GisaxStudio—An Open Platform for Analysis and Simulation of GISAXS from 3D Nanoparticle Lattices
Abstract
Featured Application
Abstract
1. Introduction
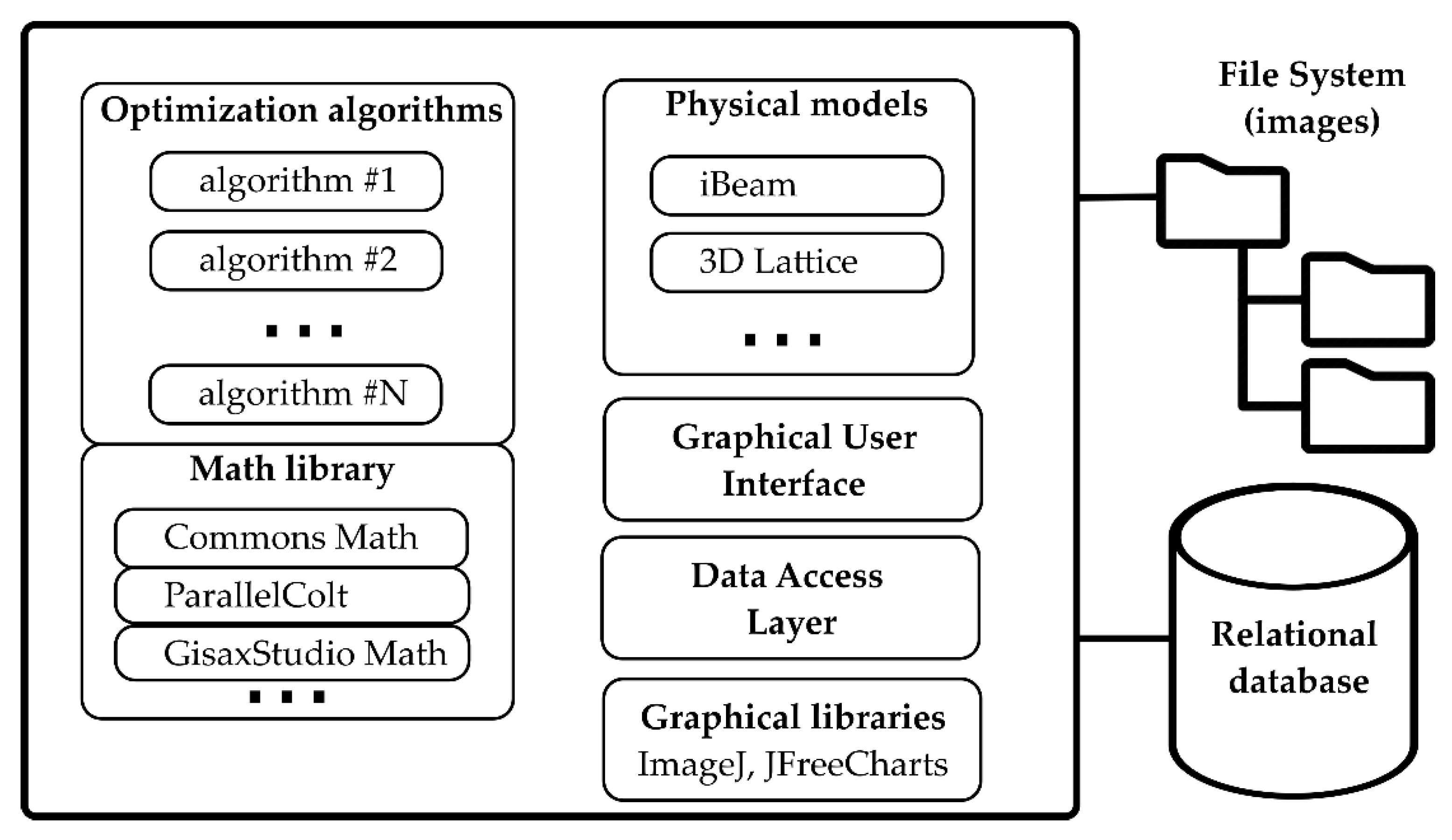
2. Materials and Methods
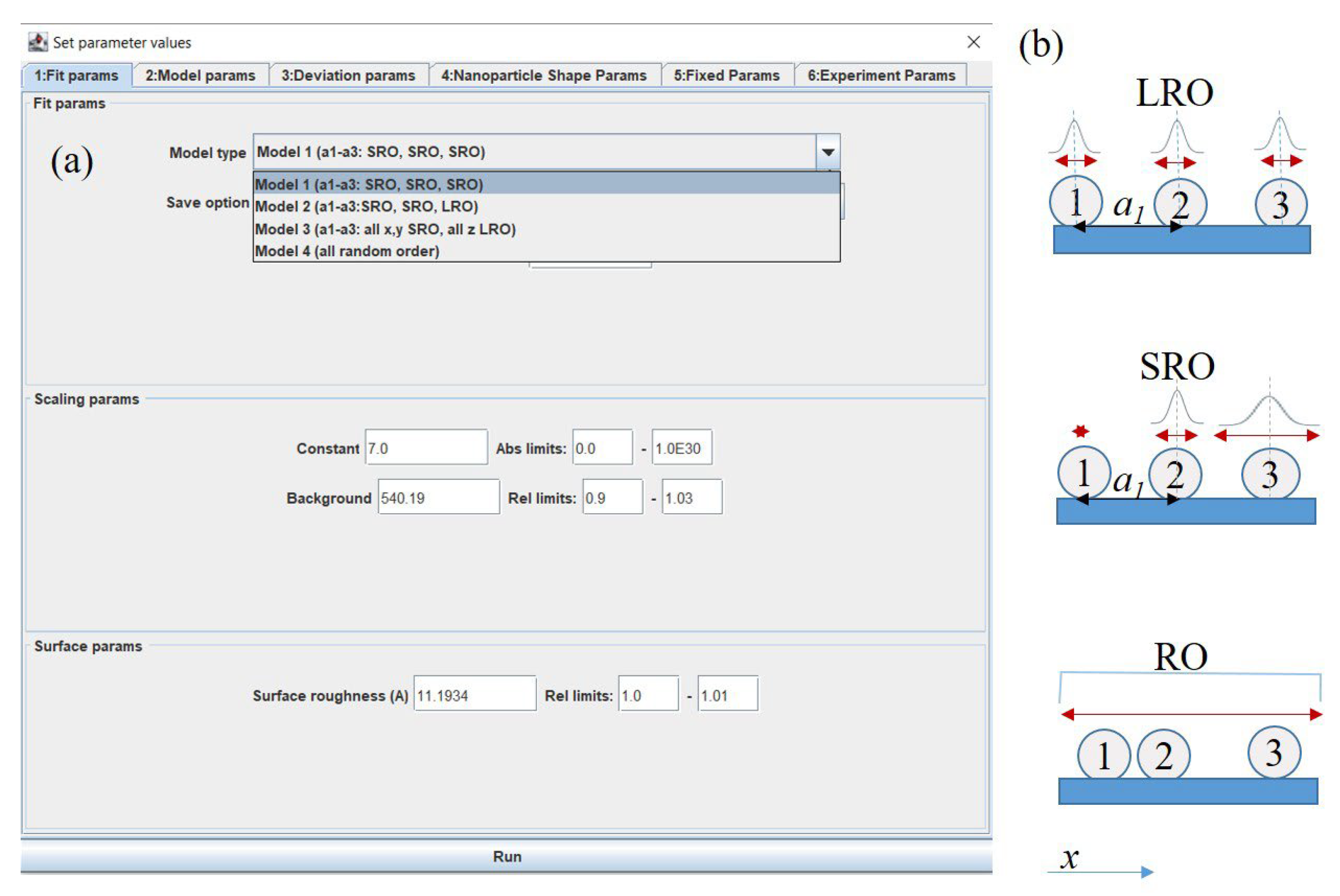
- Parameters class which extends AbstractGisaxsParam and describes the model’s parameters. For instance, instead of the existing iBeam model, suppose our model is a line y = ax + b. The parameter class would then have only two double members: x and y. Members must be annotated in order to expose them to the user via GUI, so that the parameters class would contain simply:public class LinearModelParams extends AbstractGisaxsParam {@GxParam(defval = 1.00, absLimits = {-1e5, 1e30}, name = "Slope (a)", group= "Slope and intercept", tab = "First tab", ordinal = 1)public double a;@GxParam(defval = 0, absLimits = {-1e30, 1e5}, name = "y-intercept (b)",group = "Slope and intercept", tab = "First tab", ordinal = 2)public double b;}Through the use of reflection, GisaxStudio will analyze its own code at runtime and dynamically build the GUI dialog for the selected model (e.g., Figure 5a for the iBeam model). Besides primitive types, the parameter class also supports complex numbers, lists of values (manifested as dropdowns for the user), etc. Of course, besides fittable parameters, the parameters class can also include constants and other non-fittable parameters that are easily assigned by the user.
- Model class, which implements the IGisaxsModel interface, defines 16 methods, the most important being compute(double[]), which computes the model given an array of fittable parameters. In our example, compute would receive a and b via the double array and simply return a*x + b, where x is a vector that has already been assigned:@Overridepublic double[] compute(double[] params) {LinearModelParams pObj = (LinearModelParams)fitVectorToParamObject(params);return _xFit.times(pObj.a).plus(pObj.b).toArray();}LinearModel is actually included as a proof-of-concept in the GisaxStudio code, and the code shown above is taken from those two classes (although it is not shown in Figure 1). Helper method fitVectorToParamObject is used to instantiate and populate the params object so that the programmer could use meaningful variable names (a, b) in the code and autocompletion features instead of working with params[0] and params[1].
