Characterization of Lacticaseibacillus rhamnosus, Levilactobacillus brevis and Lactiplantibacillus plantarum Metabolites and Evaluation of Their Antimicrobial Activity against Food Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures
2.2. Pathogenic Microorganisms
2.3. Preparation of Standards
2.4. Extraction Procedures
2.5. Liquid Liquid Extraction (LLE)
2.6. Instrumentation and Analytical Conditions
2.6.1. Chromatographic Conditions (HPLC-UV/DAD)
2.6.2. Precision of Standard Calibration Curves
2.7. Antimicrobial Activity Assay
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Morita, H.; Toh, H.; Oshima, K.; Murakami, M.; Taylor, T.D.; Igimi, S.; Hattori, M. Complete genome sequence of the probiotic Lactobacillus rhamnosus ATCC 53103. J. Bacteriol. 2009, 191, 7630–7631. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, D.; Sun, Z.; Wu, R.; Chen, X.; Chen, W.; Meng, H.; Hu, S.; Zhang, H. Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in inner Mongolia, China. J. Bacteriol. 2010, 192, 5268–5269. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Guiné, R.P.F.; Florença, S.G.; Barroca, M.J.; Anjos, O. The Link between the Consumer and the Innovations in Food Product Development. Foods 2020, 9, 1317. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, X.; Shan, C.; Xia, X.; Wang, Y.; Dong, M.; Zhou, J. Novel Bacteriocin Produced by Lactobacillus Alimentarius FM-MM4 from a Traditional Chinese Fermented Meat Nanx Wudl: Purification, Identification and Antimicrobial Characteristics. Food Control 2017, 77, 290–297. [Google Scholar] [CrossRef]
- Wakil, S.M.; Laba, S.A.; Fasiku, S.A. Isolation and Identification of Antimicrobial-Producing Lactic Acid Bacteria from Fermented Cucumber. Afr. J. Biotechnol. 2014, 13, 2556–2564. [Google Scholar] [CrossRef]
- Ryan, L.A.M.; Zannini, E.; Dal Bello, F.; Pawlowska, A.; Koehler, P.; Arendt, E.K. Lactobacillus Amylovorus DSM 19280 as a Novel Food-Grade Antifungal Agent for Bakery Products. Int. J. Food Microbiol. 2011, 146, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Gajbhiye, M.H.; Kapadnis, B.P. Antifungal-activity-producing lactic acid bacteria as biocontrol agents in plants. Biocontrol. Sci. Technol. 2016, 26, 1451–1470. [Google Scholar] [CrossRef]
- Oyetayo, V.O. Phenotypic Characterisation and Assessment of the Inhibitory Potential of Lactobacillus Isolates from Different Sources. Afr. J. Biotechnol. 2004, 3, 355–357. [Google Scholar] [CrossRef][Green Version]
- Crowley, S.; Mahony, J.; Van Sinderen, D. Current Perspectives on Antifungal Lactic Acid Bacteria as Natural Bio-Preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Leyva Salas, M.; Mounier, J.; Valence, F.; Coton, M.; Thierry, A.; Coton, E. Antifungal Microbial Agents for Food Biopreservation—A Review. Microorganisms 2017, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Balti, R.; Neves, A.R. Bioprotective Mechanisms of Lactic Acid Bacteria against Fungal Spoilage of Food. Curr. Opin. Biotechnol. 2019, 56, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Karami, S.; Roayaei, M.; Hamzavi, H.; Bahmani, M.; Hassanzad-Azar, H.; Leila, M.; Rafieian-Kopaei, M. Isolation and Identification of Probiotic Lactobacillus from Local Dairy and Evaluating Their Antagonistic Effect on Pathogens. Int. J. Pharm. Investig. 2017, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the Gut Microbiota in Nutrition and Health. BMJ 2018, 361, 36–44. [Google Scholar] [CrossRef]
- Bruni, N.; Capucchio, M.T.; Biasibetti, E.; Pessione, E.; Cirrincione, S.; Giraudo, L.; Corona, A.; Dosio, F. Antimicrobial Activity of Lactoferrin-Related Peptides and Applications in Human and Veterinary Medicine. Molecules 2016, 21, 752. [Google Scholar] [CrossRef]
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial Peptides: Premises and Promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef]
- Oliveira, P.M.; Zannini, E.; Arendt, E.K. Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: From crop farming to cereal products. Food Microbiol. 2014, 37, 78–95. [Google Scholar] [CrossRef]
- Agriopoulou, S.; Stamatelopoulou, E.; Sachadyn-Król, M.; Varzakas, T. Lactic Acid Bacteria as Antibacterial Agents to Extend the Shelf Life of Fresh and Minimally Processed Fruits and Vegetables: Quality and Safety Aspects. Microorganisms 2020, 8, 952. [Google Scholar] [CrossRef]
- Food and Agriculture Organization/World Health Organization. Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria: Report of a Joint FAO/WHO Expert Consultation 2006. 25 November 2008. Available online: http://pc.ilele.hk/public/pdf/20190225/bd3689dfc2fd663bb36def1b672ce0a4.pdf (accessed on 28 October 2021).
