Spatial Evaluation of Machine Learning-Based Species Distribution Models for Prediction of Invasive Ant Species Distribution
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
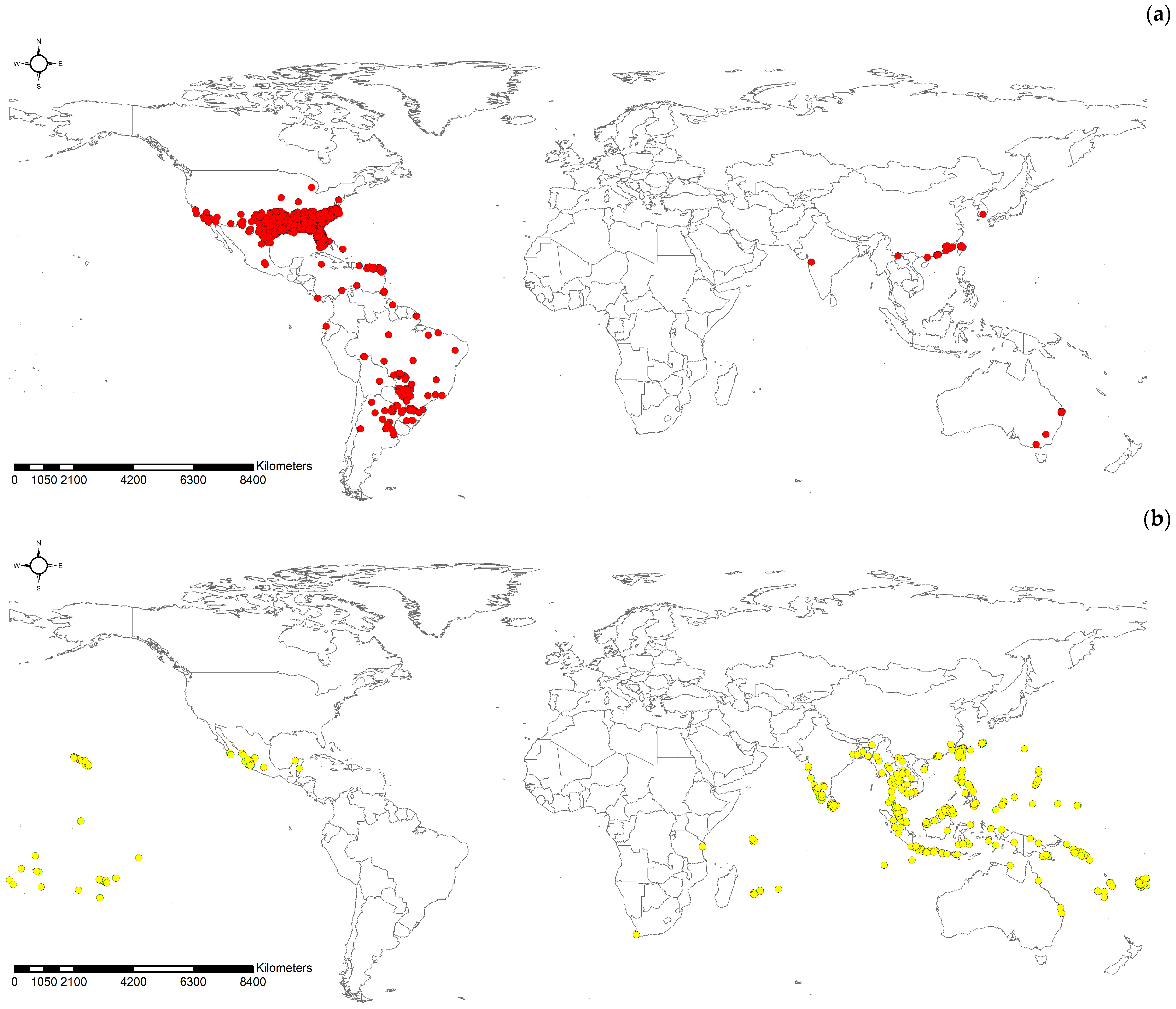
2.1. Data Acquisition and Spatial Processing
2.2. Climate Data
2.3. Bioclimatic Variable Selection
2.4. MaxEnt Operation
2.5. Random Forest Operation
2.6. Artificial Neural Network Construction
3. Results
3.1. Performance Comparison by Models
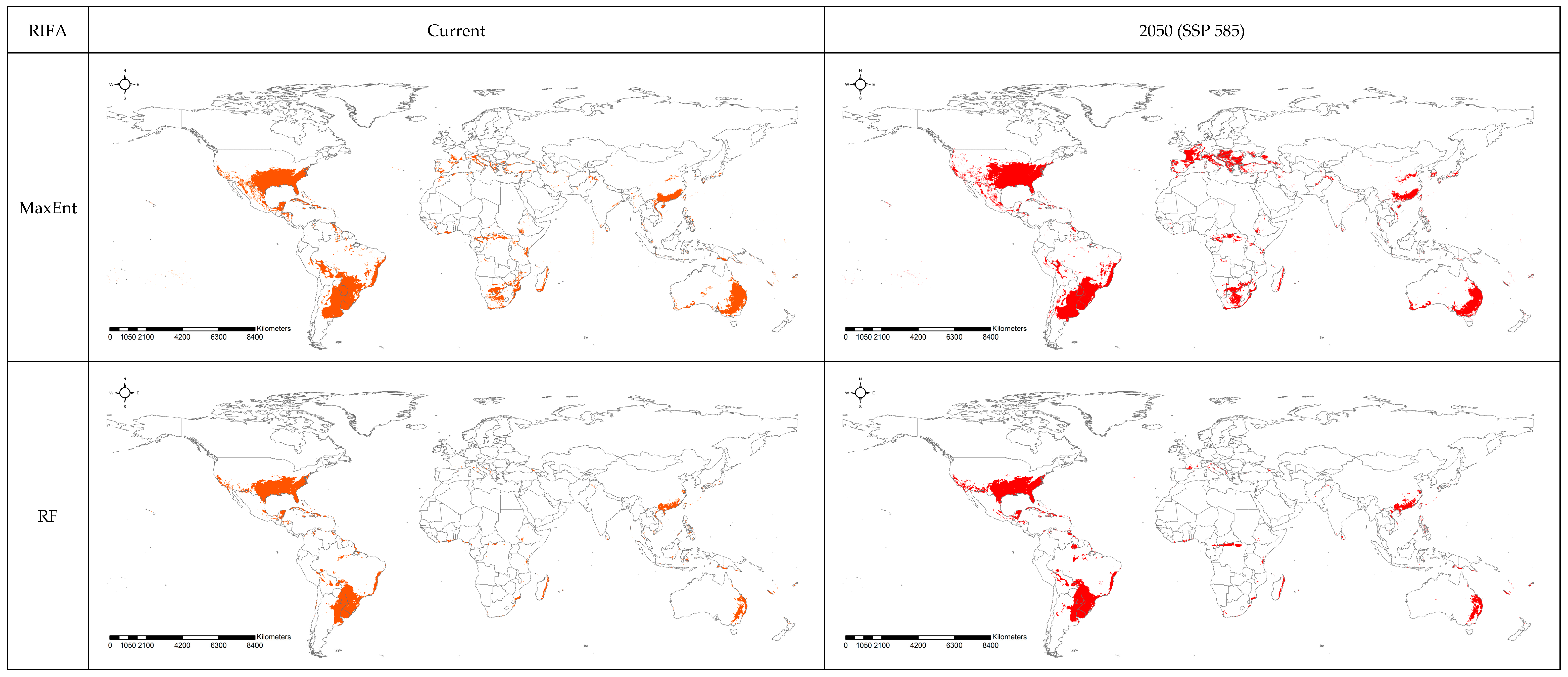
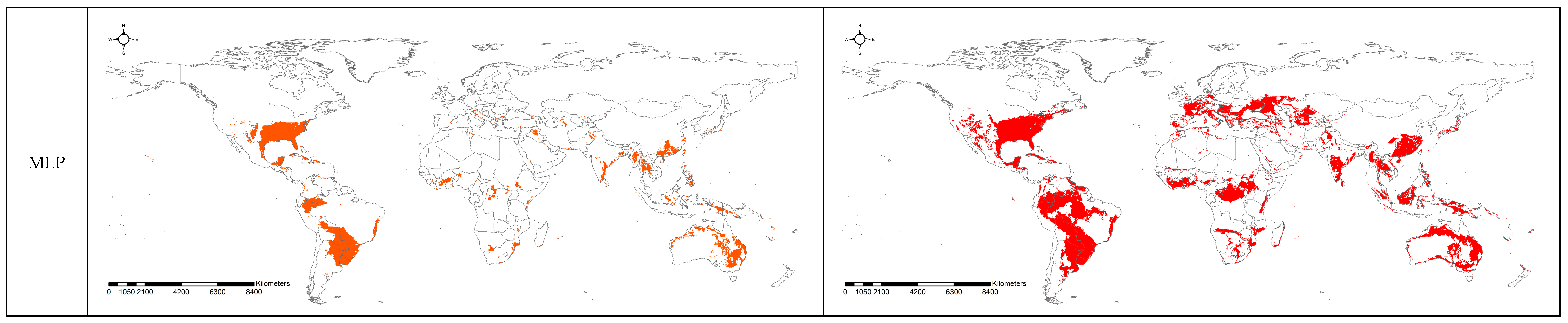
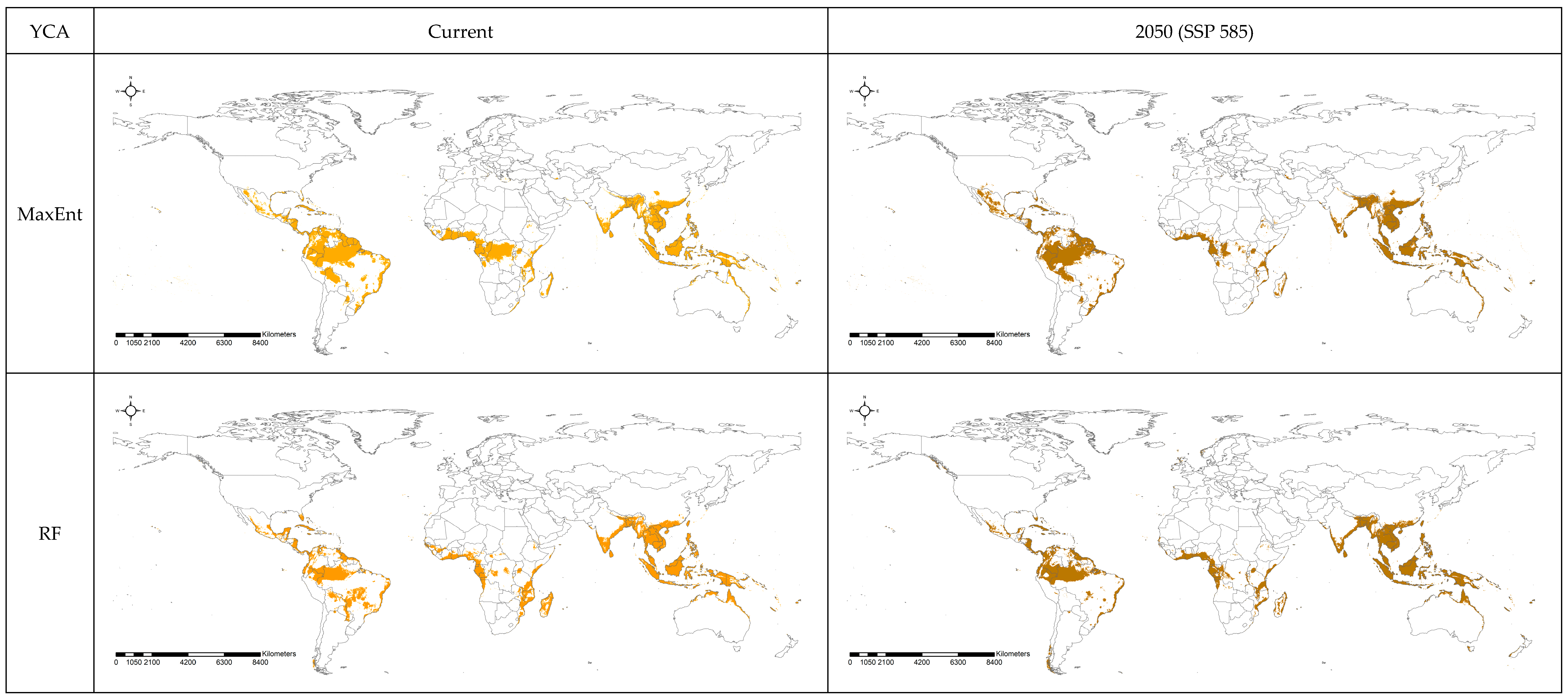
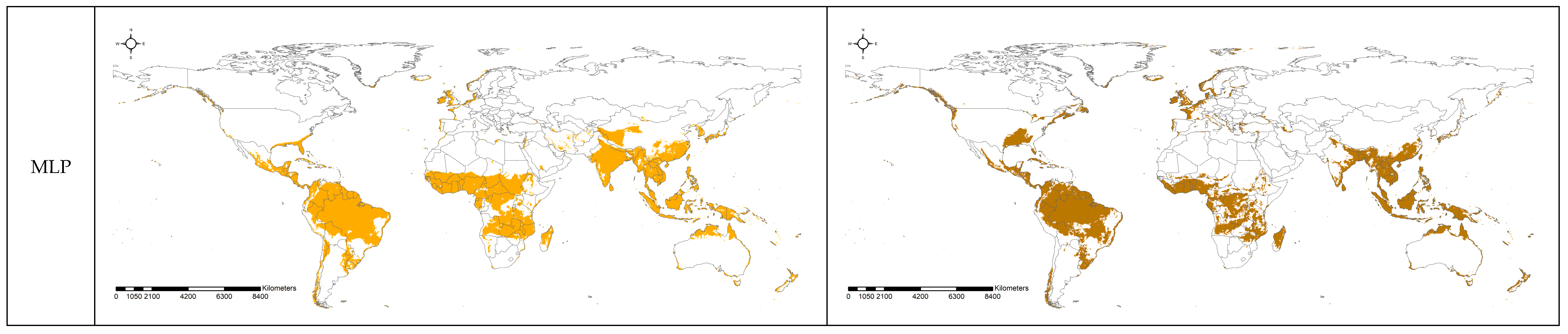
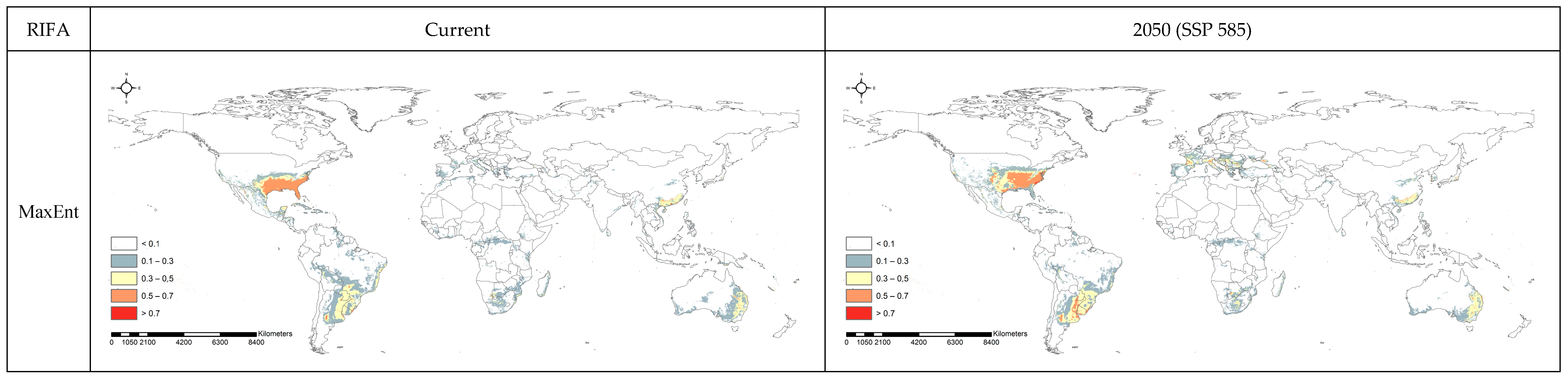
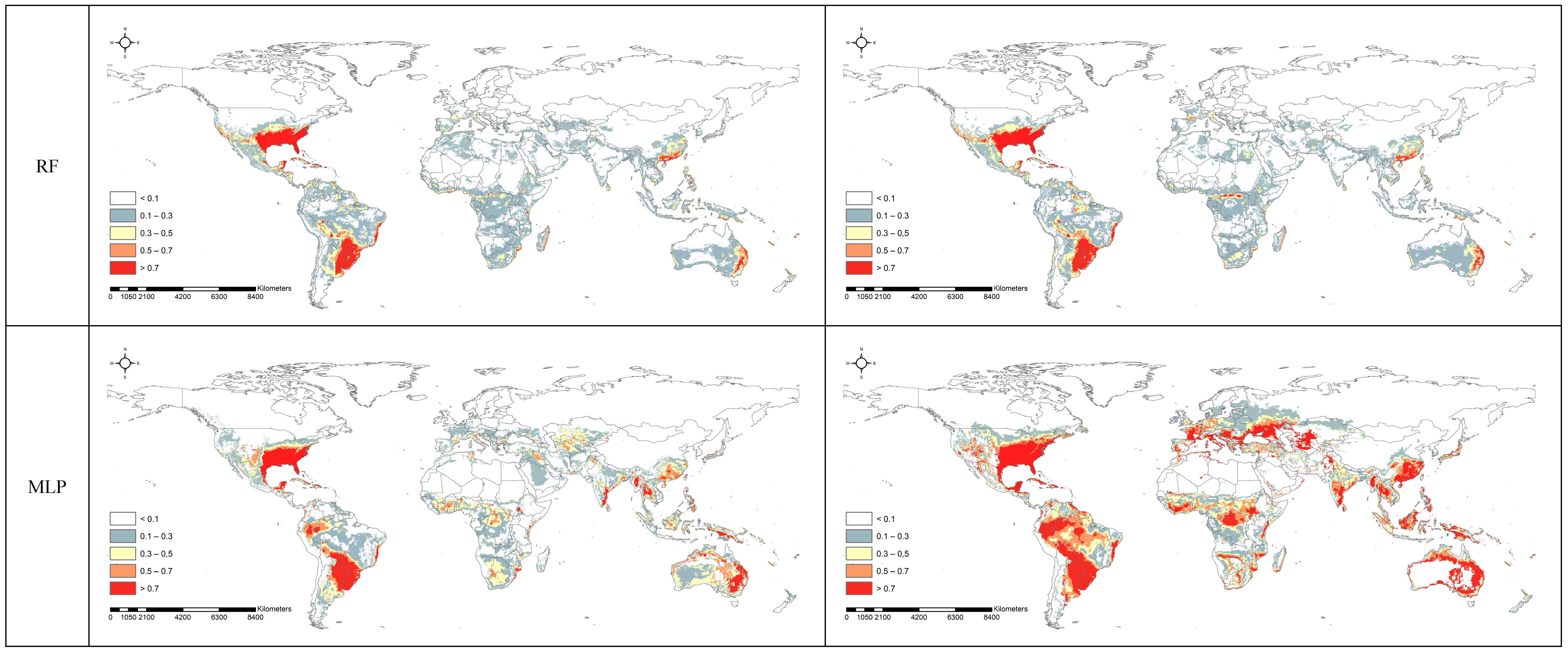
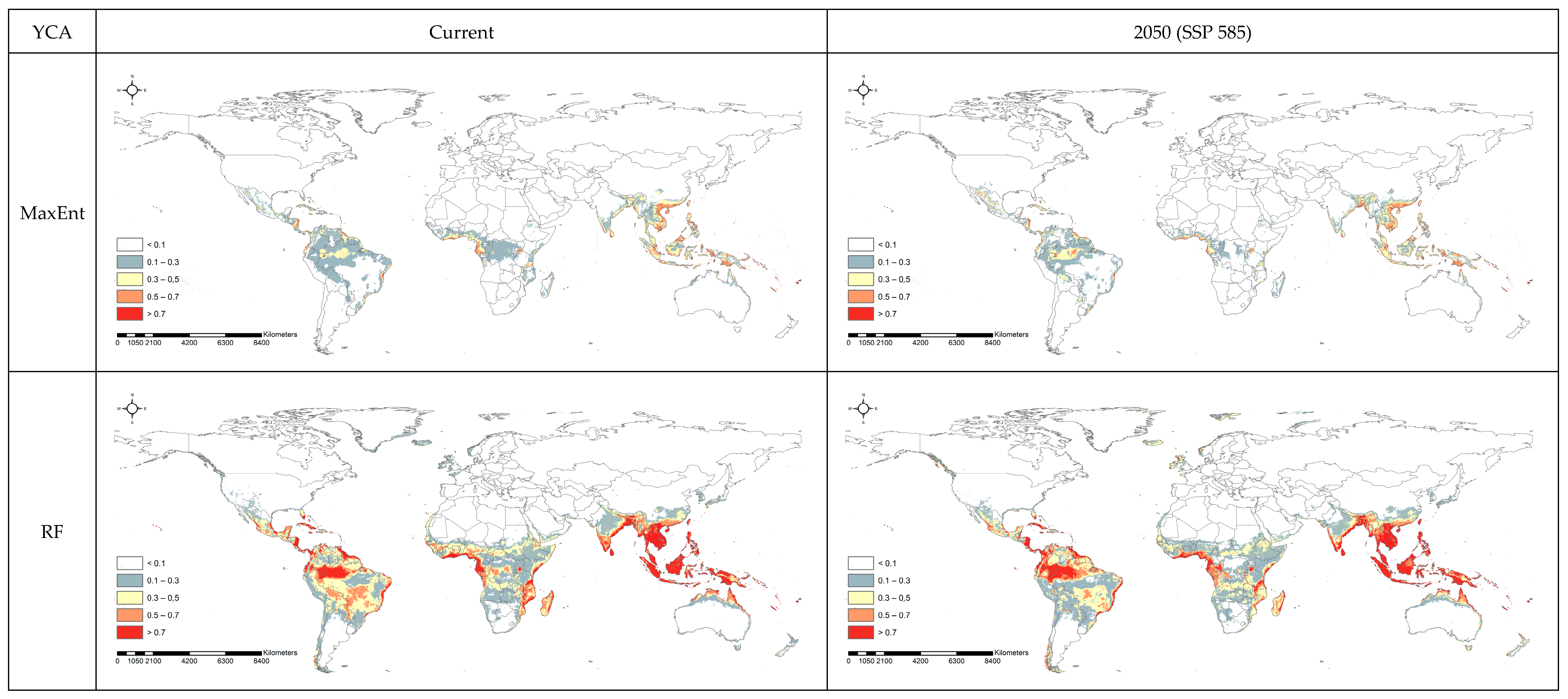
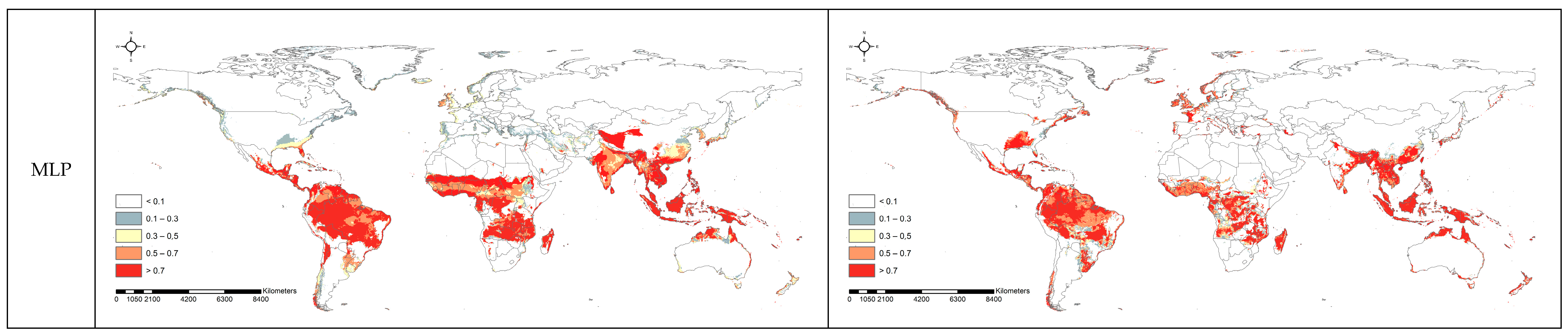
3.2. Spatial Projection Comparison by Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gobeyn, S.; Mouton, A.M.; Cord, A.F.; Kaim, A.; Volk, M.; Goethals, P.L. Evolutionary algorithms for species distribution modelling: A review in the context of machine learning. Ecol. Model. 2019, 392, 179–195. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Evans, J.S.; Murphy, M.A.; Holden, Z.A.; Cushman, S.A. Modeling Species Distribution and Change Using Random Forest; Springer: New York, NY, USA, 2011; pp. 139–159. [Google Scholar] [CrossRef]
