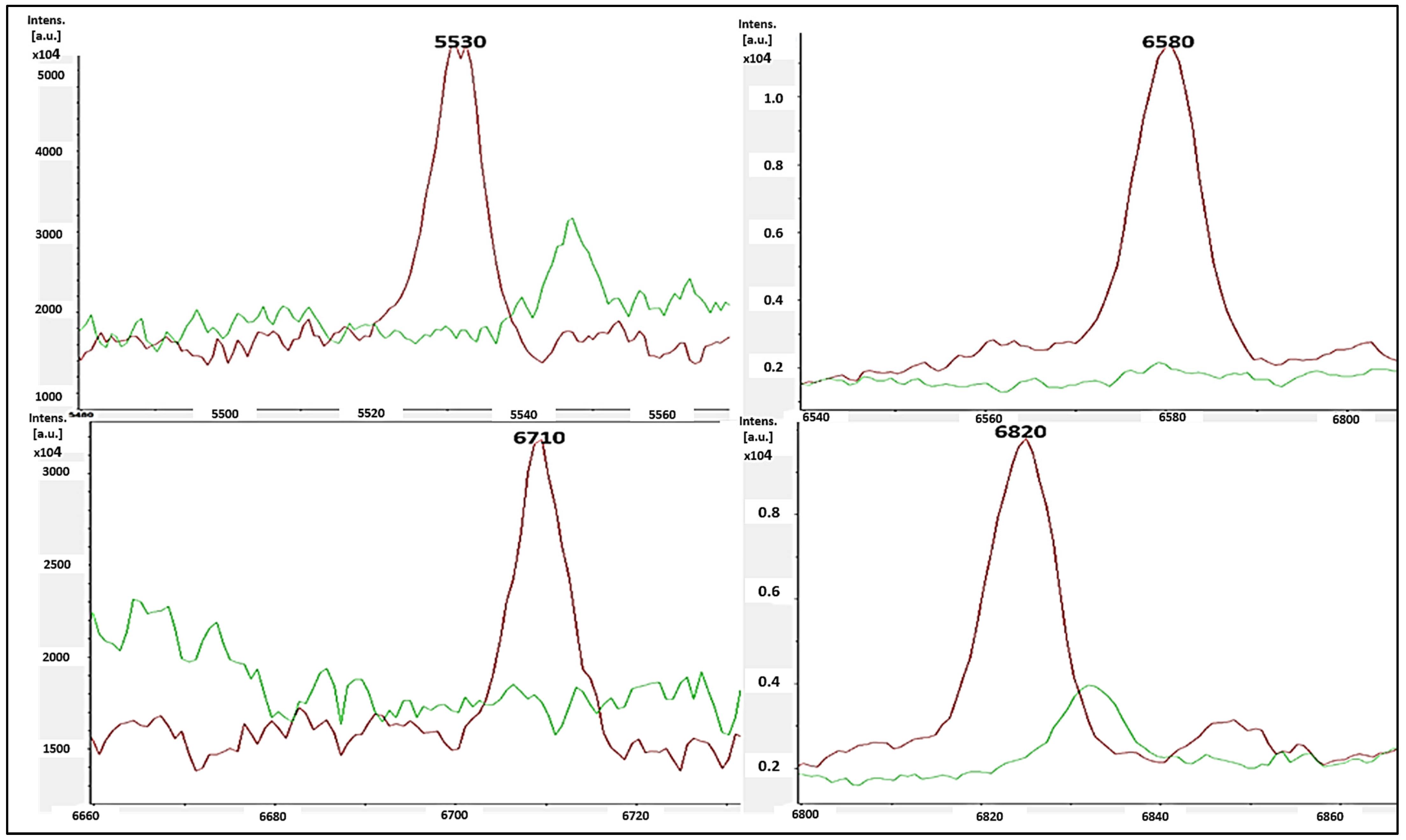
The single-peak screening of several mass regions revealed regular divergences that might be used to classify MSSA and MRSA strains. We discovered a plethora of single-peak values in the region of 2770 to 6310 Da, revealing a wide variety of intensities between both the reported MSSA and MRSA strains. As a result, the single peak is a useful tool in differentiating MRSA from MSSA. The precise discoveries were constantly expanded into the areas of 2771 Da, 2996 Da, 3720 Da, 4480 Da, 4540 Da, and 6310 Da. In such circumstances, there were several differences in the strengths of the single peaks of concentration between MRSA and MSSA. MSSA (green color) had significant strength concentrations in the mass ranges of 2771 Da, 2996 Da, 3720 Da, 4480 Da, 4540 Da, and 6310 Da (Figure 3), whereas MRSA had none (red color). MRSA (red color) had higher intensity peaks in the mass ranges of 5530 Da, 6580 Da, 6710 Da, and 6820 Da (Figure 4) that were not seen in MSSA (green color).
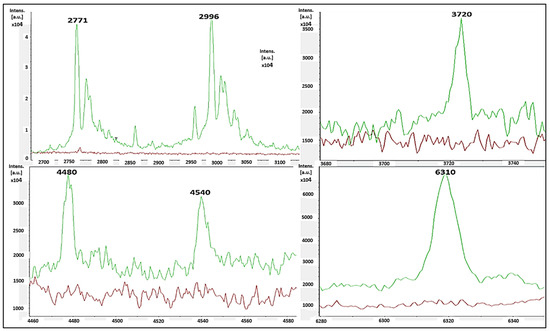
Figure 3.
MSSA (green color) had single-peak intensities (2771 Da, 2996 Da, 3720 Da, 4480 Da, 4540 Da, and 6310 Da), whereas MRSA (red color) did not (red color).
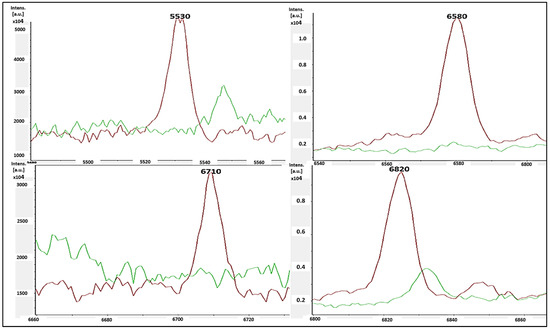
Figure 4.
MRSA (red color) had single-peak intensities (5530 Da, 6580 Da, 6710 Da, and 6820 Da), whereas MSSA (blue color) did not (green color).