3. Results
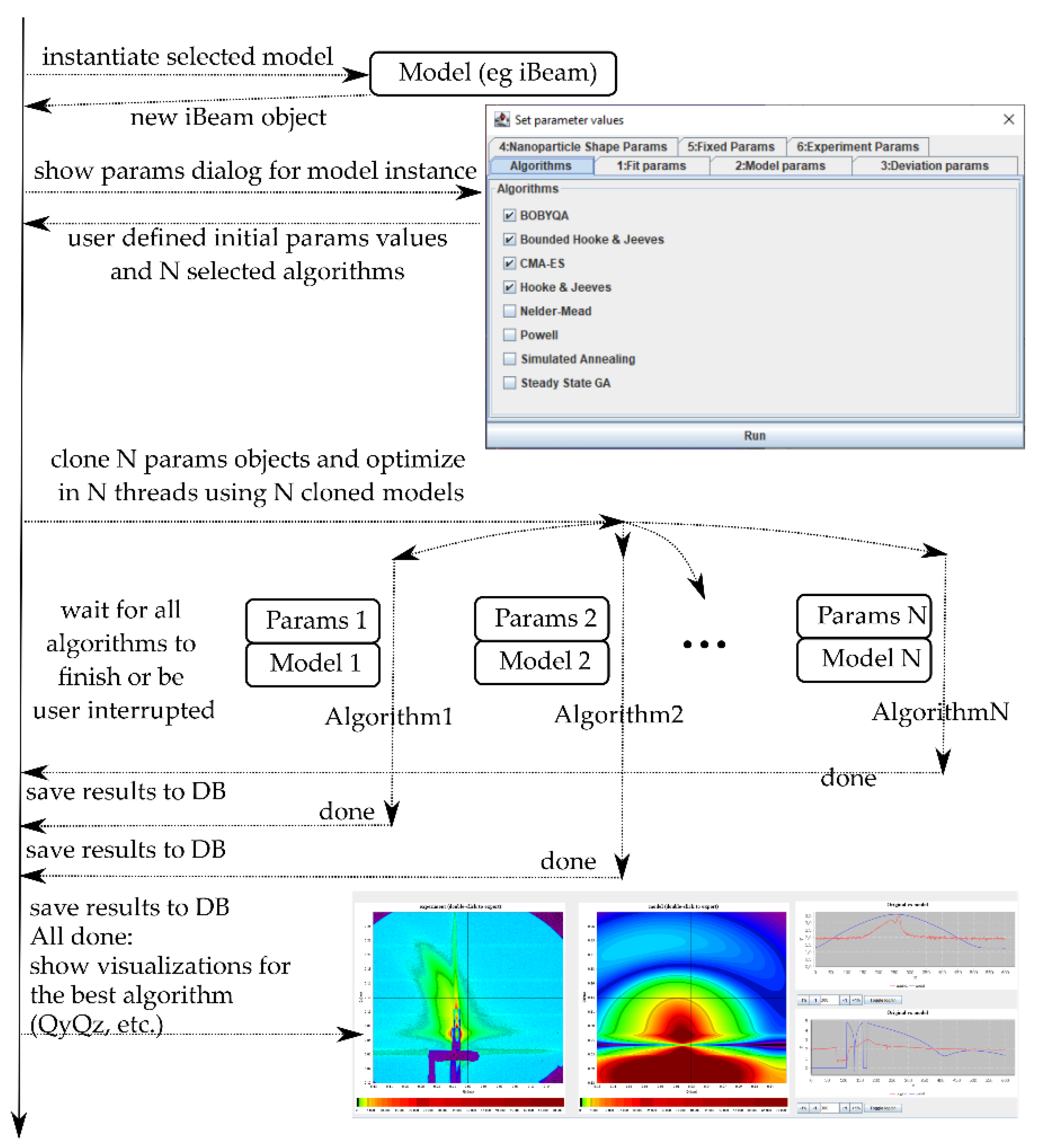
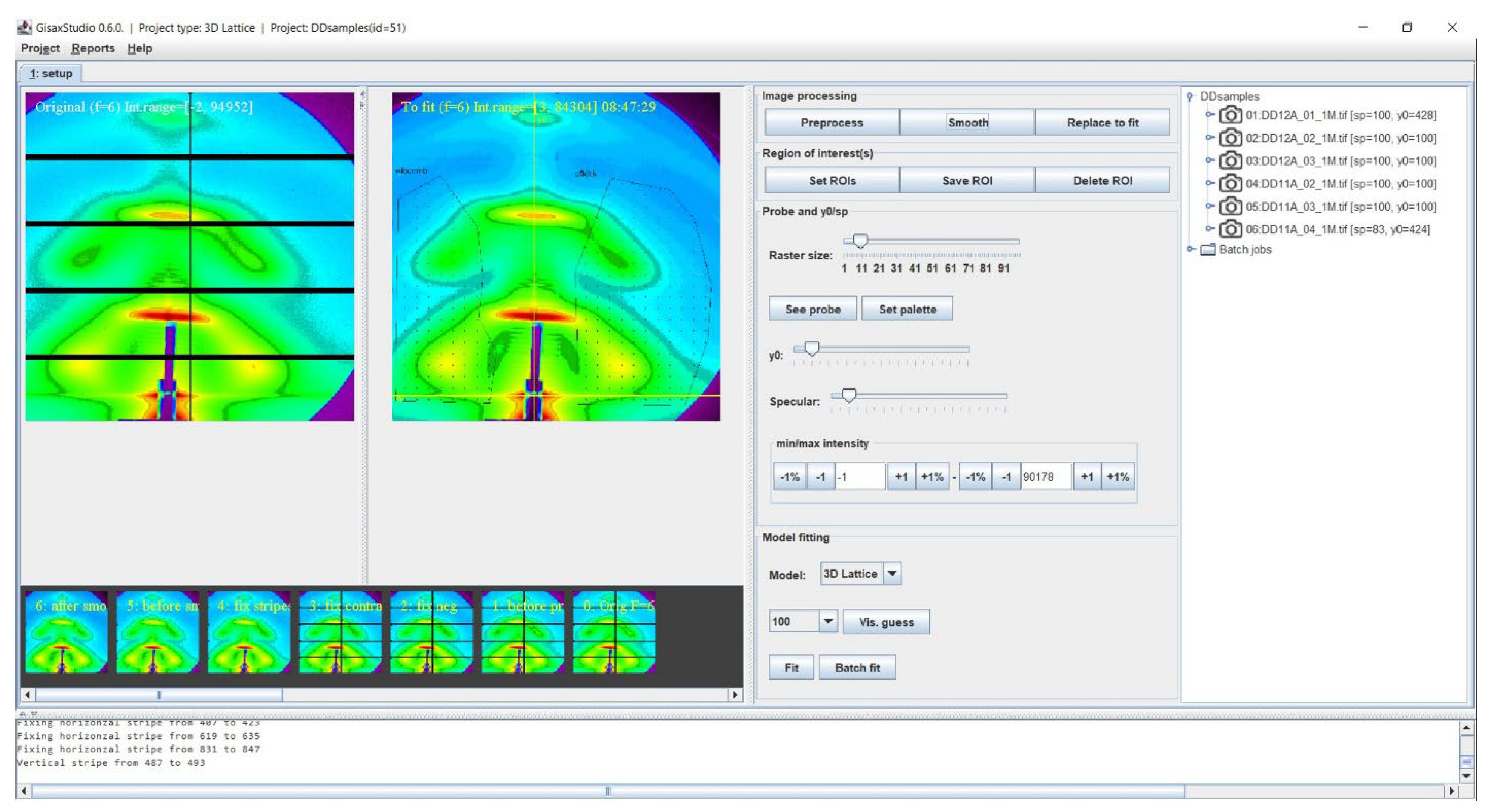
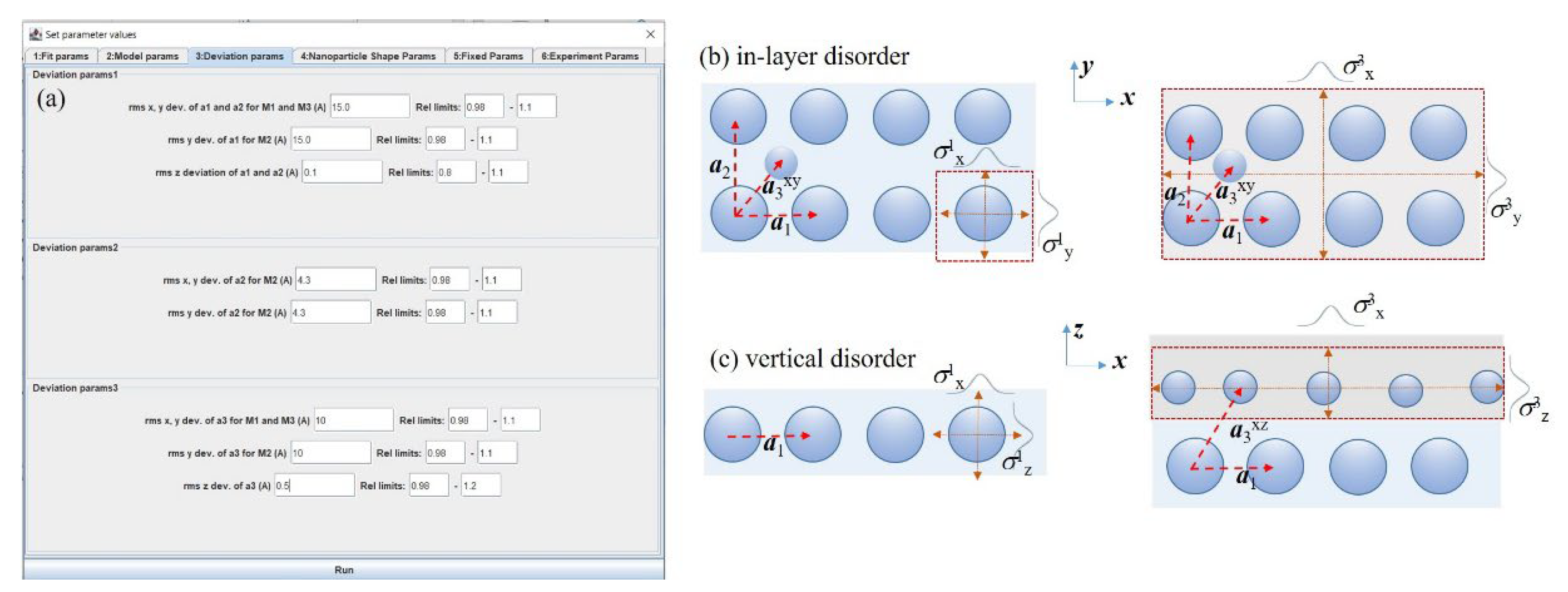
3.1. Main Features
3.2. Visual Estimate Option
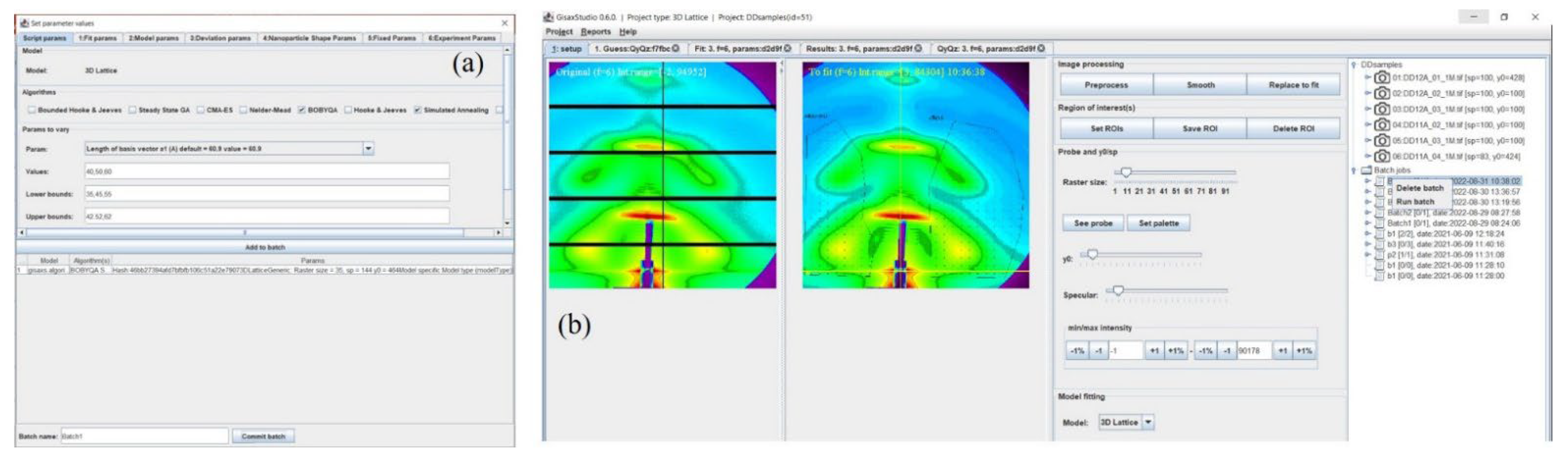
3.3. Fitting Options
- ○
- load the first unprocessed batch item (parameter set) from list
- ○
- execute the batch item (fit)
- ○
- store the results to the database
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baig, N.; Kammakakam, I.; Falath, W. Nanomaterials: A review of synthesis methods, properties, recent progress, and challenges. Mater. Adv. 2021, 2, 1821–1871. [Google Scholar] [CrossRef]
- Chandrakala, V.; Aruna, V.; Angajala, G. Review on metal nanoparticles as nanocarriers: Current challenges and perspectives in drug delivery systems. Emergent Mater. 2022. [Google Scholar] [CrossRef] [PubMed]
- de Arquer, F.P.G.; Talapin, D.V.; Klimov, V.I.; Arakawa, Y.; Bayer, M.; Sargent, E.H. Semiconductor quantum dots: Technological progress and future challenges. Science 2021, 373, eaaz8541. [Google Scholar] [CrossRef]
- Ossicini, S.; Amato, M.; Guerra, R.; Palummo, M.; Pulci, O. Silicon and Germanium Nanostructures for Photovoltaic Applications: Ab-Initio Results. Nanoscale Res. Lett. 2010, 5, 1637–1649. [Google Scholar] [CrossRef][Green Version]
- Nekić, N.; Šarić, I.; Salamon, K.; Basioli, L.; Sancho-Parramon, J.; Grenzer, J.; Hübner, R.; Bernstorff, S.; Petravić, M.; Mičetić, M. Preparation of non-oxidized Ge quantum dot lattices in amorphous Al2O3, Si3N4 and SiC matrices. Nanotechnology 2019, 30, 335601. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, E.L.; Albuquerque, E.L.; de Sousa, J.S.; Farias, G.A.; Peeters, F.M. Configuration-Interaction Excitonic Absorption in Small Si/Ge and Ge/Si Core/Shell Nanocrystals. J. Phys. Chem. C 2012, 116, 4399–4407. [Google Scholar] [CrossRef]