- Muñoz-Mas, R.; Martínez-Capel, F.; Alcaraz-Hernández, J.D.; Mouton, A.M. On species distribution modelling, spatial scales and environmental flow assessment with Multi–Layer Perceptron Ensembles: A case study on the redfin barbel (Barbus haasi; Mertens, 1925). Limnologica 2017, 62, 161–172. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y. Applying various algorithms for species distribution modelling. Integr. Zool. 2013, 8, 124–135. [Google Scholar] [CrossRef]
- Araújo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Siddiqui, J.A.; Bamisile, B.S.; Khan, M.M.; Islam, W.; Hafeez, M.; Bodlah, I.; Xu, Y. Impact of invasive ant species on native fauna across similar habitats under global environmental changes. Environ. Sci. Pollut. Res. 2021, 28, 54362–54382. [Google Scholar] [CrossRef] [PubMed]
- Wetterer, J.K. Worldwide distribution and potential spread of the long-legged ant, Anoplolepis gracilipes (Hymenoptera: Formicidae). Sociobiology 2005, 45, 77–97. [Google Scholar]
- Ascunce, M.S.; Yang, C.C.; Oakey, J.; Calcaterra, L.; Wu, W.J.; Shih, C.J.; Goudet, J.; Ross, K.G.; Shoemaker, D. Global invasion history of the fire ant Solenopsis invicta. Science 2011, 331, 1066–1068. [Google Scholar] [CrossRef]
