Celector®: An Innovative Technology for Quality Control of Living Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Cell Culture
2.2. Instrumentation
2.2.1. Fractionation Principle and Procedure
2.2.2. Optical Analysis
2.3. Cell Analysis and Collection
Immunofluorescence Staining and Flow Cytometry Analysis
2.4. Celector® Test Procedure
2.5. Statistical Analysis
3. Results and Discussion
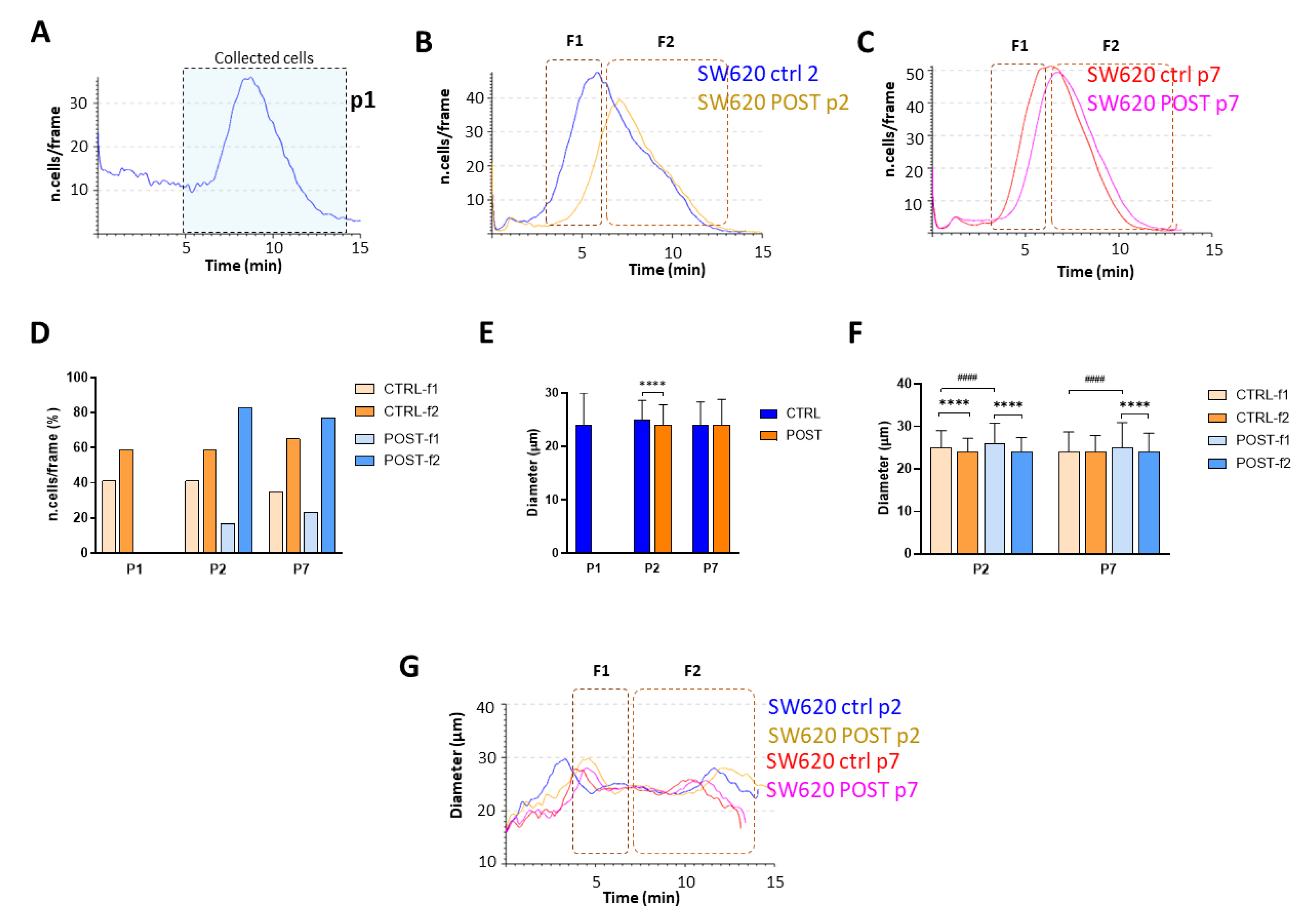
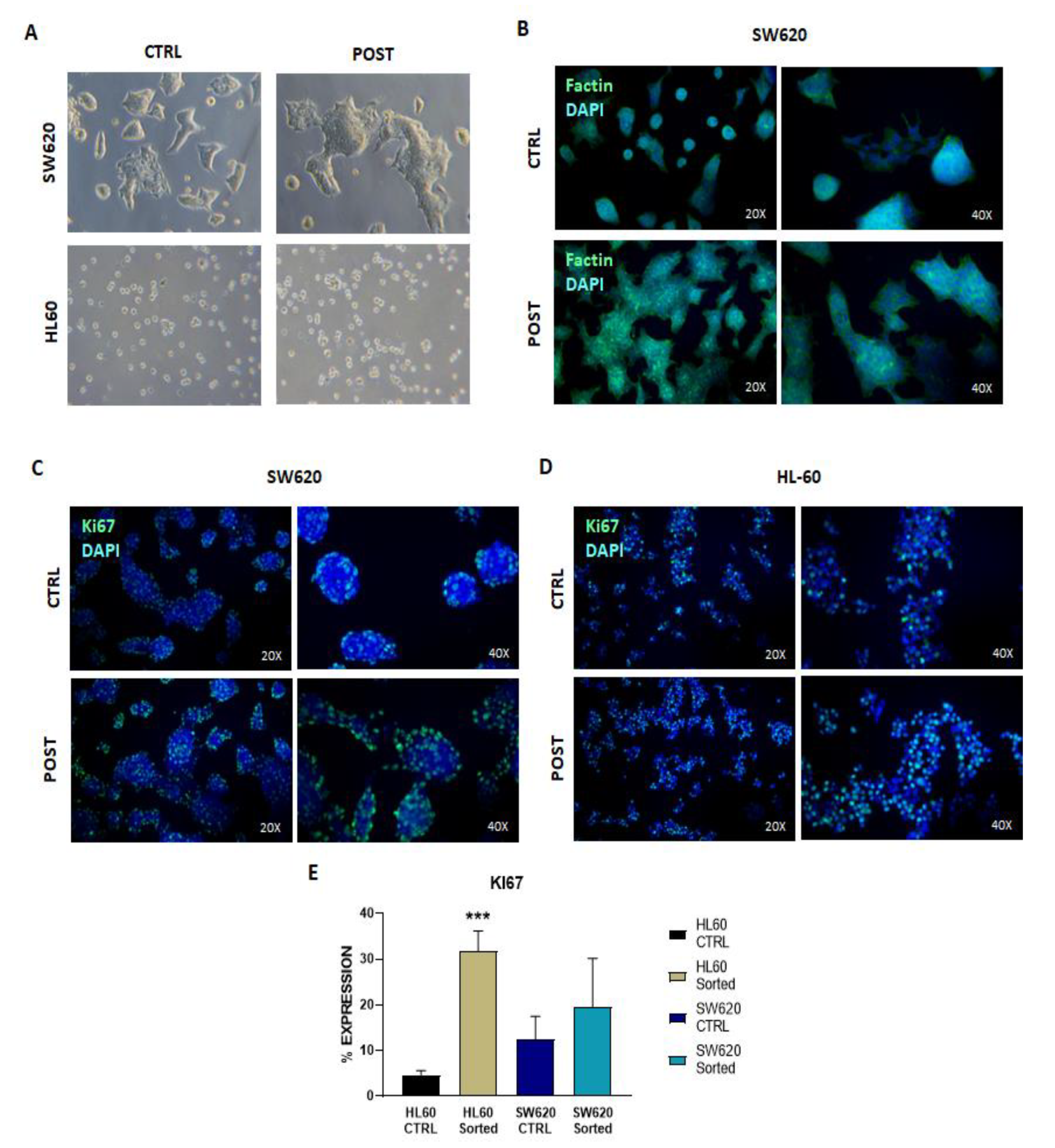
3.1. Characterization of Expanded Cell Line
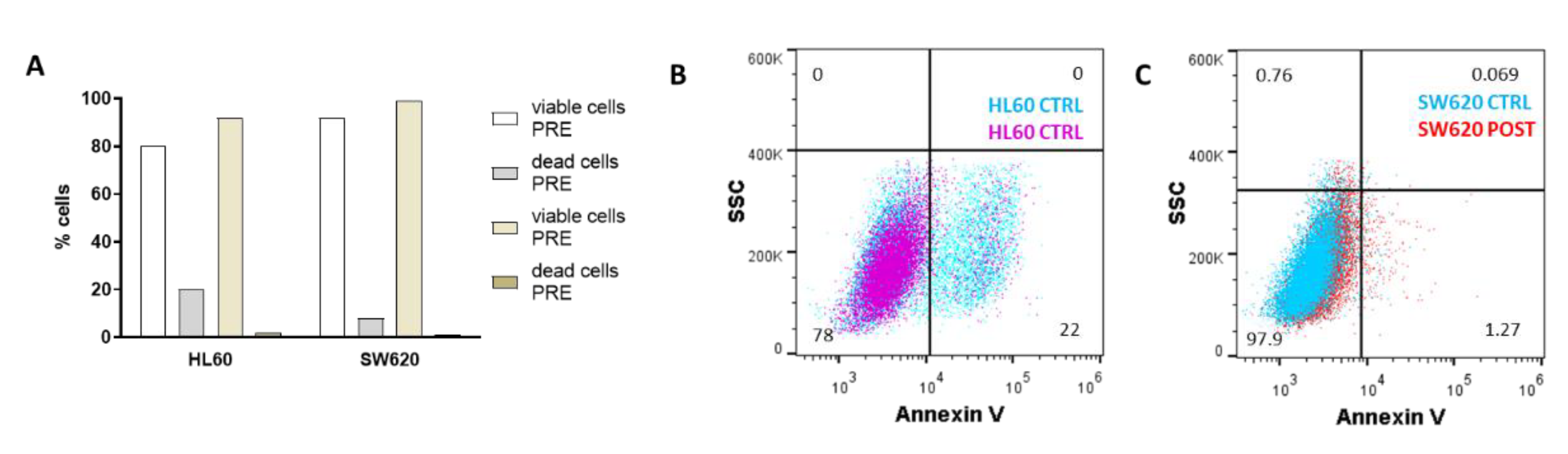
3.2. Cell Analysis Post-Selection
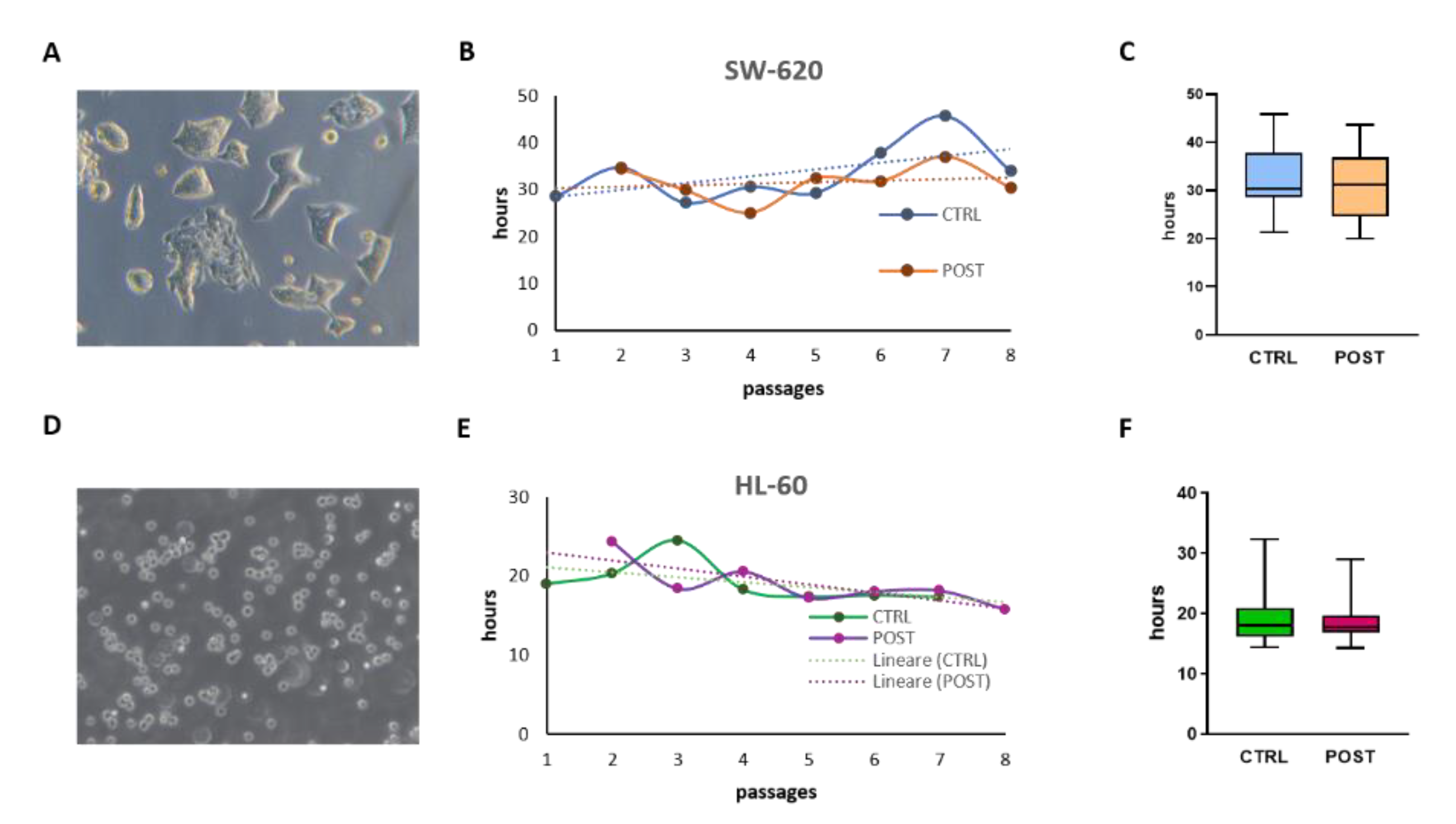
3.3. In Vitro Expansion of Tumor Cell Line
3.4. Analysis of Cell Cryo-Preservation
4. Conclusions
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cimino, M.; Gonçalves, R.M.; Barrias, C.C.; Martins, M.C.L. Xeno-Free Strategies for Safe Human Mesenchymal Stem/Stromal Cell Expansion: Supplements and Coatings. Stem Cells Int. 2017, 2017, 6597815. [Google Scholar] [CrossRef] [PubMed]
- de Peppo, G.M. GMP-Compatible, Xeno-Free Culture of Human Induced Mesenchymal Stem Cells. Methods Mol. Biol. 2021, 2286, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Aghayan, H.R.; Payab, M.; Mohamadi-Jahani, F.; Aghayan, S.S.; Larijani, B.; Arjmand, B. GMP-Compliant Production of Human Placenta-Derived Mesenchymal