- Hawaz, E. Isolation and Identification of Probiotic Lactic Acid Bacteria from Curd and in Vitro Evaluation of Its Growth Inhibition Activities against Pathogenic Bacteria. Afr. J. Microbiol. Res. 2014, 8, 1419–1425. [Google Scholar] [CrossRef]
- Jabbari, V.; Khiabani, M.S.; Mokarram, R.R.; Hassanzadeh, A.M.; Ahmadi, E.; Gharenaghadeh, S.; Karimi, N.; Kafil, H.S. Lactobacillus Plantarum as a Probiotic Potential from Kouzeh Cheese (Traditional Iranian Cheese) and Its Antimicrobial Activity. Probiotics Antimicrob. Proteins 2017, 9, 189–193. [Google Scholar] [CrossRef]
- Brosnan, B.; Coffey, A.; Arendt, E.K.; Furey, A.A. Comprehensive Investigation into Sample Extraction and Method Validation for the Identification of Antifungal Compounds Produced by Lactic Acid Bacteria Using HPLC-UV/DAD. Anal. Methods 2014, 6, 5331–5344. [Google Scholar] [CrossRef]
- Brosnan, B.; Coffey, A.; Arendt, E.K.; Furey, A. Rapid Identification, by Use of the LTQ Orbitrap Hybrid FT Mass Spectrometer, of Antifungal Compounds Produced by Lactic Acid Bacteria. Anal. Bioanal. Chem. 2012, 403, 2983–2995. [Google Scholar] [CrossRef]