- Holway, D.A.; Lach, L.; Suarez, A.V.; Tsutsui, N.D.; Case, T.J. The causes and consequences of ant invasions. Annu. Rev. Ecol. Syst. 2002, 33, 181–233. [Google Scholar] [CrossRef]
- Morrison, L.W.; Porter, S.D.; Daniels, E.; Korzukhin, M.D. Potential global range expansion of the invasive fire ant, Solenopsis invicta. Biol. Invasions 2004, 6, 183–191. [Google Scholar] [CrossRef]
- Sutherst, R.W.; Maywald, G. A climate model of the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae): Implications for invasion of new regions, particularly Oceania. Environ. Entomol. 2005, 34, 317–335. [Google Scholar] [CrossRef]
- Chen, Y. Global potential distribution of an invasive species, the yellow crazy ant (Anoplolepis gracilipes) under climate change. Integr. Zool. 2008, 3, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.M.; Byeon, D.H.; Jung, S.H.; Yu, Y.M.; Yasunaga-Aoki, C.; Lee, W.H. Global Prediction of Geographical Change of Yellow Crazy Ant (Anoplolepis gracilipes) Distribution in Response to Climate Change Scenario. J. Fac. Agric. Kyushu Univ. 2017, 62, 403–410. [Google Scholar] [CrossRef]