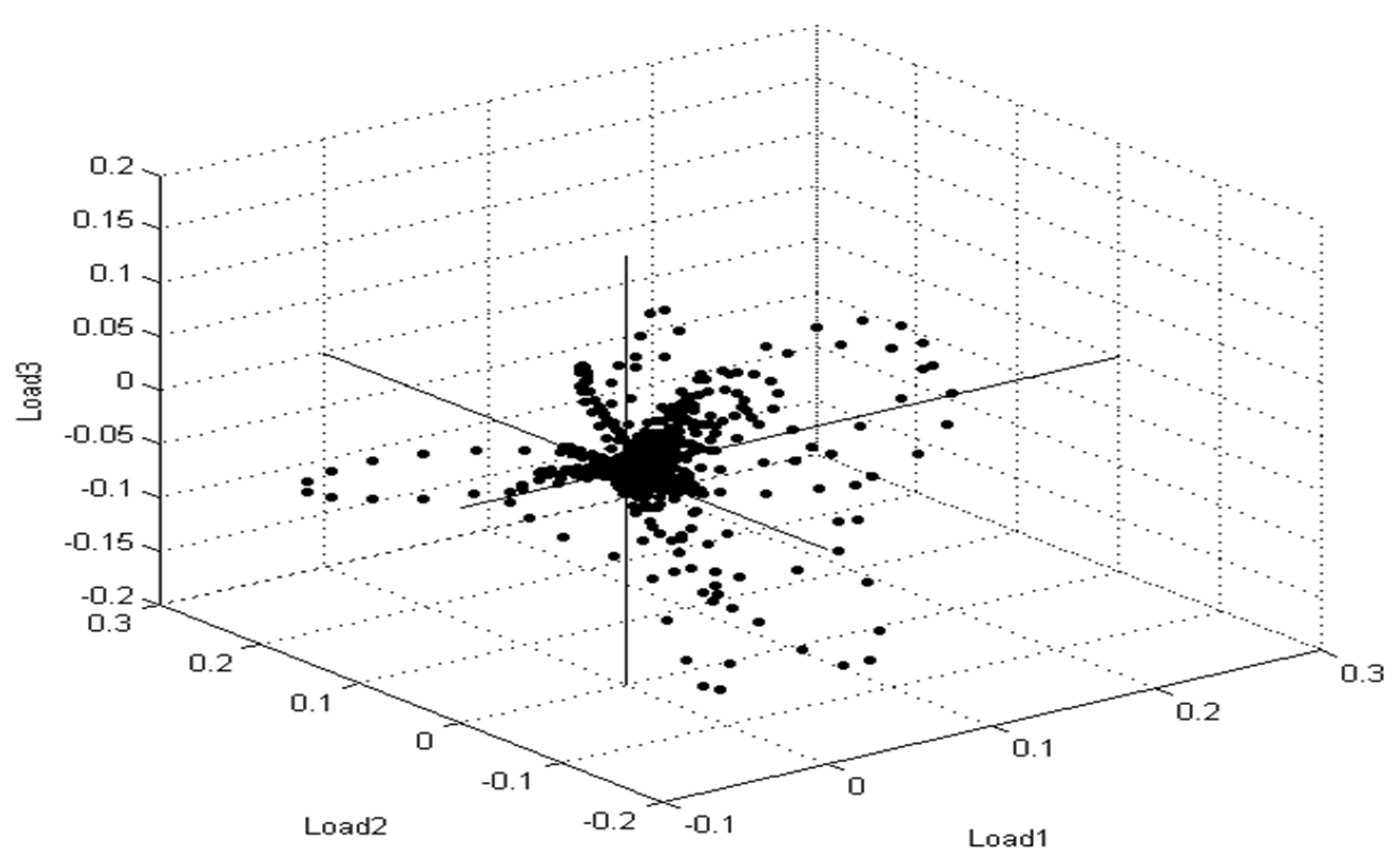
Furthermore, principal component analysis (PCA) is a supplementary mathematical technique taken from the MBT device’s Compass program for evaluating statistical models to show the degree of likeness and diversity of diverse protein profile spectra. Similarly, as mentioned in the various mathematical evaluations, PCA decreases the variance of a complicated database. In three-dimensional (3d) PCA, several spectral proteins for MSSA and MRSA isolates were identified (Figure 5). Each spectrum is symbolized by a patch, and the varied colors show the mirrored subgroup contributions, with each dot representing the side view of one of the protein’s parts of the spectrum.
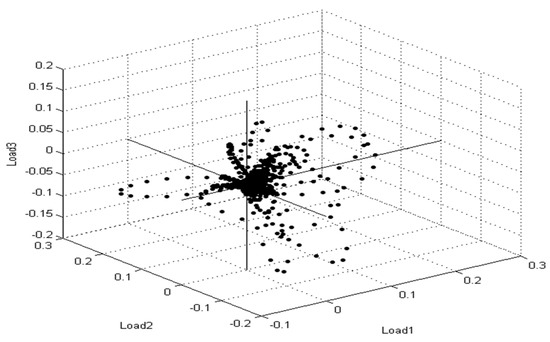
Figure 5.
Multiple spectra for S. aureus strains collected from patient cases are depicted in the PCA density. The intensity rate of single signals is shown by a single point. The signals were changed until they reached the loading value that was compatible with loading 1, loading 2, and loading 3.