- Ström, K.; Sjögren, J.; Broberg, A.; Schnürer, J. Lactobacillus Plantarum MiLAB 393 Produces the Antifungal Cyclic Dipeptides Cyclo(L-Phe-L-Pro) and Cyclo(L-Phe-Trans-4-OH-L-Pro) and 3-Phenyllactic Acid. Appl. Environ. Microbiol. 2002, 68, 4322–4327. [Google Scholar] [CrossRef]
- Valerio, F.; Lavermicocca, P.; Pascale, M.; Visconti, A. Production of Phenyllactic Acid by Lactic Acid Bacteria: An Approach to the Selection of Strains Contributing to Food Quality and Preservation. FEMS Microbiol. Lett. 2004, 233, 289–295. [Google Scholar] [CrossRef]
- Ming, L.C.; Halim, M.; Rahim, R.A.; Wan, H.Y.; Ari, A. Bin Strategies in fed-batch cultivation on the production performance of Lactobacillus salivarius I 24 viable cells. Food Sci. Biotechnol. 2016, 25, 1393–1398. [Google Scholar] [CrossRef]
- Axel, C.; Brosnan, B.; Zannini, E.; Peyer, L.C.; Furey, A.; Coffey, A.; Arendt, E.K. Antifungal Activities of Three Different Lactobacillus Species and Their Production of Antifungal Carboxylic Acids in Wheat Sourdough. Appl. Microbiol. Biotechnol. 2015, 100, 1701–1711. [Google Scholar] [CrossRef]
- Klaenhammer, T.R.; Kullen, M.J. Selection and design of probiotics. Int. J. Food Microbiol. 1999, 50, 45–57. [Google Scholar] [CrossRef]