- Sung, S.; Kwon, Y.S.; Lee, D.K.; Cho, Y. Predicting the potential distribution of an invasive species, Solenopsis invicta Buren (Hymenoptera: Formicidae), under climate change using species distribution models. Entomol. Res. 2018, 48, 505–513. [Google Scholar] [CrossRef]
- Byeon, D.H.; Lee, J.H.; Lee, H.S.; Park, Y.; Jung, S.; Lee, W.H. Prediction of spatiotemporal invasive risk by the red imported fire ant (Hymenoptera: Formicidae) in South Korea. Agronomy 2020, 10, 875. [Google Scholar] [CrossRef]
- Chen, S.; Ding, F.; Hao, M.; Jiang, D. Mapping the potential global distribution of red imported fire ant (Solenopsis invicta Buren) based on a machine learning method. Sustainability 2020, 12, 10182. [Google Scholar] [CrossRef]
- CABI (Centre for Agriculture and Bioscience International). Available online: www.cabi.org/isc/datasheet/5575 (accessed on 30 June 2020).
- GBIF (Global Biodiversity Information Facility). Available online: https://doi.org/10.15468/dl.6zb5ah (accessed on 30 June 2020).
- CABI (Centre for Agriculture and Bioscience International). Available online: www.cabi.org/isc/datasheet/50569 (accessed on 28 June 2021).
- GBIF (Global Biodiversity Information Facility). Available online: https://doi.org/10.15468/dl.8q7ydm (accessed on 28 June 2021).