Stem Cells. Methods Mol. Biol. 2021, 2286, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, U.; Spindler, R.; Wolkers, W.F.; Glasmacher, B.; Müller, T. Cryopreservation and quality control of mouse embryonic feeder cells. Cryobiology 2011, 63, 104–110. [Google Scholar] [CrossRef]
- Yuan, Z.; Lourenco Sda, S.; Sage, E.K.; Kolluri, K.K.; Lowdell, M.W.; Janes, S.M. Cryopreservation of human mesenchymal stromal cells expressing TRAIL for human anti-cancer therapy. Cytotherapy 2016, 18, 860–869. [Google Scholar] [CrossRef]
- Ornelas-González, A.; González-González, M.; Rito-Palomares, M. Microcarrier-based stem cell bioprocessing: GMP-grade culture challenges and future trends for regenerative medicine. Crit. Rev. Biotechnol. 2021, 41, 1081–1095. [Google Scholar] [CrossRef]
- Wartmann, D.; Rothbauer, M.; Kuten, O.; Barresi, C.; Visus, C.; Felzmann, T.; Ertl, P. Automated, Miniaturized, and Integrated Quality Control-on-Chip (QC-on-a-Chip) for Cell-Based Cancer Therapy Applications. Front. Mater. 2015, 2, 60. [Google Scholar] [CrossRef]
- Reschiglian, P.; Zattoni, A.; Roda, B.; Casolari, S.; Moon, M.H.; Lee, J.; Jung, J.; Rodmalm, K.; Cenacchi, G. Bacteria sorting by field-flow fractionation. Application to whole-cell Escherichia coil vaccine strains. Anal. Chem. 2002, 74, 4895–4904. [Google Scholar] [CrossRef]