- Barcenilla, C.; Ducic, M.; López, M.; Prieto, M.; Álvarez-Ordóñez, A. Application of lactic acid bacteria for the biopreservation of meat products: A systematic review. Meat Sci. 2022, 183, 108661. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.J.; Yang, S.Y.; Yoon, K.S. Lactic acid bacteria starter in combination with sodium chloride controls pathogenic Escherichia coli (EPEC, ETEC, and EHEC) in kimchi. Food Microbiol. 2021, 100, 103868. [Google Scholar] [CrossRef] [PubMed]
- Messens, W.; De Vuyst, L. Inhibitory Substances Produced by Lactobacilli Isolated from Sourdoughs-A Review. Int. J. Food Microbiol. 2002, 72, 31–43. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Valerio, F.; Evidente, A.; Lazzaroni, S.; Corsetti, A.; Gobbetti, M. Purification and Characterization of Novel Antifungal Compounds from the Sourdough Lactobacillus Plantarum Strain 21B. Appl. Environ. Microbiol. 2010, 66, 4084–4090. [Google Scholar] [CrossRef] [PubMed]
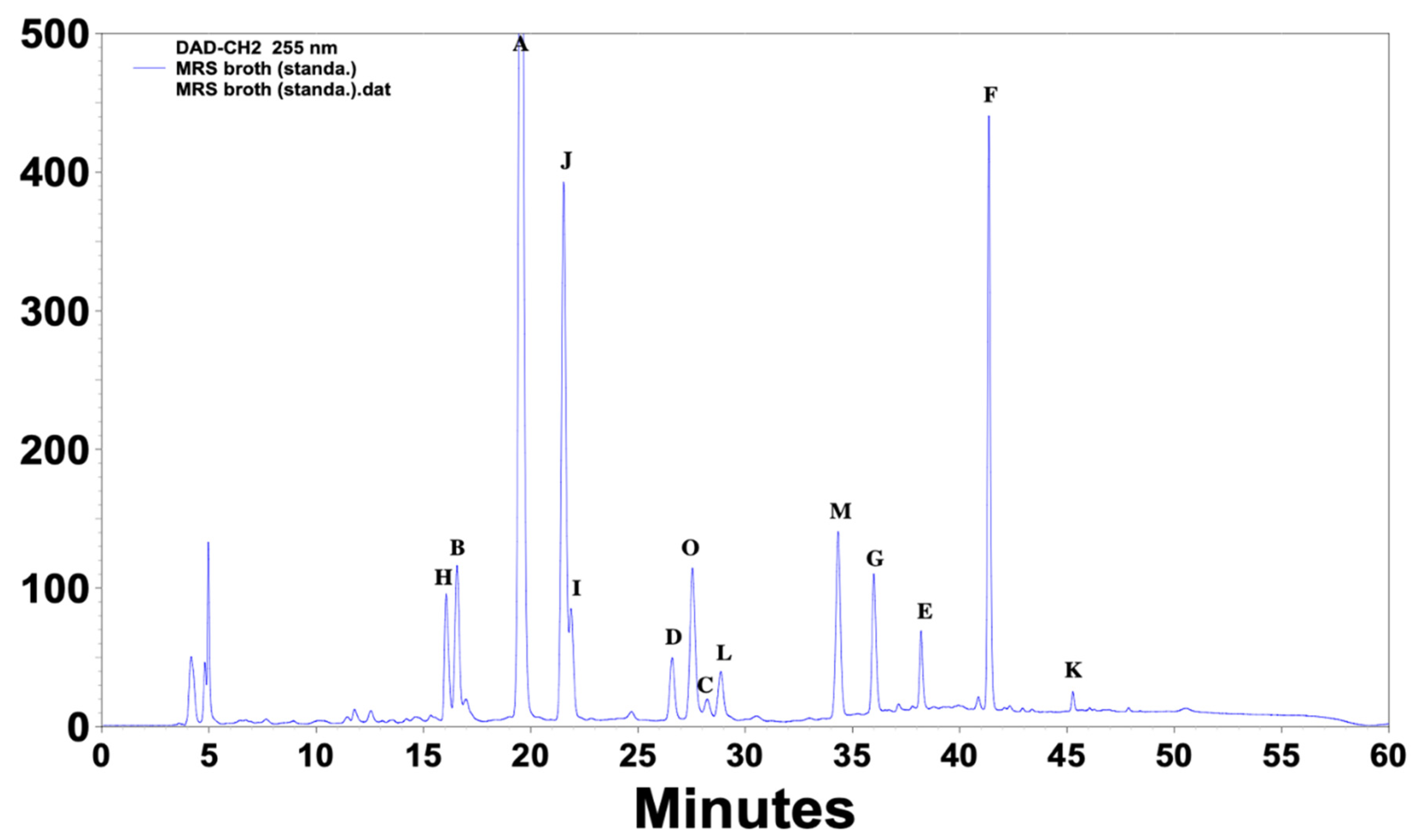
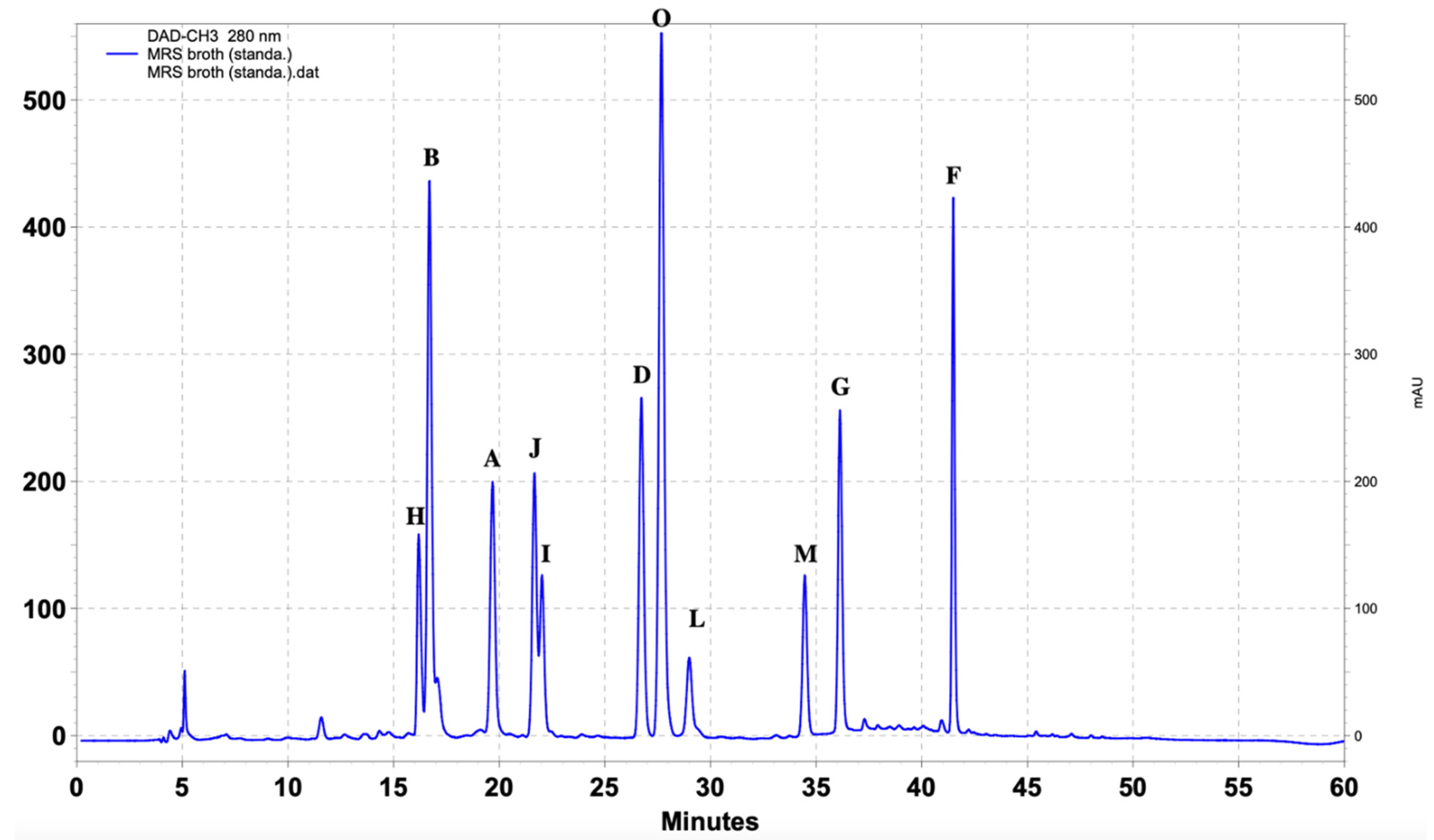
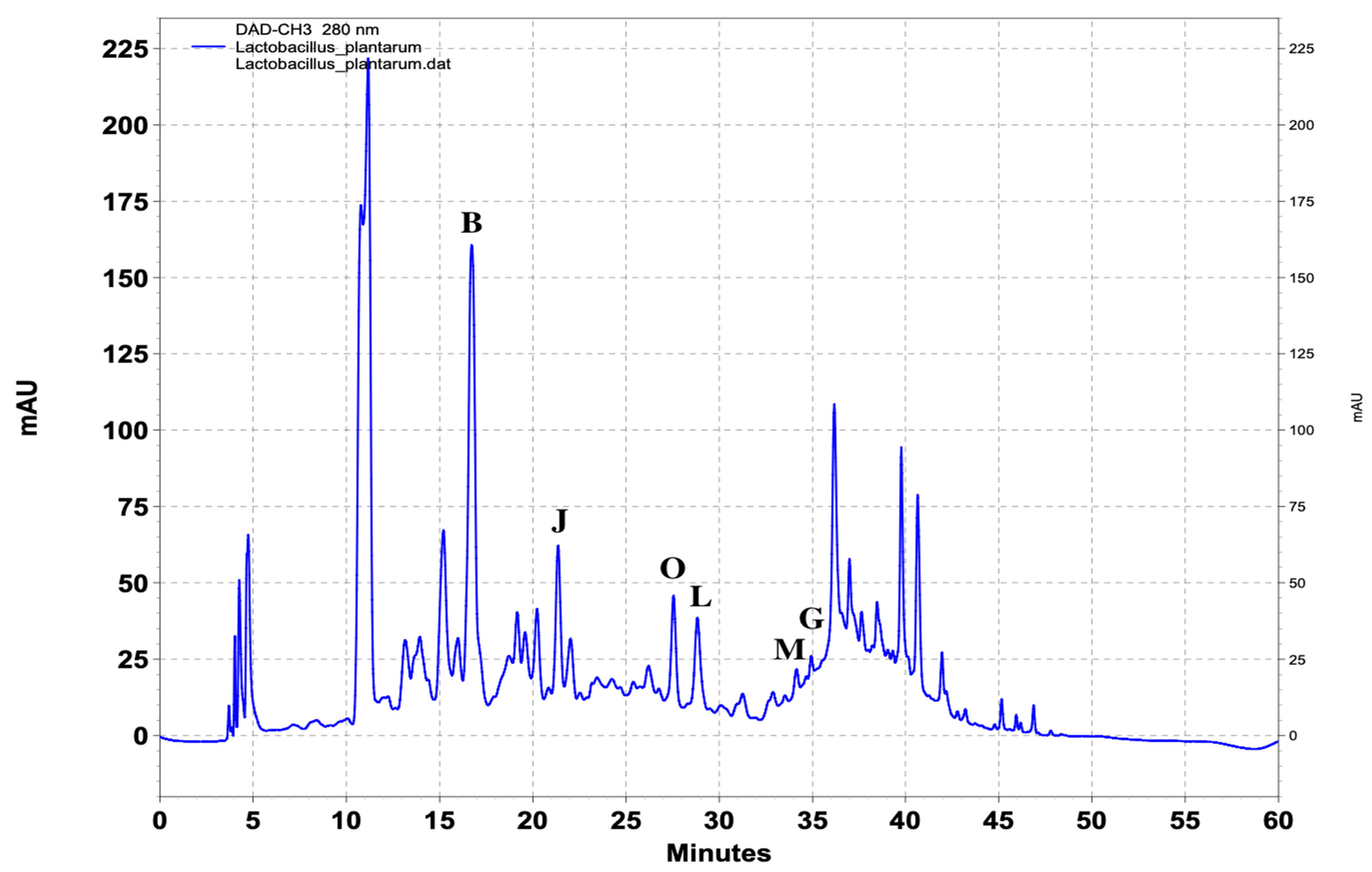


| Standard Compounds | UV (nm) | Retention Time (min) |
|---|---|---|
| (A) 4-Hydroxybenzoic acid | 251, 269, 344 | 19.3 |