- Nekić, N.; Sancho-Parramon, J.; Radovic, I.B.; Grenzer, J.; Huebner, R.; Bernstorff, S.; Ivanda, M.; Buljan, M. Ge/Si core/shell quantum dots in alumina: Tuning the optical absorption by the core and shell size. Nanophotonics 2017, 6, 1055–1062. [Google Scholar] [CrossRef][Green Version]
- Buljan, M.; Radić, N.; Sancho-Paramon, J.; Janicki, V.; Grenzer, J.; Radovic, I.B.; Siketić, Z.; Ivanda, M.; Utrobičić, A.; Hübner, R.; et al. Production of three-dimensional quantum dot lattice of Ge/Si core–shell quantum dots and Si/Ge layers in an alumina glass matrix. Nanotechnology 2015, 26, 065602. [Google Scholar] [CrossRef]
- Basioli, L.; Sancho-Parramon, J.; Despoja, V.; Fazinić, S.; Radović, I.B.; Mihalić, I.B.; Salamon, K.; Nekić, N.; Ivanda, M.; Dražić, G.; et al. Ge Quantum Dots Coated with Metal Shells (Al, Ta, and Ti) Embedded in Alumina Thin Films for Solar Energy Conversion. ACS Appl. Nano Mater. 2020, 3, 8640–8650. [Google Scholar] [CrossRef]
- Kes, H.; Okan, S.; Aktas, S. The excitons in infinite potential centered multilayered coaxial quantum wire and the magnetic field effects on their properties. Superlattices Microstruct. 2020, 139, 106421. [Google Scholar] [CrossRef]
- Basioli, L.; Tkalčević, M.; Bogdanović-Radović, I.; Dražić, G.; Nadazdy, P.; Siffalovic, P.; Salamon, K.; Mičetić, M. 3D Networks of Ge Quantum Wires in Amorphous Alumina Matrix. Nanomaterials 2020, 10, 1363. [Google Scholar] [CrossRef] [PubMed]