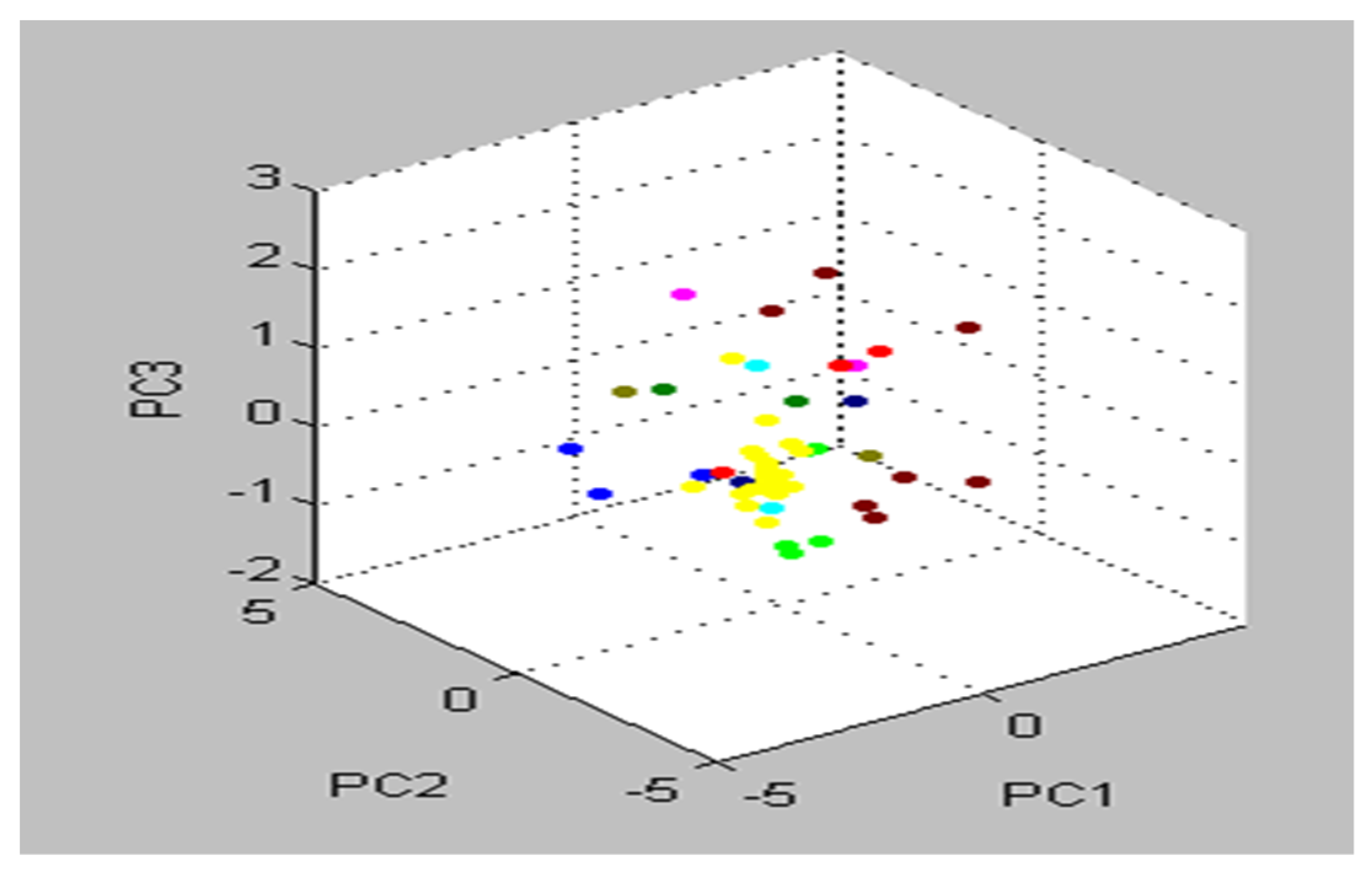
The preponderance of peaks, including all S. aureus isolates, were tightly related and matched together, as shown by the cluster view of the 3d PCA (Figure 6). When it comes to PCA assessment groups, each peak may generate loading values produced from the PCs computation. During our experiment, each signal was assigned loadings of 1, 2, and 3, values derived from calculations of PC 1, 2, and 3. To illustrate the protein profiles for the 22 detected MRSA isolates, the Compass software programed in the MBT gadget provided an actual gel image. The spectra ranged from 3000 Da to 11,000 Da, with prominent peaks between 3000 and 8000 Da (Figure 7).
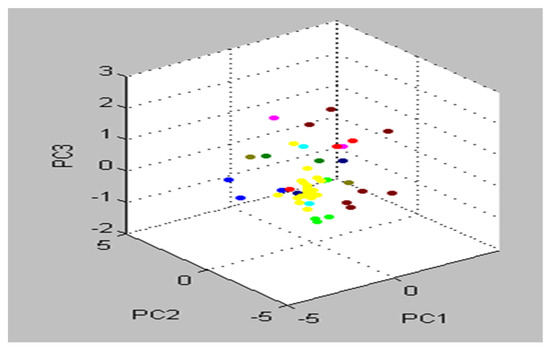
Figure 6.
Almost all S. aureus strains were categorized as one group in the cluster view of the 3d PCA (strictly correlated and harmonized together).
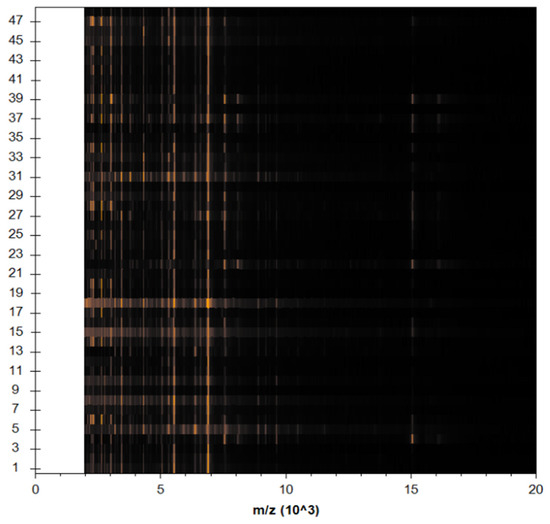
Figure 7.
View of protein on a Spectra gel for 22 MRSA strains. The yellow dots represent a collection of diverse protein spectra with varying interiors.
It was established that the detection and differentiation of various strains of S. aureus via MBT are more specific, accurate, and rapid than other classical methods, with accuracy reaching 100%. Comparing the protein fingerprint device with the conventional methods for the identification of various types of microorganisms in terms of time, we find that the protein fingerprint device is very fast and takes a little time—about an hour and a half—to identify 96 samples from the preparation of the sample to obtaining the results.
3.5. Molecular Identification of MRSA Strains
The MBT results were confirmed using SYBR® Green qPCR. MRSA was thought to be recognized by the sequences of the mecA and PVL genes. The replicating occurrences of the predicted base pairs were produced via PCR amplification with these target DNA sequences. Each genome was expanded separately, and the size of each possible product had been determined for a long time. The mecA gene was found in 22 MRSA strains, according to our findings. In contrast, the PVL gene was not detected in all the tested isolates. To assess the amount of the mecA gene, the qPCR outputs were passed through a LabChip GXII Automatic electrophoresis device, which revealed that the size was 310 bp. When the MBT results were compared to the qPCR results, they were found to be completely consistent; as a result, qPCR is currently used as an MBT validation process.
3.6. Antimicrobial Susceptibility Testing of MRSA Using AST GP71 Cards
A total of 22 isolates of MRSA were tested against antimicrobial drugs (AST-GP71 cards) purchased from BioMérieux, Craponne, France. As shown in Table 3, of the 22 MRSA isolates, 20/22 (90.91%), 19/22 (86.36%), 18/22 (81.82%), 17/22 (77.27%), 15/22 (68.18%), 13/22 (59.1%), and 12/22 (54.55%) MRSA strains were resistant to cefoxitin, daptomycin, erythromycin, benzylpenicillin, ciprofloxacin, oxacillin, and clindamycin, respectively, whereas all 22 MRSA strains were strongly susceptible (100%) to linezolid, nitrofurantoin, quinupristin–dalfopristin, tigecycline, and vancomycin. Moreover, 20/22 (90.91%), 18/22 (81.82%), and 17/22 (77.27%) of the MRSA strains were highly active against rifampin, trimethoprim–sulfamethoxazole, and gentamicin, respectively.