| (Β) DL-p-Hydroxyphenyllactic acid | 228, 274, 395 | 16.3 |
| (C) Phenyllactic acid | 216, 255, 394 | 28 |
| (D) 3-(4-Hydroxyphenyl) propionic acid | 235, 277, 342 | 26.4 |
| (E) Hydrocinnamic acid | 220, 255, 315 | 38 |
| (F) Methylcinnamic acid | 251, 270, 293 | 41.3 |
| (G) Salicylic acid | 235, 302, 386 | 36 |
| (H) 1,2-Dihydroxybenzene | 229, 271, 390 | 15.7 |
| (I) 3,4-Dihydrocinnamic acid | 222, 324, 314 | 21.7 |
| (J) Vanillic acid | 253, 268, 294 | 21.3 |
| (K) 3,4-Dihydroxyhydrocinnamic acid | 222, 255, 323 | 45.3 |
| (L) Ferulic acid | 225, 234, 322 | 28.7 |
| (M) Benzoic acid | 230, 273, 388 | 34.2 |
| (O) 4-Hydrocinnamic acid | 227, 239, 310 | 27.4 |
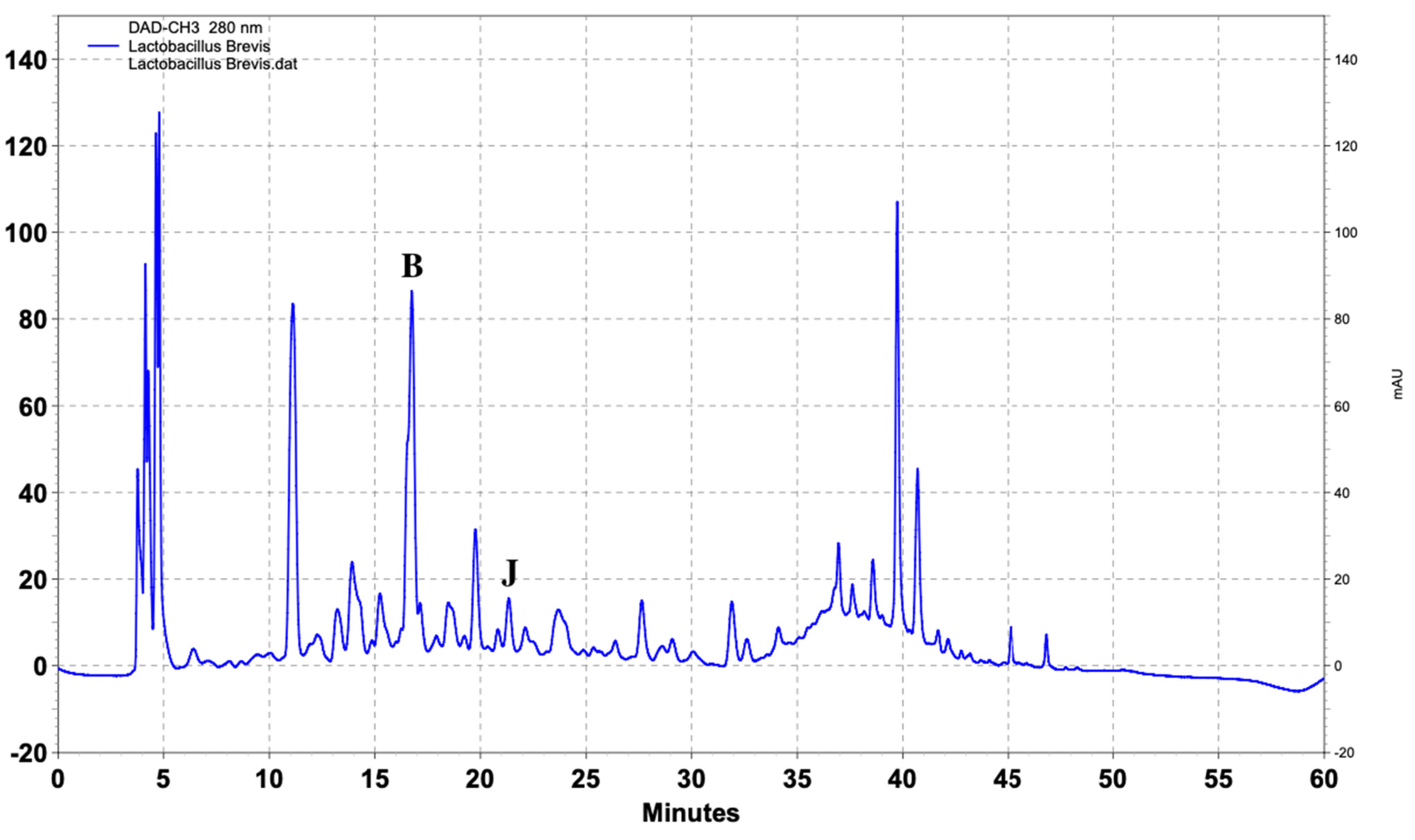
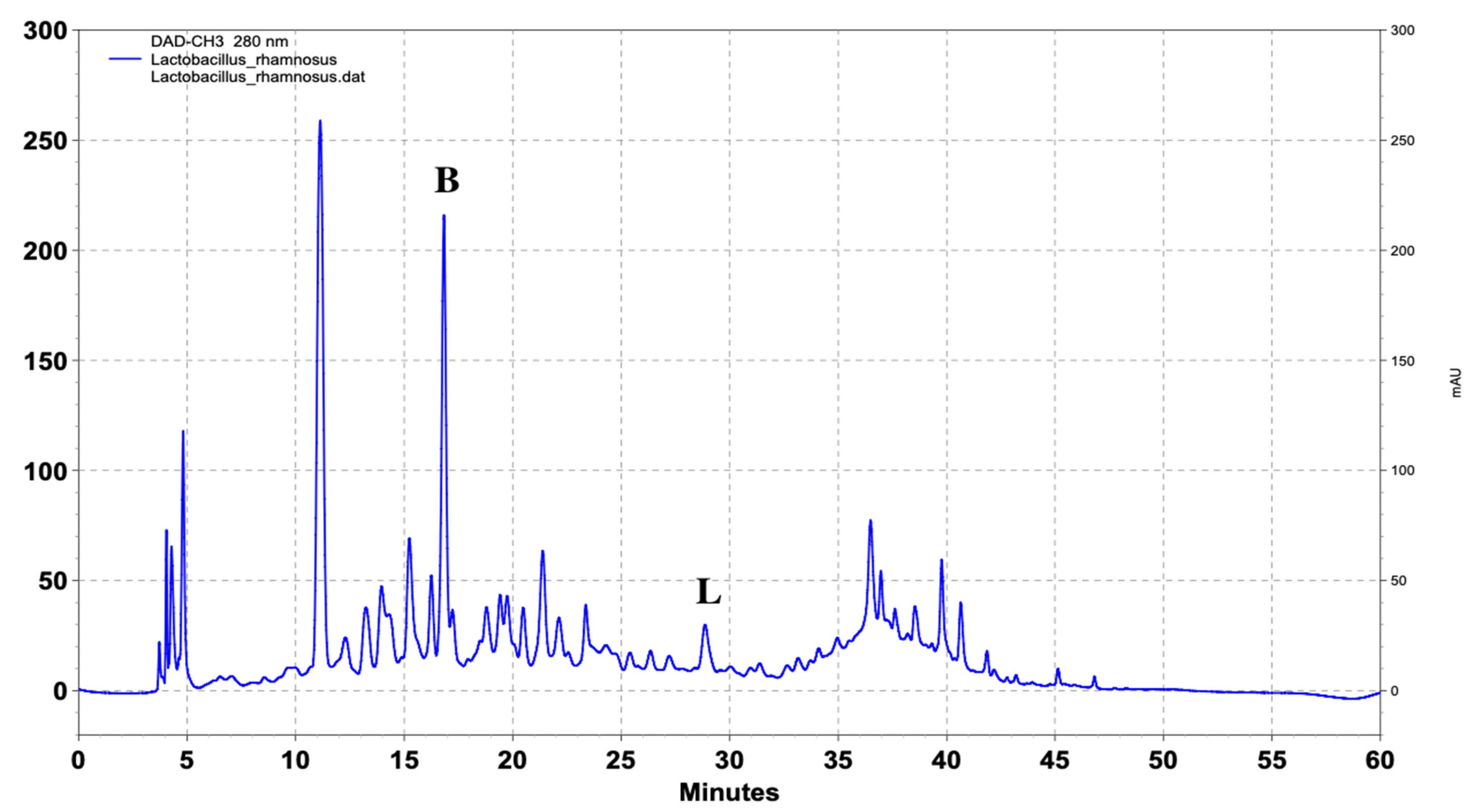
| Name of Standard | Levilactobacillusbrevis (ppm) | Lactiplantibacillusplantarum (ppm) | Lacticaseibacillusrhamnosus (ppm) |
|---|---|---|---|
| (A) 4-Hydroxybenzoic acid | - | - | - |
| (Β) DL-p-Hydroxyphenyllactic acid | 103.4 | 217.8 | 147.6 |
| (C) Phenyllactic acid | - | - | - |
| (D) 3-(4-Hydroxyphenyl) propionic acid | - | - | - |
| (E) Hydrocinnamic acid | - | - | - |
| (F) Methylcinnamic acid | - | - | - |
| (G) Salicylic acid | - | 1.8 | - |