- Lazarenkova, O.L.; Balandin, A.A. Miniband formation in a quantum dot crystal. J. Appl. Phys. 2001, 89, 5509–5515. [Google Scholar] [CrossRef]
- Buljan, M.; Roshchupkina, O.; Šantić, A.; Holý, V.; Baehtz, C.; Mücklich, A.; Horák, L.; Valeš, V.; Radić, N.; Bernstorff, S.; et al. Growth of a three-dimensional anisotropic lattice of Ge quantum dots in an amorphous alumina matrix. J. Appl. Crystallogr. 2013, 46, 709–715. [Google Scholar] [CrossRef]
- Müller-Buschbaum, P. A Basic Introduction to Grazing Incidence Small-Angle X-Ray Scattering. In Applications of Synchrotron Light to Scattering and Diffraction in Materials and Life Sciences; Lecture Notes in Physics; Chapter 3; Springer: Berlin/Heidelberg, Germany, 2009; pp. 61–89. [Google Scholar] [CrossRef]
- Renaud, G.; Lazzari, R.; Leroy, F. Probing surface and interface morphology with Grazing Incidence Small Angle X-Ray Scattering. Surf. Sci. Rep. 2009, 64, 255–380. [Google Scholar] [CrossRef]
- Cristofolini, L. Synchrotron X-ray techniques for the investigation of structures and dynamics in interfacial systems. Curr. Opin. Colloid Interface Sci. 2014, 19, 228–241. [Google Scholar] [CrossRef]
- Hexemer, A.; Müller-Buschbaum, P. Advanced grazing-incidence techniques for modern soft-matter materials analysis. IUCrJ 2015, 2, 106–125. [Google Scholar] [CrossRef]
- Müller-Buschbaum, P. GISAXS and GISANS as metrology technique for understanding the 3D morphology of block copolymer thin films. Eur. Polym. J. 2016, 81, 470–493. [Google Scholar] [CrossRef]