Table 3.
Presentation of AST-GP71 card for MRSA. Vitek 2 Compact AST-GP71 cards were used to determine the susceptibility of MRSA recovered from patients suffering from DFIs to antimicrobial agents.
4. Discussion
An increase in morbidity and death is associated with DFI when blood sugar is uncontrolled and poor self-care is performed [2,38]. Diabetic foot infection (DFI) is caused by one or more bacteria and is one of the most notable consequences of diabetes, according to Inzucchi et al. [1] and Hinojosa et al. [3]. Acute DFI is caused by Gram-positive cocci that live in an aerobic environment, such as S. aureus. Diabetic foot ulcers (DFUs) are commonly caused by S. aureus. In diabetic foot infection, S. aureus is an important pathogen, either alone or as part of an integrated infection [39]. MRSA infection is becoming more common in both hospitals and the population [40,41,42]. In diabetic patients, swabs taken from foot ulcers are usually infected with MRSA [43,44].
Based on the results obtained in the current study, out of 250 patients with DM (type II), 48 (19.2%) S. aureus isolates were isolated; 26/250 (10.4%) were classified as MSSA isolates and 22/250 (8.8%) as MRSA isolates. S. aureus is the predominant causal bacterium in diabetic foot ulcers, according to multiple studies, and Anwar et al. [2] found S. aureus in 38.7% of patients suffering from DFIs. A high incidence of Gram-positive bacteria has been seen in DFI in numerous studies [45,46]. S. aureus is the most common cause of skin infections in particular, and is actually a normal skin flora. There has been evidence of Gram-negative microorganisms, Enterobacteriaceae such as E. coli, Proteus species, and anaerobes in other research [47,48].
Among the four bacteria evaluated by Cervantes-Garca et al. [49], S. aureus accounted for 42%, followed by Escherichia coli (36%) and coagulase-negative S. aureus (25%), and Pseudomonas aeruginosa and Klebsiella pneumonia each accounted for 7%. Additionally, MRSA’s presence in 6% of S. aureus isolates and its representation of 31.6% of them is reason for concern, considering the pathogen’s clinical relevance. This incidence is lower than the 15–30% rate commonly used to describe MRSA in DFUs [50], which may be explained by the low prevalence of MRSA in the country [51,52,53].
Several alternative MRSA investigative techniques have been established, including selective chromogenic media, PCR tests, and, more recently, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) [54,55]. MRSA identification can be divided into two main categories: performance/efficacy criteria and convenience/efficiency requirements. High performance and efficacy are benefits of PCR, and qPCR techniques take only a few hours [56,57]. MALDI-TOF MS, on the other hand, has the possibility to be an effective and easy type of screening test for MRSA characterization, because it is already regularly used in several microbiology laboratories; undoubtedly, the discovery of resistance to antibiotics using MALDI-TOF MS is receiving more and more attention [55].
The MALDI-TOF MS approach for identifying microorganisms mainly depends on the analysis of mass spectra that indicate particular molecular fingerprints of microbes, typically proteins [13]. The mass spectrum profile (MSP), a collection of signals utilized for the detection of S. aureus, is mostly composed of conserved ribosomal proteins, which represent approximately 50% of the total number of signals [58]. The high percentage of cellular proteins suggested that S. aureus identification was very repeatable and reliable, as evidenced by the score values seen in our investigations. In this investigation, the MALDI-TOF MS methodology validated higher scores (≥2.0) for Staphylococcus spp. isolates in under 2 min, indicating that it has several advantages over traditional diagnostic techniques, such as saving time, being more cost-effective, and being >99% more sensitive [59]. Prior studies also reported similar results in which 93–100% of S. aureus isolates could be correctly identified using MALDI-TOF MS with high score values [17,60,61]. In all of these studies, there is no indication of the S. aureus subspecies, but considering the clinical origin of the isolates, it is reasonable to assume that all were S. aureus subsp. aureus.
The efficacy of the MALDI-TOF MS approach to successfully differentiate MRSA and MSSA strains was demonstrated in this investigation through the identification of distinct new peaks. Earlier research has also suggested unique MRSA peaks, but a systematic discrimination strategy focusing on these peaks in hospital studies remains unavailable. Furthermore, the observed peaks have a variety of patterns [17,55,62,63] or are only relevant to a subset of MRSA strains [64,65]. The discriminating techniques discussed here, on the other hand, offer great reliability and predictive ability, and can be used in ordinary clinical settings.
Despite phenotypic and MALDI-TOF MS techniques being used for the identification of MRSA, the identification of mecA currently plays a crucial role as a confirmatory method of MRSA. Because of its precision and accuracy, real-time PCR amplification of mecA to confirm MRSA has been deemed the gold standard approach [2,66]. In this investigation, 22 of the isolated bacteria screened positive for mecA. Despite the fact that two strains were morphologically tolerant to cefoxitin, there was no evidence of mecA amplification, suggesting that it could be carrying another gene, such as mecC, instead of mecA [67]. The MecA gene has been found in cefoxitin-sensitive bacteria, which contradicts this finding [68]. This discrepancy between genotypic approaches could be due to differences in culture conditions, temperature, culture media composition, inoculum volume, growth conditions, or the medical team’s physical expertise [69].
The existence of PVL is one of the characteristics that contributes to S. aureus virulence by causing excessive inflammatory reactions and tissue destruction, and ultimately, overriding the host’s defenses [70]. The PVL gene, on the other hand, is less common throughout the world, with important spatial variability in frequency (5% in France; 4.9% in the United Kingdom) [71]. PVL screening was unsuccessful in this investigation, even when employing standard and real-time PCR procedures. PVL-positive isolates have indeed been reported to be less common within DFI strains [71]. Additional explanations could be that our investigation had a small sample size, and some specimens were acquired from healthcare supplies, despite the fact that this gene has mostly been found in domestic animals [72].
For better control of DFIs, awareness of the isolates’ local antimicrobial sensitivity profiles is critical. Antimicrobial drugs such as linezolid, nitrofurantoin, quinupristin–dalfopristin, tigecycline, and vancomycin demonstrated 100% sensitivity to MRSA in this study. These results are in accordance with earlier investigations [35,73,74]. Numerous worldwide surveys, nonetheless, have found a rise in vancomycin-resistant S. aureus and E. faecalis strains [34,38]. Even though MRSA strains are completely responsive to vancomycin, treatment failure, according to Sharma and Hammerschlag [75], is not unheard of. A vancomycin prescription necessitates cautious drug-level monitoring due to the possibility of neurotoxic effects. As a result, prudent administration of this antibiotic is essential to prevent the emergence of vancomycin-resistant strains in Saudi Arabia in the future.