| (H) 1,2-Dihydroxybenzene | - | - | - |
| (I) 3,4-Dihydrocinnamic acid | - | - | - |
| (J) Vanillic acid | 2.6 | 3.4 | - |
| (K) 3,4-Dihydroxyhydrocinnamic acid | - | - | - |
| (L) Ferulic acid | - | 6.4 | 4.9 |
| (M) Benzoic acid | - | 4.6 | - |
| (O) 4-Hydrocinnamic acid | - | 1.4 | - |
| Name Strain | Cfu/mL | Incubation Time (Days) | DL-p-Hydroxy-Phenyllactic Acid | Salicylic Acid | Vanillic Acid | Ferulic Acid | Benzoic Acid | 4-Hydro-Cinnamic Acid |
|---|---|---|---|---|---|---|---|---|
| Levilactobacillus brevis (ppm) | 108 | 5 | 80.2 | - | 1.5 | - | - | - |
| 105 | 10 | 98.3 | - | 2.4 | - | - | - | |
| 105 | 15 | 100.2 | - | 2.4 | - | - | - | |
| 104 | 20 | 102.3 | - | 2.5 | - | - | - | |
| 104 | 25 | 103.4 | - | 2.6 | - | - | - | |
| 102 | 30 | 103.4 | - | 2.6 | - | - | - | |
| Lactiplantibacillus plantarum (ppm) | 108 | 5 | 150.3 | 1.0 | 2.2 | 4.8 | 3.2 | 1.0 |
| 105 | 10 | 200.10 | 1.6 | 3.2 | 6.05 | 3.8 | 1.25 | |
| 105 | 15 | 214.3 | 1.6 | 3.3 | 6.35 | 3.9 | 1.3 | |
| 103 | 20 | 216.8 | 1.72 | 3.35 | 6.40 | 4.4 | 1.35 | |
| 102 | 25 | 217.5 | 1.8 | 3.4 | 6.40 | 4.55 | 1.40 | |
| 101 | 30 | 217.8 | 1.8 | 3.4 | 6.4 | 4.6 | 1.4 | |