- Smilgies, D. GISAXS: A versatile tool to assess structure and self-assembly kinetics in block copolymer thin films. J. Appl. Polym. Sci. 2021, 60, 1023–1041. [Google Scholar] [CrossRef]
- Buljan, M.; Radić, N.; Bernstorff, S.; Dražić, G.; Bogdanović-Radović, I.; Holý, V. Grazing-incidence small-angle X-ray scattering: Application to the study of quantum dot lattices. Acta Crystallogr. Sect. A Found. Crystallogr. 2012, 68, 124–138. [Google Scholar] [CrossRef]
- Basioli, L.; Salamon, K.; Tkalčević, M.; Mekterović, I.; Bernstorff, S.; Mičetić, M. Application of GISAXS in the Investigation of Three-Dimensional Lattices of Nanostructures. Crystals 2019, 9, 479. [Google Scholar] [CrossRef]
- Buljan, M.; Karlušić, M.; Nekić, N.; Jerčinović, M.; Bogdanović-Radović, I.; Bernstorff, S.; Radić, N.; Mekterović, I. GISAXS analysis of ion beam modified films and surfaces. Comput. Phys. Commun. 2017, 212, 69–81. [Google Scholar] [CrossRef]
- Lazzari, R. IsGISAXS: A program for grazing-incidence small-angle X-ray scattering analysis of supported islands. J. Appl. Crystallogr. 2002, 35, 406–421. [Google Scholar] [CrossRef]
- Tate, M.P.; Urade, V.N.; Kowalski, J.D.; Wei, T.-C.; Hamilton, B.D.; Eggiman, B.W.; Hillhouse, H.W. Simulation and Interpretation of 2D Diffraction Patterns from Self-Assembled Nanostructured Films at Arbitrary Angles of Incidence: From Grazing Incidence (Above the Critical Angle) to Transmission Perpendicular to the Substrate. J. Phys. Chem. B 2006, 110, 9882–9892. [Google Scholar] [CrossRef]
- Babonneau, D. FitGISAXS: Software package for modelling and analysis of GISAXS data using IGOR Pro. J. Appl. Crystallogr. 2010, 43, 929–936. [Google Scholar] [CrossRef]