Sekhar et al. [76] found 100% sensitivity to cotrimoxazole in S. aureus and 100% tolerance to ciprofloxacin. Other investigations, on the other hand, have indicated that this prevalent strain is susceptible to ciprofloxacin to a certain degree [77]. Antibiotic sensitivities to cotrimoxazole, linezolid, and doxycycline were discovered in the research of Bansal et al. [77] and Gadepalli et al. [78] against MRSA strains. Amazing resistance to vancomycin, the major anti-staphylococcal medication, was highlighted in several studies [77,78]. Perim et al. [79] conducted a study that was similar to ours and showed that MRSA bacteria reacted effectively to vancomycin, while resistance was also discovered. They also said gentamicin was one of the medications that had a good response to therapy against both Gram-positive and Gram-negative diabetic foot ulcer pathogens.
MRSA occurrence and the ability to determine which patients are most likely to be infected with MRSA could aid clinicians in making more informed decisions about which treatments to use, and which ones are unnecessary in low-risk patients [80]. The Centers for Disease Control and Prevention estimate that up to half of all antibiotics are inappropriate, making this topic particularly important to those involved in antimicrobial stewardship efforts. It is dangerous for the patient to be exposed to possible antibiotic side-effects if antibiotics are misused, and if antibiotics misuse is correlated with antimicrobial resistance, the cost of healthcare will certainly increase. Clinicians should keep an eye on MRSA prevalence in order to make informed decisions regarding DFI treatment [80]. There is a link between unduly vigorous antibiotic treatments for multi-drug-resistant organisms and higher mortality rates in patients with other diseases, which is why it is imperative to identify patients at high and low risk for MRSA DFI so that personalized treatment can be provided [81].
The study’s limitations include difficulty in extrapolating it to all of Saudi Arabia, because it was primarily focused on the study site. Furthermore, because of the historical study design and the shortage of supporting documentation in medical files, assessing the clinical picture of the participants was not possible. Furthermore, similar to Hassan et al. [82] and Ahmadishooli et al. [23], indicating the employed groups of empirical prescriptions over the antimicrobial sensitivity test could be useful in creating presumptive therapeutic strategies for DFIs. More progressive studies are needed to evaluate the clinical aspects of DFIs, as well as their responsiveness to antibiotic therapy.
In conclusion, MALDI-TOF MS, compared with other methods, is a powerful and accurate tool for the identification and discrimination both MRSA and MSSA isolates recovered from diabetic foot patients. MRSA prevalence was alarmingly high in DFIs, which is a cause for concern because antibiotic options are limited and could result in a worst-case scenario. A high level of antimicrobial resistance was found in MRSA isolates, and antibiotic therapy based on antibiotic susceptibility patterns is essential for a successful outcome.
Author Contributions
Conceptualization, A.A. and AE.; methodology, A.A. and A.E.; software, A.A. and A.E.; validation, A.A.; formal analysis, A.A. and A.E.; investigation, A.A. and A.E.; resources, A.A. and A.E.; data curation, A.A. and A.E.; writing—original draft preparation, A.A. and A.E.; writing—review and editing, A.A. and A.E.; visualization, A.A. and A.E.; supervision, A.A. and A.E.; project administration, A.A.; funding acquisition, A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Qassim University, represented by the Deanship of Scientific Research, grant number 20004-bhsc-2020-1-1-W and the APC was funded by Qassim University, represented by the Deanship of Scientific Research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
The authors gratefully acknowledge Qassim University, represented by the Deanship of Scientific Research, on the financial support for this research under the number (20004-bhsc-2020-1-1-W) during the academic year 1442H/2020 AD. The researchers would like to thank the Deanship of Scientific Research, Qassim University for support of this project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Inzucchi, S.E.; Bergenstal, R.M.; Buse, J.B.; Diamant, M.; Ferrannini, E.; Nauck, M.; Peters, A.L.; Tsapas, A.; Wender, R.; Matthews, D.R. Management of hyperglycemia in type 2 diabetes, 2015: A patient-centered approach: Update to a position statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care 2015, 38, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Anwar, K.; Hussein, D.; Salih, J. Antimicrobial susceptibility testing and phenotypic detection of MRSA isolated from diabetic foot infection. Int. J. Gen. Med. 2020, 13, 1349. [Google Scholar] [CrossRef]
- Hinojosa, C.A.; Boyer-Duck, E.; Anaya-Ayala, J.E.; Nunez-Salgado, A.; Laparra-Escareno, H.; Torres-Machorro, A.; Lizola, R. Impact of the bacteriology of diabetic foot ulcers in limb loss. Wound Repair Regen. 2016, 24, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Uçkay, I.; Aragon-Sanchez, J.; Lew, D.; Lipsky, B.A. Diabetic foot infections: What have we learned in the last 30 years? Int. J. Infect. Dis. 2015, 40, 81–91. [Google Scholar] [CrossRef]
- Karmaker, M.; Sanyal, S.K.; Sultana, M.; Hossain, M. Association of bacteria in diabetic and non-diabetic foot infection–An investigation in patients from Bangladesh. J. Infect. Public Health 2016, 9, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Ornskov, D.; Kolmos, B.; Horn, P.B.; Nielsen, J.N.; Brandslund, I.; Schouenborg, P. Screening for methicillin-resistant Staphylococcus aureus in clinical swabs using a high-throughput real-time PCR-based method. Clin. Microbiol. Infect. 2008, 14, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Mutonga, D.M.; Mureithi, M.W.; Ngugi, N.N.; Otieno, F.C. Bacterial isolation and antibiotic susceptibility from diabetic foot ulcers in Kenya using microbiological tests and comparison with RT-PCR in detection of S. aureus and MRSA. BMC Res. Notes 2019, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mendes, J.; Marques-Costa, A.; Vilela, C.; Neves, J.; Candeias, N.; Cavaco-Silva, P.; Melo-Cristino, J. Clinical and bacteriological survey of diabetic foot infections in Lisbon. Diabetes Res. Clin. Pract. 2012, 95, 153–161. [Google Scholar] [CrossRef]