- Brown, J.L.; Anderson, B. SDMtoolbox: A python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol. 2014, 5, 694–700. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- van Vuuren, D.P.; Carter, T.R. Climate and socio-economic scenarios for climate change research and assessment: Reconciling the new with the old. Clim. Chang. 2014, 122, 415–429. [Google Scholar] [CrossRef]
- Schandl, H.; Lu, Y.; Che, N.; Newth, D.; West, J.; Frank, S.; Obersteiner, M.; Rendall, A.; Hatfield-Dodds, S. Shared socio-economic pathways and their implications for global materials use. Resour. Conserv. Recycl. 2020, 160, 104866. [Google Scholar] [CrossRef]
- Yoon, S.; Lee, W.H. Methodological analysis of bioclimatic variable selection in species distribution modeling with application to agricultural pests (Metcalfa pruinosa and Spodoptera litura). Comput. Electron. Agric. 2021, 190, 106430. [Google Scholar] [CrossRef]
- Byeon, D.H.; Kim, S.H.; Jung, J.M.; Jung, S.; Kim, K.H.; Lee, W.H. Climate-based ensemble modelling to evaluate the global distribution of Anoplophora glabripennis (Motschulsky). Agric. For. Entomol. 2021, 23, 569–583. [Google Scholar] [CrossRef]