| Lacticaseibacillus rhamnosus (ppm) | 108 | 5 | 123.2 | - | - | 3.85 | - | - |
| 105 | 10 | 138.0 | - | - | 4.45 | - | - | |
| 105 | 15 | 142.3 | - | - | 4.55 | - | - | |
| 104 | 20 | 145.4 | - | - | 4.85 | - | - | |
| 104 | 25 | 147.6 | - | - | 4.90 | - | - | |
| 102 | 30 | 147.6 | - | - | 4.90 | - | - |
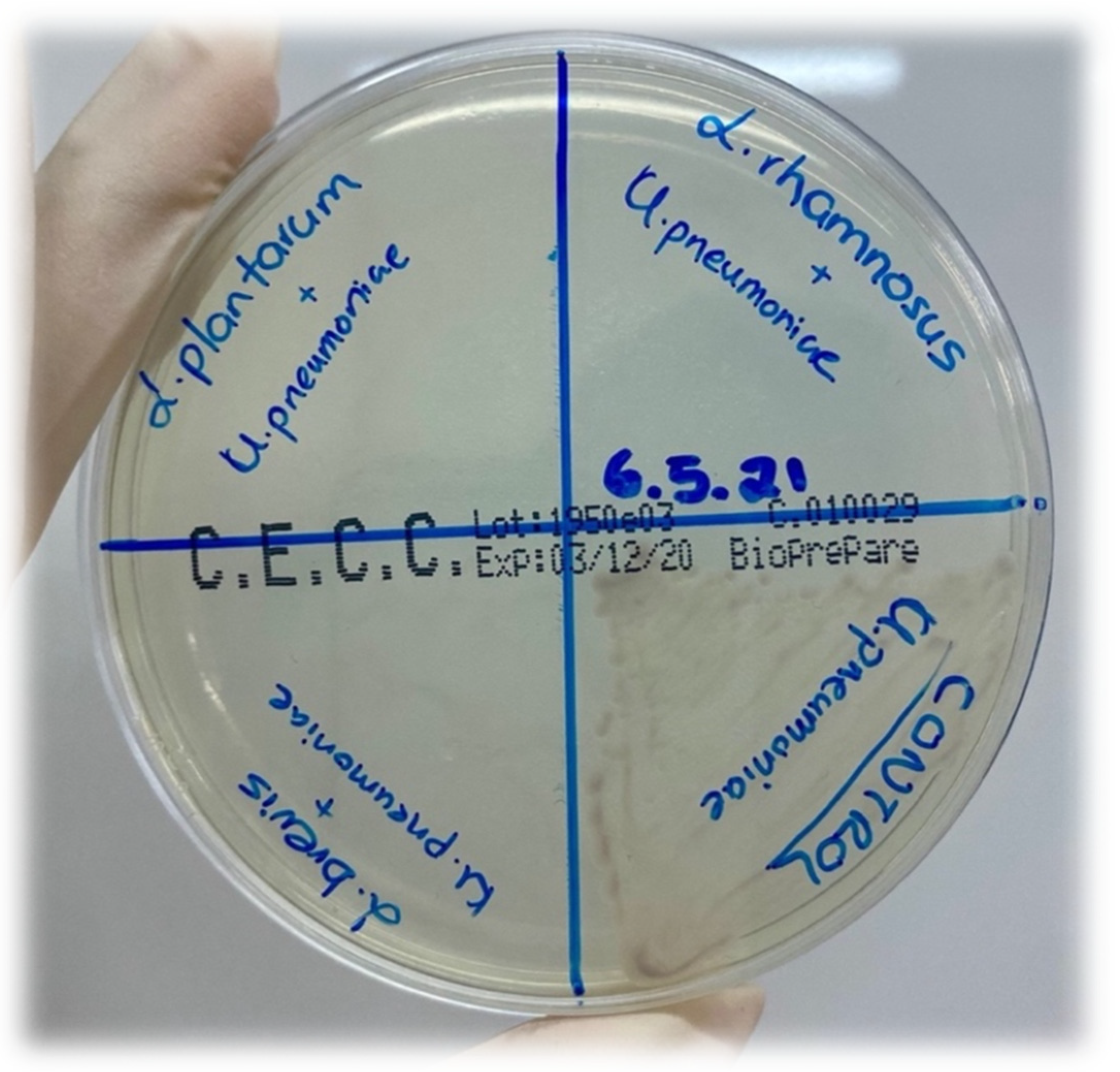
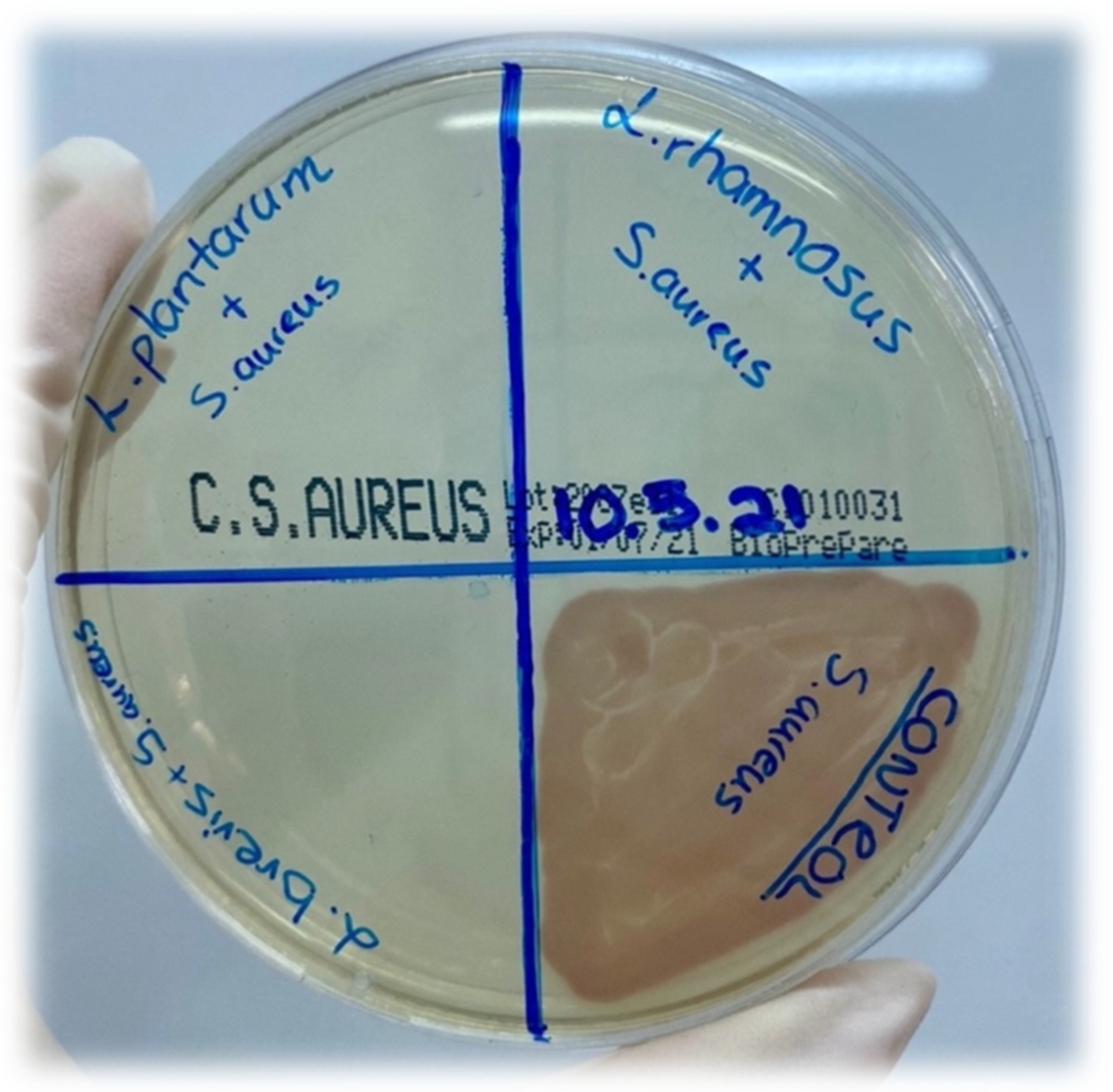
| Name of Pathogen Strain | Metabolites of L. brevis | Metabolites of L. plantarum | Metabolites of L. rhamnosus | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| Salmonella enterica ATCC14028 (cfu/mL) | 108 | 103 | 0 | 108 | 102 | 0 | 108 | 103 | 0 |
| Staphylococcus aureus ATCC29213 (cfu/mL) | 108 | 103 | 0 | 108 | 102 | 0 | 108 | 103 | 0 |
| Escherichia coli ATCC25922 (cfu/mL) | 108 | 103 | 0 | 108 | 102 | 0 | 108 | 103 | 0 |
| Klebsiella pneumoniae ATCC700603 (cfu/mL) | 108 | 103 | 0 | 108 | 102 | 0 | 108 | 103 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vougiouklaki, D.; Tsironi, T.; Papaparaskevas, J.; Halvatsiotis, P.; Houhoula, D. Characterization of Lacticaseibacillus rhamnosus, Levilactobacillus brevis and Lactiplantibacillus plantarum Metabolites and Evaluation of Their Antimicrobial Activity against Food Pathogens. Appl. Sci. 2022, 12, 660. https://doi.org/10.3390/app12020660