- Anafo, R.B.; Atiase, Y.; Dayie, N.T.; Kotey, F.C.; Tetteh-Quarcoo, P.B.; Duodu, S.; Osei, M.-M.; Alzahrani, K.J.; Donkor, E.S. Methicillin-resistant Staphylococcus aureus (MRSA) infection of diabetic foot ulcers at a tertiary care hospital in Accra, Ghana. Pathogens 2021, 10, 937. [Google Scholar] [CrossRef]
- Ogba, O.M.; Nsan, E.; Eyam, E.S. Aerobic bacteria associated with diabetic foot ulcers and their susceptibility pattern. Biomed. Dermatol. 2019, 3, 1–6. [Google Scholar] [CrossRef]
- Reina-Bueno, M.; Palomo-Toucedo, I.C.; Castro-Méndez, A.; Domínguez-Maldonado, G.; del Carmen Vázquez-Bautista, M. Methicillin-Resistant Staphylococcus aureus Diabetic Foot Crossed Infection: A Case Report. Pathogens 2020, 9, 549. [Google Scholar] [CrossRef]
- Jneid, J.; Lavigne, J.; La Scola, B.; Cassir, N. The diabetic foot microbiota: A review. Hum. Microbiome J. 2017, 5, 1–6. [Google Scholar] [CrossRef]
- Złoch, M.; Maślak, E.; Kupczyk, W.; Jackowski, M.; Pomastowski, P.; Buszewski, B. Culturomics Approach to Identify Diabetic Foot Infection Bacteria. Int. J. Mol. Sci. 2021, 22, 9574. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Moussa, I.M.; Alenzi, A.; Al-Maary, K.S.; Mubarak, A.S.; Alshammari, H.D.; Al-Sarar, D.; Alsubki, R.A.; Hemeg, H.A. Multidrug-resistant Escherichia coli in Raw Milk: Molecular Characterization and the potential impact of camel’s Urine as an Antibacterial Agent. Saudi J. Biol. Sci. 2021, 28, 2091–2097. [Google Scholar] [CrossRef] [PubMed]
- Nicoleta, M.; Gheorghe, I.; Popa, M.; LAZĂR, V.; Banu, O.; Bolocan, A.; Grigore, R.; Berteșteanu, Ș.V.; Octav, P. Phenotypic and genotypic investigation of resistance and virulence features of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients. Rom. Biotechnol. Lett. 2016, 21, 11591. [Google Scholar]
- Malachowa, N.; DeLeo, F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010, 67, 3057–3071. [Google Scholar] [CrossRef]
- Elbehiry, A.; Al-Dubaib, M.; Marzouk, E.; Osman, S.; Edrees, H. Performance of MALDI biotyper compared with Vitek™ 2 compact system for fast identification and discrimination of Staphylococcus species isolated from bovine mastitis. MicrobiologyOpen 2016, 5, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Guo, J.; Chen, R.; Hu, L.; Xia, Q.; Wu, W.; Wang, J.; Hu, F. Multicenter evaluation of three different MALDI-TOF MS systems for identification of clinically relevant filamentous fungi. Med. Mycol. 2021, 59, 81–86. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 2011, 49, 3321–3324. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Qian, J.; Ge, Y.; Ye, K.; Zhou, C.; Zhang, H. Principal component analysis of MALDI-TOF MS of whole-cell foodborne pathogenic bacteria. Anal. Biochem. 2020, 592, 113582. [Google Scholar] [CrossRef]
- Boudreau, M.A.; Fishovitz, J.; Llarrull, L.I.; Xiao, Q.; Mobashery, S. Phosphorylation of BlaR1 in manifestation of antibiotic resistance in methicillin-resistant Staphylococcus aureus and its abrogation by small molecules. ACS Infect. Dis. 2015, 1, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Armstrong, D.G.; Lipsky, B.A. Preventing foot ulcers in patients with diabetes. JAMA 2005, 293, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Ahmadishooli, A.; Davoodian, P.; Shoja, S.; Ahmadishooli, B.; Dadvand, H.; Hamadiyan, H.; Shahriarirad, R. Frequency and antimicrobial susceptibility patterns of diabetic foot infection of patients from Bandar Abbas District, Southern Iran. J. Pathog. 2020, 2020, 1057167. [Google Scholar] [CrossRef] [PubMed]
- Han, L.L.; McDougal, L.K.; Gorwitz, R.J.; Mayer, K.H.; Patel, J.B.; Sennott, J.M.; Fontana, J.L. High frequencies of clindamycin and tetracycline resistance in methicillin-resistant Staphylococcus aureus pulsed-field type USA300 isolates collected at a Boston ambulatory health center. J. Clin. Microbiol. 2007, 45, 1350–1352. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Akhi, M.T.; Ghotaslou, R.; Memar, M.Y.; Asgharzadeh, M.; Varshochi, M.; Pirzadeh, T.; Alizadeh, N. Frequency of MRSA in diabetic foot infections. Int. J. Diabetes Dev. Ctries. 2017, 37, 58–62. [Google Scholar] [CrossRef]
- Mota-Meira, M.; Lapointe, G.; Lacroix, C.; Lavoie, M.C. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 2000, 44, 24–29. [Google Scholar] [CrossRef]
- Renneberg, J.; Rieneck, K.; Gutschik, E. Evaluation of Staph ID 32 system and Staph-Zym system for identification of coagulase-negative staphylococci. J. Clin. Microbiol. 1995, 33, 1150–1153. [Google Scholar] [CrossRef]
- Kateete, D.P.; Kimani, C.N.; Katabazi, F.A.; Okeng, A.; Okee, M.S.; Nanteza, A.; Joloba, M.L.; Najjuka, F.C. Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 1–7. [Google Scholar] [CrossRef]
- Bauer, A.; Kirby, W.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493. [Google Scholar] [CrossRef] [PubMed]
- Spanu, T.; Sanguinetti, M.; Ciccaglione, D.; D’Inzeo, T.; Romano, L.; Leone, F.; Fadda, G. Use of the VITEK 2 system for rapid identification of clinical isolates of staphylococci from bloodstream infections. J. Clin. Microbiol. 2003, 41, 4259–4263. [Google Scholar] [CrossRef] [PubMed]
- Sastry, A.S.; Bhat, S. Essentials of Medical Microbiology; JP Medical Ltd.: Puducherry, India, 2018. [Google Scholar]
- Bhatta, D.R.; Cavaco, L.M.; Nath, G.; Kumar, K.; Gaur, A.; Gokhale, S.; Bhatta, D.R. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: A matter of concern for community infections (a hospital based prospective study). BMC Infect. Dis. 2016, 16, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, A.; Jana, D.; Dutta, K.; Dua, P.; Ghosh, C. Prevalence of Panton-Valentine leukocidin gene among community acquired Staphylococcus aureus: A real-time PCR study. J. Pathog. 2018, 2018, 4518541. [Google Scholar] [CrossRef] [PubMed]