- Drees, B.; Summerlin, B.; Vinson, S.B. Foraging activity and temperature relationship for the red imported fire ant. Southwest. Entomol. 2007, 32, 149. [Google Scholar] [CrossRef]
- Bos, M.M.; Tylianakis, J.M.; Steffan-Dewenter, I.; Tscharntke, T. The invasive Yellow Crazy Ant and the decline of forest ant diversity in Indonesian cacao agroforests. Biol. Invasions. 2008, 10, 1399–1409. [Google Scholar] [CrossRef]
- Hoffmann, B.D. Integrating biology into invasive species management is a key principle for eradication success: The case of yellow crazy ant Anoplolepis gracilipes in northern Australia. Bull. Entomol. Res. 2015, 105, 141–151. [Google Scholar] [CrossRef]
- Muscarella, R.; Galante, P.J.; Soley-Guardia, M.; Boria, R.A.; Kass, J.M.; Uriarte, M.; Anderson, R.P. ENM eval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- Kramer-Schadt, S.; Niedballa, J.; Pilgrim, J.D.; Schröder, B.; Lindenborn, J.; Reinfelder, V.; Stillfried, M.; Heckmann, I.; Scharf, A.K.; Augeri, D.M.; et al. The importance of correcting for sampling bias in MaxEnt species distribution models. Divers. Distrib. 2013, 19, 1366–1379. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: https://www.R-project.org/ (accessed on 17 December 2021).