- Marassi, V.; Maggio, S.; Battistelli, M.; Stocchi, V.; Zattoni, A.; Reschiglian, P.; Guescini, M.; Roda, B. An ultracentrifugation—hollow-fiber flow field-flow fractionation orthogonal approach for the purification and mapping of extracellular vesicle subtypes. J. Chromatogr. A 2021, 1638, 461861. [Google Scholar] [CrossRef]
- Marrazzo, P.; Pizzuti, V.; Zia, S.; Sargenti, A.; Gazzola, D.; Roda, B.; Bonsi, L.; Alviano, F. Microfluidic Tools for Enhanced Characterization of Therapeutic Stem Cells and Prediction of Their Potential Antimicrobial Secretome. Antibiotics 2021, 10, 750. [Google Scholar] [CrossRef]
- Marassi, V.; Roda, B.; Casolari, S.; Ortelli, S.; Blosi, M.; Zattoni, A.; Costa, A.L.; Reschiglian, P. Hollow-fiber flow field-flow fractionation and multi-angle light scattering as a new analytical solution for quality control in pharmaceutical nanotechnology. Microchem. J. 2018, 136, 149–156. [Google Scholar] [CrossRef]
- Roda, B.; Marassi, V.; Zattoni, A.; Borghi, F.; Anand, R.; Agostoni, V.; Gref, R.; Reschiglian, P.; Monti, S. Flow field-flow fractionation and multi-angle light scattering as a powerful tool for the characterization and stability evaluation of drug-loaded metal-organic framework nanoparticles. Anal. Bioanal. Chem. 2018, 410, 5245–5253. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-B.; Yang, J.; Huang, Y.; Vykoukal, J.; Becker, F.F.; Gascoyne, P.R.C. Cell Separation by Dielectrophoretic Field-flow-fractionation. Anal. Chem. 2000, 72, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Hervieu, C.; Verdier, M.; Barthout, E.; Begaud, G.; Christou, N.; Sage, M.; Pannequin, J.; Battu, S.; Mathonnet, M. A Label-Free Cell Sorting Approach to Highlight the Impact of Intratumoral Cellular Heterogeneity and Cancer Stem Cells on Response to Therapies. Cells 2022, 11, 2264. [Google Scholar] [CrossRef] [PubMed]
- Faye, P.A.; Vedrenne, N.; De la Cruz-Morcillo, M.A.; Barrot, C.C.; Richard, L.; Bourthoumieu, S.; Sturtz, F.; Funalot, B.; Lia, A.S.; Battu, S. New Method for Sorting Endothelial and Neural Progenitors from Human Induced Pluripotent Stem Cells by Sedimentation Field Flow Fractionation. Anal. Chem. 2016, 88, 6696–6702. [Google Scholar] [CrossRef]
- Cailleteau, C.; Micallef, L.; Lepage, C.; Cardot, P.J.; Beneytout, J.L.; Liagre, B.; Battu, S. Investigating the relationship between cell cycle stage and diosgenin-induced megakaryocytic differentiation of HEL cells using sedimentation field-flow fractionation. Anal. Bioanal. Chem. 2010, 398, 1273–1283. [Google Scholar] [CrossRef]
- Melucci, D.; Roda, B.; Zattoni, A.; Casolari, S.; Reschiglian, P.; Roda, A. Field-flow fractionation of cells with chemiluminescence detection. J. Chromatogr. A 2004, 1056, 229–236. [Google Scholar] [CrossRef]