- Bobenchik, A.M.; Hindler, J.A.; Giltner, C.L.; Saeki, S.; Humphries, R.M. Performance of Vitek 2 for antimicrobial susceptibility testing of Staphylococcus spp. and Enterococcus spp. J. Clin. Microbiol. 2014, 52, 392–397. [Google Scholar] [CrossRef]
- Cockerill, F.R.; Wikler, M.; Bush, K.; Dudley, M.; Eliopoulos, G.; Hardy, D. Clinical and laboratory standards institute. In Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Karadağ, F.Y.; Saltoğlu, N.; Ak, Ö.; Aydın, G.Ç.; Şenbayrak, S.; Erol, S.; Özatağ, D.M.; Kadanalı, A.; Küçükardalı, Y.; Çomoğlu, Ş. Foot self-care in diabetes mellitus: Evaluation of patient awareness. Prim. Care Diabetes 2019, 13, 515–520. [Google Scholar] [CrossRef]
- Lipsky, B.A.; Pecoraro, R.E.; Wheat, L.J. The diabetic foot: Soft tissue and bone infection. Infect. Dis. Clin. N. Am. 1990, 4, 409–432. [Google Scholar] [CrossRef]
- Reacher, M.H.; Shah, A.; Livermore, D.M.; Wale, M.C.; Graham, C.; Johnson, A.P.; Heine, H.; Monnickendam, M.A.; Barker, K.F.; James, D. Bacteraemia and antibiotic resistance of its pathogens reported in England and Wales between 1990 and 1998: Trend analysis. BMJ 2000, 320, 213–216. [Google Scholar] [CrossRef]
- Al-Moyed, K.A.; Al-Haddad, A.M.; Al-Areqi, B.A.; Al-Danani, D.A. Prevalence of Staphylococcus aureus infection among diabetic foot patients in Sana’a city-Yemen. Alandalus J. Appl. Sci. 2014, 6, 58–77. [Google Scholar] [CrossRef][Green Version]
- Lawes, T.; López-Lozano, J.-M.; Nebot, C.; Macartney, G.; Subbarao-Sharma, R.; Dare, C.R.; Edwards, G.F.; Gould, I.M. Turning the tide or riding the waves? Impacts of antibiotic stewardship and infection control on MRSA strain dynamics in a Scottish region over 16 years: Non-linear time series analysis. BMJ Open 2015, 5, e006596. [Google Scholar] [CrossRef]
- Tentolouris, N.; Jude, E.; Smirnof, I.; Knowles, E.; Boulton, A. Methicillin-resistant Staphylococcus aureus: An increasing problem in a diabetic foot clinic. Diabet. Med. 1999, 16, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.J.; Lipsky, B.A. Diagnosis and management of infection in the diabetic foot. Med. Clin. 2013, 97, 911–946. [Google Scholar] [CrossRef] [PubMed]
- Citron, D.M.; Goldstein, E.J.; Merriam, C.V.; Lipsky, B.A.; Abramson, M.A. Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J. Clin. Microbiol. 2007, 45, 2819–2828. [Google Scholar] [CrossRef]
- Anvarinejad, M.; Pouladfar, G.; Japoni, A.; Bolandparvaz, S.; Satiary, Z.; Abbasi, P.; Mardaneh, J. Isolation and antibiotic susceptibility of the microorganisms isolated from diabetic foot infections in Nemazee Hospital, Southern Iran. J. Pathog. 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hatipoglu, M.; Mutluoglu, M.; Turhan, V.; Uzun, G.; Lipsky, B.A.; Sevim, E.; Demiraslan, H.; Eryilmaz, E.; Ozuguz, C.; Memis, A. Causative pathogens and antibiotic resistance in diabetic foot infections: A prospective multi-center study. J. Diabetes Its Complicat. 2016, 30, 910–916. [Google Scholar] [CrossRef]
- Katz, D.E.; Friedman, N.D.; Ostrovski, E.; Ravid, D.; Amrami, N.; Avivi, D.; Mengesha, B.; Zaidenstein, R.; Lazarovitch, T.; Dadon, M. Diabetic foot infection in hospitalized adults. J. Infect. Chemother. 2016, 22, 167–173. [Google Scholar] [CrossRef]
- Cervantes-García, E.; García-González, R.; Reséndiz-Albor, A.; Salazar-Schettino, P.M. Infections of diabetic foot ulcers with methicillin-resistant Staphylococcus aureus. Int. J. Low. Extrem. Wounds 2015, 14, 44–49. [Google Scholar] [CrossRef]
- Eleftheriadou, I.; Tentolouris, N.; Argiana, V.; Jude, E.; Boulton, A.J. Methicillin-resistant Staphylococcus aureus in diabetic foot infections. Drugs 2010, 70, 1785–1797. [Google Scholar] [CrossRef]
- Egyir, B.; Guardabassi, L.; Esson, J.; Nielsen, S.S.; Newman, M.J.; Addo, K.K.; Larsen, A.R. Insights into nasal carriage of Staphylococcus aureus in an urban and a rural community in Ghana. PLoS ONE 2014, 9, e96119. [Google Scholar] [CrossRef]
- Donkor, E.S.; Kotey, F.C.; Dayie, N.T.; Duodu, S.; Tetteh-Quarcoo, P.B.; Osei, M.-M.; Tette, E.M. Colonization of HIV-infected children with methicillin-resistant Staphylococcus aureus. Pathogens 2019, 8, 35. [Google Scholar] [CrossRef]
- Dayie, N.T.; Sekoh, D.N.; Kotey, F.C.; Egyir, B.; Tetteh-Quarcoo, P.B.; Adutwum-Ofosu, K.K.; Ahenkorah, J.; Osei, M.-M.; Donkor, E.S. Nasopharyngeal Carriage of Methicillin-Resistant Staphylococcus aureus (MRSA) among Sickle Cell Disease (SCD) Children in the Pneumococcal Conjugate Vaccine Era. Infect. Dis. Rep. 2021, 13, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Grmek-Kosnik, I.; Dermota, U.; Ribic, H.; Storman, A.; Petrovic, Z.; Zohar-Cretnik, T. Evaluation of single vs pooled swab cultures for detecting MRSA colonization. J. Hosp. Infect. 2018, 98, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Kim, I.; Chung, S.H.; Chung, Y.; Han, M.; Kim, J.-S. Rapid discrimination of methicillin-resistant Staphylococcus aureus by MALDI-TOF MS. Pathogens 2019, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Li, J.; Yang, H.; Yu, J.; Wei, H. Accurate detection of methicillin-resistant Staphylococcus aureus in mixtures by use of single-bacterium duplex droplet digital PCR. J. Clin. Microbiol. 2017, 55, 2946–2955. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A.; Rochas, O. Laboratory-based and point-of-care testing for MSSA/MRSA detection in the age of whole genome sequencing. Front. Microbiol. 2018, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Pomastowski, P.; Szultka, M.; Kupczyk, W.; Jackowski, M.; Buszewski, B. Evaluation of intact cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for capillary electrophoresis detection of controlled bacterial clumping. Anal. Bioanal. Tech. 2015, 13, 1–7. [Google Scholar]