- Chourou, S.T.; Sarje, A.; Li, X.S.; Chan, E.R.; Hexemer, A. HipGISAXS: A high-performance computing code for simulating grazing-incidence X-ray scattering data. J. Appl. Crystallogr. 2013, 46, 1781–1795. [Google Scholar] [CrossRef]
- Benecke, G.; Wagermaier, W.; Li, C.; Schwartzkopf, M.; Flucke, G.; Hoerth, R.; Zizak, I.; Burghammer, M.; Metwalli, E.; Müller-Buschbaum, P.; et al. A customizable software for fast reduction and analysis of large X-ray scattering data sets: Applications of the new DPDAK package to small-angle X-ray scattering and grazing-incidence small-angle X-ray scattering. J. Appl. Crystallogr. 2014, 47, 1797–1803. [Google Scholar] [CrossRef]
- Durniak, C.; Ganeva, M.; Pospelov, G.; Van Herck, W.; Wuttke, J. BornAgain—Software for Simulating and Fitting X-ray and Neutron Small-Angle Scattering at Grazing Incidence. In Proceedings of the 26th International Conference on Amorphous and Nanocrystalline Semiconductors, Aachen, Germany, 13–18 September 2015; Available online: http://juser.fz-juelich.de/record/255761 (accessed on 28 August 2022).
- Pospelov, G.; Van Herck, W.; Burle, J.; Loaiza, J.M.C.; Durniak, C.; Fisher, J.M.; Ganeva, M.; Yurov, D.; Wuttke, J. BornAgain: Software for simulating and fitting grazing-incidence small-angle scattering. J. Appl. Crystallogr. 2020, 53, 262–276. [Google Scholar] [CrossRef]
- Available online: http://gisaxs.com/index.php/Software/ (accessed on 21 September 2022).
- GisaxStudio. Available online: http://homer.zpr.fer.hr/gisaxstudio/ (accessed on 30 August 2022).