- Lattuada, D.; Roda, B.; Pignatari, C.; Magni, R.; Colombo, F.; Cattaneo, A.; Zattoni, A.; Cetin, I.; Reschiglian, P.; Bolis, G. A tag-less method for direct isolation of human umbilical vein endothelial cells by gravitational field-flow fractionation. Anal. Bioanal. Chem. 2013, 405, 977–984. [Google Scholar] [CrossRef]
- Roda, B.; Lanzoni, G.; Alviano, F.; Zattoni, A.; Costa, R.; Di Carlo, A.; Marchionni, C.; Franchina, M.; Ricci, F.; Tazzari, P.L.; et al. A novel stem cell tag-less sorting method. Stem Cell Rev. Rep. 2009, 5, 420–427. [Google Scholar] [CrossRef]
- Casciaro, F.; Zia, S.; Forcato, M.; Zavatti, M.; Beretti, F.; Bertucci, E.; Zattoni, A.; Reschiglian, P.; Alviano, F.; Bonsi, L.; et al. Unravelling Heterogeneity of Amplified Human Amniotic Fluid Stem Cells Sub-Populations. Cells 2021, 10, 158. [Google Scholar] [CrossRef]
- Zia, S.; Cavallo, C.; Vigliotta, I.; Parisi, V.; Grigolo, B.; Buda, R.; Marrazzo, P.; Alviano, F.; Bonsi, L.; Zattoni, A.; et al. Effective Label-Free Sorting of Multipotent Mesenchymal Stem Cells from Clinical Bone Marrow Samples. Bioengineering 2022, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Diab, K.A.; Fahmy, M.A.; Hassan, E.M.; El-Toumy, S.A. Evaluation of the cytotoxic, anticancer, and genotoxic activities of Acacia nilotica flowers and their effects on N-methyl-N-nitrosourea-induced genotoxicity in mice. Mol. Biol. Rep. 2022, 49, 8439–8448. