Combining UNet 3+ and Transformer for Left Ventricle Segmentation via Signed Distance and Focal Loss
Abstract
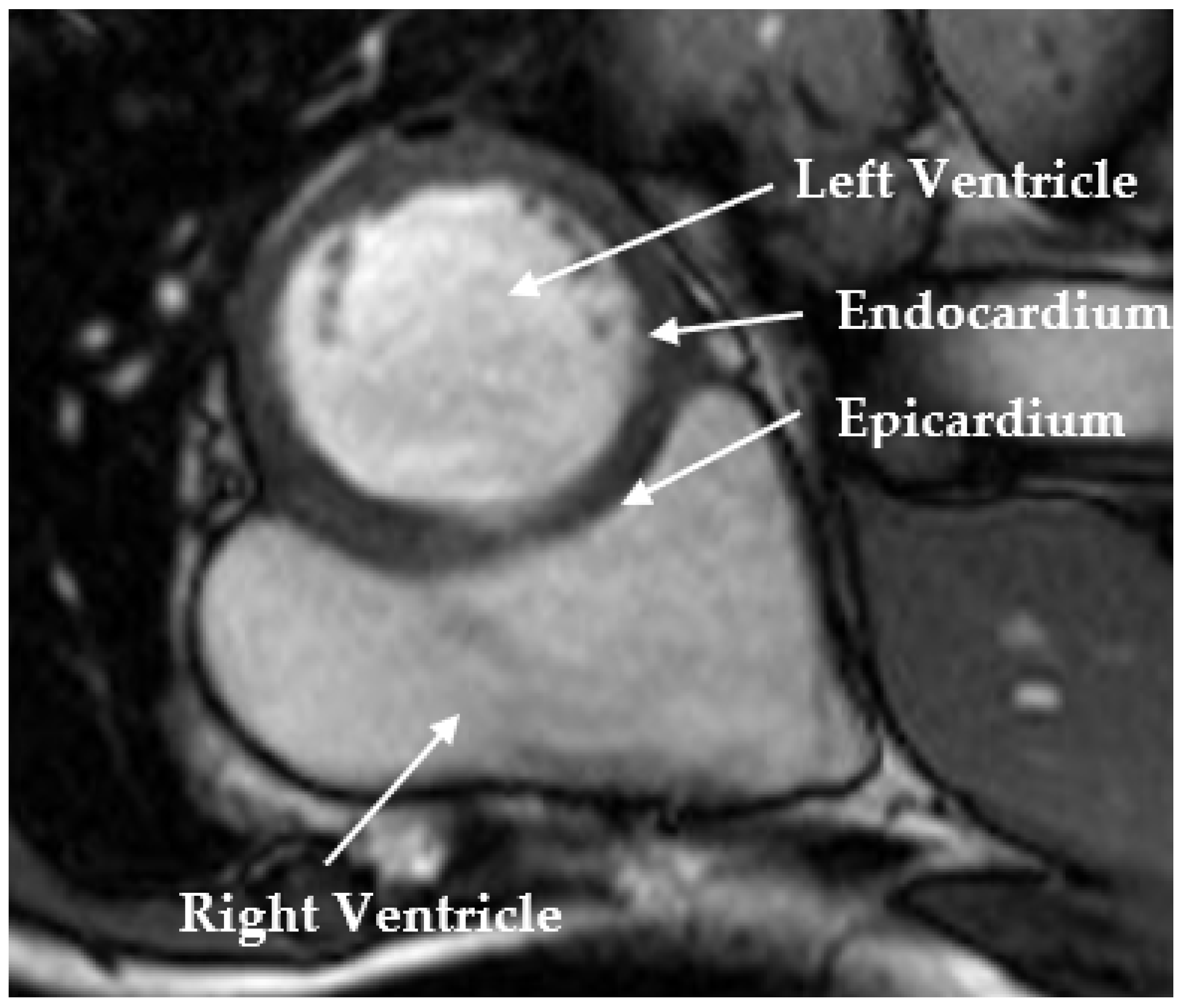
:1. Introduction