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Srivastava, V.; Griess, V.C.; Keena, M.A. Assessing the potential distribution of Asian gypsy moth in Canada: A comparison of two methodological approaches. Sci. Rep. 2020, 10, 22. [Google Scholar] [CrossRef]
- Liu, C.; Newell, G.; White, M. On the selection of thresholds for predicting species occurrence with presence-only data. Ecol. Evol. 2016, 6, 337–348. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News. 2002, 2, 18–22. [Google Scholar]
- Cutler, D.R.; Edwards Jr, T.C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Watts, M.J.; Worner, S.P. Using artificial neural networks to determine the relative contribution of abiotic factors influencing the establishment of insect pest species. Ecol. Inform. 2008, 3, 64–74. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009. [Google Scholar]
- Chollet, F. Keras. GitHub. 2015. Available online: https://github.com/fchollet/keras (accessed on 20 April 2020).
- Zhang, W.; Du, Y.; Yoshida, T.; Yang, Y. DeepRec: A deep neural network approach to recommendation with item embedding and weighted loss function. Inf. Sci. 2019, 470, 121–140. [Google Scholar] [CrossRef]
- Kumar, S.; Graham, J.; West, A.M.; Evangelista, P.H. Using district-level occurrences in MaxEnt for predicting the invasion potential of an exotic insect pest in India. Comput. Electrons Agric. 2014, 103, 55–62. [Google Scholar] [CrossRef]
- Wisz, M.S.; Hijmans, R.J.; Li, J.; Peterson, A.T.; Graham, C.H.; Guisan, A. NCEAS Predicting Species Distributions Working Group. Effects of sample size on the performance of species distribution models. Divers. Distrib. 2008, 14, 763–773. [Google Scholar] [CrossRef]
- Al-Anazi, A.F.; Gates, I.D. Support vector regression to predict porosity and permeability: Effect of sample size. Comput Geosci. 2012, 39, 64–76. [Google Scholar] [CrossRef]
- Luan, J.; Zhang, C.; Xu, B.; Xue, Y.; Ren, Y. The predictive performances of random forest models with limited sample size and different species traits. Fish. Res. 2020, 227, 105534. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Leroy, B.; Delsol, R.; Hugueny, B.; Meynard, C.N.; Barhoumi, C.; Barbet-Massin, M.; Bellard, C. Without quality presence–absence data, discrimination metrics such as TSS can be misleading measures of model performance. J. Biogeogr. 2018, 45, 1994–2002. [Google Scholar] [CrossRef]
- Khan, A.M.; Li, Q.; Saqib, Z.; Khan, N.; Habib, T.; Khalid, N.; Majeed, M.; Tariq, A. MaxEnt modelling and impact of climate change on habitat suitability variations of economically important Chilgoza Pine (Pinus gerardiana Wall.) in South Asia. Forests 2022, 13, 715. [Google Scholar] [CrossRef]
- Mi, C.; Huettmann, F.; Guo, Y.; Han, X.; Wen, L. Why choose Random Forest to predict rare species distribution with few samples in large undersampled areas? Three Asian crane species models provide supporting evidence. PeerJ 2017, 5, e2849. [Google Scholar] [CrossRef] [PubMed]
- Alwosheel, A.; van Cranenburgh, S.; Chorus, C.G. Is your dataset big enough? Sample size requirements when using artificial neural networks for discrete choice analysis. J. Choice Model. 2018, 28, 167–182. [Google Scholar] [CrossRef]
- Fox, E.W.; Hill, R.A.; Leibowitz, S.G.; Olsen, A.R.; Thornbrugh, D.J.; Weber, M.H. Assessing the accuracy and stability of variable selection methods for random forest modeling in ecology. Environ Monit Assess. 2017, 189, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Van Gelder, P.H.; Vrijling, J.K.; Ma, J. Forecasting daily streamflow using hybrid ANN models. J. Hydrol. 2006, 324, 383–399. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, D.E.; Lee, H.; Jung, S.; Lee, W.H. Ensemble evaluation of the potential risk areas of yellow-legged hornet distribution. Environ Monit Assess. 2021, 193, 1–15. [Google Scholar] [CrossRef]
- Velasco, J.A.; González-Salazar, C. Akaike information criterion should not be a “test” of geographical prediction accuracy in ecological niche modelling. Ecol. Inform. 2019, 51, 25–32. [Google Scholar] [CrossRef]
- Fletcher, R.J., Jr.; Hefley, T.J.; Robertson, E.P.; Zuckerberg, B.; McCleery, R.A.; Dorazio, R.M. A practical guide for combining data to model species distributions. Ecology 2019, 100, e02710. [Google Scholar] [CrossRef]