- Manukumar, H.; Umesha, S. MALDI-TOF-MS based identification and molecular characterization of food associated methicillin-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 1–16. [Google Scholar]
- Harris, L.G.; El-Bouri, K.; Johnston, S.; Rees, E.; Frommelt, L.; Siemssen, N.; Christner, M.; Davies, A.P.; Rohde, H.; Mack, D. Rapid identification of staphylococci from prosthetic joint infections using MALDI-TOF mass-spectrometry. Int. J. Artif. Organs 2010, 33, 568–574. [Google Scholar] [CrossRef]
- Bergeron, M.; Dauwalder, O.; Gouy, M.; Freydiere, A.-M.; Bes, M.; Meugnier, H.; Benito, Y.; Etienne, J.; Lina, G.; Vandenesch, F. Species identification of staphylococci by amplification and sequencing of the tuf gene compared to the gap gene and by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 343–354. [Google Scholar] [CrossRef]
- Lasch, P.; Fleige, C.; Stämmler, M.; Layer, F.; Nübel, U.; Witte, W.; Werner, G. Insufficient discriminatory power of MALDI-TOF mass spectrometry for typing of Enterococcus faecium and Staphylococcus aureus isolates. J. Microbiol. Methods 2014, 100, 58–69. [Google Scholar] [CrossRef]
- Østergaard, C.; Hansen, S.G.; Møller, J.K. Rapid first-line discrimination of methicillin resistant Staphylococcus aureus strains using MALDI-TOF MS. Int. J. Med. Microbiol. 2015, 305, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.D.; Wang, H.; Karichu, J.; Richter, S.S. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn. Microbiol. Infect. Dis. 2016, 86, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Víquez-Molina, G.; Aragon-Sanchez, J.; Perez-Corrales, C.; Murillo-Vargas, C.; López-Valverde, M.E.; Lipsky, B.A. Virulence factor genes in Staphylococcus aureus isolated from diabetic foot soft tissue and bone infections. Int. J. Low. Extrem. Wounds 2018, 17, 36–41. [Google Scholar] [CrossRef]
- Baddour, M.; AbuElKheir, M.; Fatani, A. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Curr. Microbiol. 2007, 55, 473–479. [Google Scholar] [CrossRef]
- Deplano, A.; Vandendriessche, S.; Nonhoff, C.; Denis, O. Genetic diversity among methicillin-resistant Staphylococcus aureus isolates carrying the mecC gene in Belgium. J. Antimicrob. Chemother. 2014, 69, 1457–1460. [Google Scholar] [CrossRef]
- Mottola, C.; Semedo-Lemsaddek, T.; Mendes, J.J.; Melo-Cristino, J.; Tavares, L.; Cavaco-Silva, P.; Oliveira, M. Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J. Biomed. Sci. 2016, 23, 1–10. [Google Scholar] [CrossRef]
- Kavitha, Y.; Mohan, S.; Moinuddin, S.K. Bacteriological profile of diabetic foot infection with special reference to ESBL and MRSA in a coastal tertiary care teaching hospital. Indian J. Microbiol. Res. 2017, 4, 68–73. [Google Scholar]
- Stappers, M.H.; Hagen, F.; Reimnitz, P.; Mouton, J.W.; Meis, J.F.; Gyssens, I.C. Direct molecular versus culture-based assessment of Gram-positive cocci in biopsies of patients with major abscesses and diabetic foot infections. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Dunyach-Remy, C.; Ngba Essebe, C.; Sotto, A.; Lavigne, J.-P. Staphylococcus aureus toxins and diabetic foot ulcers: Role in pathogenesis and interest in diagnosis. Toxins 2016, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Amin, D.H.M.; Guler, E.; Baddal, B. Prevalence of Panton-Valentine leukocidin in methicillin-resistant Staphylococcus aureus clinical isolates at a university hospital in Northern Cyprus: A pilot study. BMC Res. Notes 2020, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Saseedharan, S.; Sahu, M.; Chaddha, R.; Pathrose, E.; Bal, A.; Bhalekar, P.; Sekar, P.; Krishnan, P. Epidemiology of diabetic foot infections in a reference tertiary hospital in India. Braz. J. Microbiol. 2018, 49, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Telles, J.P.; Cieslinski, J.; Tuon, F.F. Daptomycin to bone and joint infections and prosthesis joint infections: A systematic review. Braz. J. Infect. Dis. 2019, 23, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Hammerschlag, M.R. Treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in children: A reappraisal of vancomycin. Curr. Infect. Dis. Rep. 2019, 21, 1–8. [Google Scholar] [CrossRef]
- Sekhar, S.M.; Vyas, N.; Unnikrishnan, M.; Rodrigues, G.; Mukhopadhyay, C. Antimicrobial susceptibility pattern in diabetic foot ulcer: A pilot study. Ann. Med. Health Sci. Res. 2014, 4, 742–745. [Google Scholar] [CrossRef]
- Bansal, E.; Garg, A.; Bhatia, S.; Attri, A.; Chander, J. Spectrum of microbial flora in diabetic foot ulcers. Indian J. Pathol. Microbiol. 2008, 51, 204. [Google Scholar]
- Gadepalli, R.; Dhawan, B.; Sreenivas, V.; Kapil, A.; Ammini, A.; Chaudhry, R. A clinico-microbiological study of diabetic foot ulcers in an Indian tertiary care hospital. Diabetes Care 2006, 29, 1727–1732. [Google Scholar] [CrossRef] [PubMed]
- Perim, M.C.; da Costa Borges, J.; Celeste, S.R.C.; de Freitas Orsolin, E.; Mendes, R.R.; Mendes, G.O.; Ferreira, R.L.; Carreiro, S.C.; da Silva Pranchevicius, M.C. Aerobic bacterial profile and antibiotic resistance in patients with diabetic foot infections. Rev. Da Soc. Bras. De Med. Trop. 2015, 48, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Reveles, K.R.; Duhon, B.M.; Moore, R.J.; Hand, E.O.; Howell, C.K. Epidemiology of methicillin-resistant Staphylococcus aureus diabetic foot infections in a large academic hospital: Implications for antimicrobial stewardship. PLoS ONE 2016, 11, e0161658. [Google Scholar] [CrossRef]
- Kett, D. Improving Medicine through Pathway Assessment of Critical Therapy of Hospital-Acquired Pneumonia (IMPACTT-HAP) Investigators. Implementation of guidelines for management of possible multidrug-resistant pneumonia in intensive care: An observational, multicentre cohort study. Lancet Infect. Dis. 2011, 11, 181–189. [Google Scholar]
- Hassan, M.A.; Tamer, T.M.; Rageh, A.A.; Abou-Zeid, A.M.; Abd El-Zaher, E.H.; Kenawy, E.-R. Insight into multidrug-resistant microorganisms from microbial infected diabetic foot ulcers. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 1261–1270. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).