Vougiouklaki D, Tsironi T, Papaparaskevas J, Halvatsiotis P, Houhoula D. Characterization of Lacticaseibacillus rhamnosus, Levilactobacillus brevis and Lactiplantibacillus plantarum Metabolites and Evaluation of Their Antimicrobial Activity against Food Pathogens. Applied Sciences. 2022; 12(2):660. https://doi.org/10.3390/app12020660
Chicago/Turabian StyleVougiouklaki, Despina, Theofania Tsironi, Joseph Papaparaskevas, Panagiotis Halvatsiotis, and Dimitra Houhoula. 2022. "Characterization of Lacticaseibacillus rhamnosus, Levilactobacillus brevis and Lactiplantibacillus plantarum Metabolites and Evaluation of Their Antimicrobial Activity against Food Pathogens" Applied Sciences 12, no. 2: 660. https://doi.org/10.3390/app12020660
APA StyleVougiouklaki, D., Tsironi, T., Papaparaskevas, J., Halvatsiotis, P., & Houhoula, D. (2022). Characterization of Lacticaseibacillus rhamnosus, Levilactobacillus brevis and Lactiplantibacillus plantarum Metabolites and Evaluation of Their Antimicrobial Activity against Food Pathogens. Applied Sciences, 12(2), 660. https://doi.org/10.3390/app12020660