- Özesmi, S.L.; Tan, C.O.; Özesmi, U. Methodological issues in building, training, and testing artificial neural networks in ecological applications. Ecol. Model. 2006, 195, 83–93. [Google Scholar] [CrossRef]
- Xu, Y.J.; Zeng, L.; Lu, Y.Y.; Liang, G.W. Effect of soil humidity on the survival of Solenopsis invicta Buren workers. Insect. Soc. 2009, 56, 367–373. [Google Scholar] [CrossRef]
- Vinson, S.B. Insect life: Invasion of the red imported fire ant (Hymenoptera: Formicidae). Am. Entomol. 1997, 43, 23–39. [Google Scholar] [CrossRef]
- McGlynn, T.P. The worldwide transfer of ants: Geographical distribution and ecological invasions. J. Biogeogr. 1999, 26, 535–548. [Google Scholar] [CrossRef]
- Jung, J.M.; Jung, S.; Ahmed, M.R.; Cho, B.K.; Lee, W.H. Invasion risk of the yellow crazy ant (Anoplolepis gracilipes) under the Representative Concentration Pathways 8.5 climate change scenario in South Korea. J. Asia-Pac. Biodivers. 2017, 10, 548–554. [Google Scholar] [CrossRef]
- Jung, J.M.; Lee, H.S.; Lee, J.H.; Jung, S.; Lee, W.H. Development of a predictive model for soil temperature and its application to species distribution modeling of ant species in South Korea. Ecol. Inform. 2021, 61, 101220. [Google Scholar] [CrossRef]
| Species | Variables | Description |
|---|---|---|
| S. invicta | Bio1 | Annual Mean Temperature |
| Bio2 | Mean Diurnal Range | |
| Bio3 | Isothermality | |
| Bio5 | Max Temperature of Warmest Month | |
| Bio8 | Mean Temperature of Wettest Quarter | |
| Bio9 | Mean Temperature of Driest Quarter | |
| Bio12 | Annual Precipitation | |
| Bio14 | Precipitation Seasonality | |
| Bio15 | Precipitation of Warmest Quarter | |
| Elevation | ||
| A. gracilipes | Bio1 | Annual Mean Temperature |
| Bio2 | Mean Diurnal Range | |
| Bio3 | Isothermality | |
| Bio5 | Max Temperature of Warmest Month | |
| Bio7 | Temperature Annual Range | |
| Bio15 | Precipitation Seasonality | |
| Bio16 | Precipitation of Wettest Quarter | |
| Bio18 | Precipitation of Warmest Quarter | |
| Bio19 | Precipitation of Coldest Quarter | |
| Elevation |
| Species | S. invicta | A. gracilipes | ||||
|---|---|---|---|---|---|---|
| Model | MaxEnt | RF | MLP | MaxEnt | RF | MLP |
| AUC | 0.949 | 0.939 | 0.911 | 0.967 | 0.940 | 0.894 |
| pAUC | 1.960 | 1.970 | 1.930 | 1.930 | 1.930 | 1.800 |
| TSS | 0.923 | 0.879 | 0.815 | 0.906 | 0.882 | 0.783 |
| Accuracy | 0.951 | 0.941 | 0.907 | 0.930 | 0.940 | 0.890 |
| Sensitivity | 0.973 | 0.941 | 0.906 | 0.977 | 0.910 | 0.944 |
| Specificity | 0.949 | 0.938 | 0.909 | 0.929 | 0.972 | 0.839 |
| Species | Model | MaxEnt | RF | MLP | |||
|---|---|---|---|---|---|---|---|
| Climate | Current | 2050 | Current | 2050 | Current | 2050 | |
| S. invicta | % presence | 5.19% | 5.63% | 4.04% | 4.31% | 6.41% | 16.54% |
| Mean prob. | 0.307 | 0.319 | 0.771 | 0.756 | 0.763 | 0.766 | |
| A. gracilipes | % presence | 7.28% | 6.17% | 8.35% | 7.80% | 21.76% | 17.79% |
| Mean prob. | 0.317 | 0.349 | 0.719 | 0.739 | 0.748 | 0.784 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, W.-H.; Song, J.-W.; Yoon, S.-H.; Jung, J.-M. Spatial Evaluation of Machine Learning-Based Species Distribution Models for Prediction of Invasive Ant Species Distribution. Appl. Sci. 2022, 12, 10260. https://doi.org/10.3390/app122010260
Lee W-H, Song J-W, Yoon S-H, Jung J-M. Spatial Evaluation of Machine Learning-Based Species Distribution Models for Prediction of Invasive Ant Species Distribution. Applied Sciences. 2022; 12(20):10260. https://doi.org/10.3390/app122010260
Chicago/Turabian StyleLee, Wang-Hee, Jae-Woo Song, Sun-Hee Yoon, and Jae-Min Jung. 2022. "Spatial Evaluation of Machine Learning-Based Species Distribution Models for Prediction of Invasive Ant Species Distribution" Applied Sciences 12, no. 20: 10260. https://doi.org/10.3390/app122010260
APA StyleLee, W.-H., Song, J.-W., Yoon, S.-H., & Jung, J.-M. (2022). Spatial Evaluation of Machine Learning-Based Species Distribution Models for Prediction of Invasive Ant Species Distribution. Applied Sciences, 12(20), 10260. https://doi.org/10.3390/app122010260