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zia, S.; Roda, B.; Maggio, A.; Marrazzo, P.; Pizzuti, V.; Alviano, F.; Bonsi, L.; Marassi, V.; Zattoni, A.; Reschiglian, P. Celector®: An Innovative Technology for Quality Control of Living Cells. Appl. Sci. 2022, 12, 9967. https://doi.org/10.3390/app12199967
Zia S, Roda B, Maggio A, Marrazzo P, Pizzuti V, Alviano F, Bonsi L, Marassi V, Zattoni A, Reschiglian P. Celector®: An Innovative Technology for Quality Control of Living Cells. Applied Sciences. 2022; 12(19):9967. https://doi.org/10.3390/app12199967
Chicago/Turabian StyleZia, Silvia, Barbara Roda, Alessia Maggio, Pasquale Marrazzo, Valeria Pizzuti, Francesco Alviano, Laura Bonsi, Valentina Marassi, Andrea Zattoni, and Pierluigi Reschiglian. 2022. "Celector®: An Innovative Technology for Quality Control of Living Cells" Applied Sciences 12, no. 19: 9967. https://doi.org/10.3390/app12199967
APA StyleZia, S., Roda, B., Maggio, A., Marrazzo, P., Pizzuti, V., Alviano, F., Bonsi, L., Marassi, V., Zattoni, A., & Reschiglian, P. (2022). Celector®: An Innovative Technology for Quality Control of Living Cells. Applied Sciences, 12(19), 9967. https://doi.org/10.3390/app12199967