- Tkalčević, M.; Boršćak, D.; Periša, I.; Bogdanović-Radović, I.; Janković, I.; Petravić, M.; Bernstorff, S.; Mičetić, M. Multiple Exciton Generation in 3D-Ordered Networks of Ge Quantum Wires in Alumina Matrix. Materials 2022, 15, 5353. [Google Scholar] [CrossRef]
- Shaji, A.; Micetic, M.; Halahovets, Y.; Nadazdy, P.; Matko, I.; Jergel, M.; Majkova, E.; Siffalovic, P. Real-time tracking of the self-assembled growth of a 3D Ge quantum dot lattice in an alumina matrix. J. Appl. Crystallogr. 2020, 53, 1029–1038. [Google Scholar] [CrossRef]
- Tkalčević, M.; Basioli, L.; Salamon, K.; Šarić, I.; Parramon, J.S.; Bubaš, M.; Bogdanović-Radović, I.; Bernstorff, S.; Fogarassy, Z.; Balázsi, K.; et al. Ge quantum dot lattices in alumina prepared by nitrogen assisted deposition: Structure and photoelectric conversion efficiency. Sol. Energy Mater. Sol. Cells 2020, 218, 110722. [Google Scholar] [CrossRef]
- Krause, M.; Buljan, M.; Mücklich, A.; Möller, W.; Fritzsche, M.; Facsko, S.; Heller, R.; Zschornak, M.; Wintz, S.; Endrino, J.L.; et al. Compositionally modulated ripples during composite film growth: Three-dimensional pattern formation at the nanoscale. Phys. Rev. B 2014, 89, 085418. [Google Scholar] [CrossRef]
- Karlušić, M.; Mičetić, M.; Kresić, M.; Jakšić, M.; Šantić, B.; Bogdanović-Radović, I.; Bernstorff, S.; Lebius, H.; Ban-D’Etat, B.; Rožman, K.; et al. Nanopatterning surfaces by grazing incidence swift heavy ion irradiation. Appl. Surf. Sci. 2021, 541, 148467. [Google Scholar] [CrossRef]
- Karlušić, M.; Bernstorff, S.; Siketić, Z.; Šantić, B.; Bogdanović-Radović, I.; Jakšić, M.; Schleberger, M.; Buljan, M. Formation of swift heavy ion tracks on a rutile TiO2 (001) surface. J Appl Crystallogr. 2016, 49, 1704–1712. [Google Scholar] [CrossRef] [PubMed]
- Radović, I.B.; Buljan, M.; Karlušić, M.; Jerčinović, M.; Dražič, G.; Bernstorff, S.; Boettger, R. Modification of semiconductor or metal nanoparticle lattices in amorphous alumina by MeV heavy ions. New J. Phys. 2016, 18, 093032. [Google Scholar] [CrossRef]
- Karlušić, M.; Kozubek, R.; Lebius, H.; Ban-D’Etat, B.; Wilhelm, R.A.; Buljan, M.; Siketić, Z.; Scholz, F.; Meisch, T.; Jakšić, M.; et al. Response of GaN to energetic ion irradiation: Conditions for ion track formation. J. Phys. D: Appl. Phys. 2015, 48, 325304. [Google Scholar] [CrossRef]
- Commons Math: The Apache Commons Mathematics Library. Available online: https://commons.apache.org/proper/commons-math/ (accessed on 30 August 2022).
- Parallel Colt. A Multithreaded Version of Colt—A library for High Performance Scientific Computing in JAVA. Available online: https://github.com/rwl/ParallelColt (accessed on 30 August 2022).
- H2 Database Engine. Available online: https://www.h2database.com/ (accessed on 30 August 2022).




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mekterović, I.; Svalina, G.; Isaković, S.; Mičetić, M. GisaxStudio—An Open Platform for Analysis and Simulation of GISAXS from 3D Nanoparticle Lattices. Appl. Sci. 2022, 12, 9773. https://doi.org/10.3390/app12199773
Mekterović I, Svalina G, Isaković S, Mičetić M. GisaxStudio—An Open Platform for Analysis and Simulation of GISAXS from 3D Nanoparticle Lattices. Applied Sciences. 2022; 12(19):9773. https://doi.org/10.3390/app12199773
Chicago/Turabian StyleMekterović, Igor, Gabrijela Svalina, Senad Isaković, and Maja Mičetić. 2022. "GisaxStudio—An Open Platform for Analysis and Simulation of GISAXS from 3D Nanoparticle Lattices" Applied Sciences 12, no. 19: 9773. https://doi.org/10.3390/app12199773
APA StyleMekterović, I., Svalina, G., Isaković, S., & Mičetić, M. (2022). GisaxStudio—An Open Platform for Analysis and Simulation of GISAXS from 3D Nanoparticle Lattices. Applied Sciences, 12(19), 9773. https://doi.org/10.3390/app12199773