1.1. Traditional Segmentation Methods
1.2. Deep Learning
1.3. Transformers
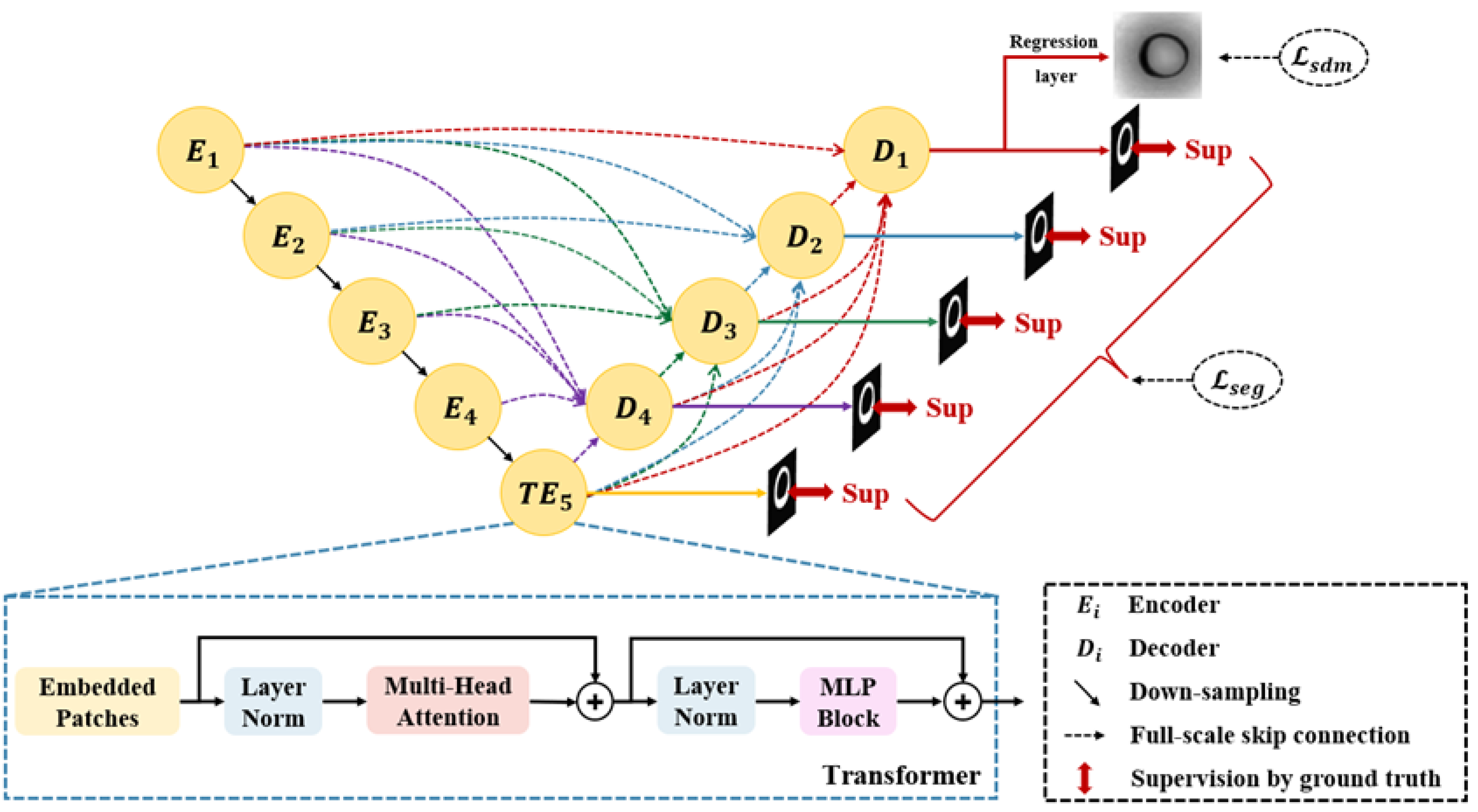
2. Method
2.1. Segmentation Network
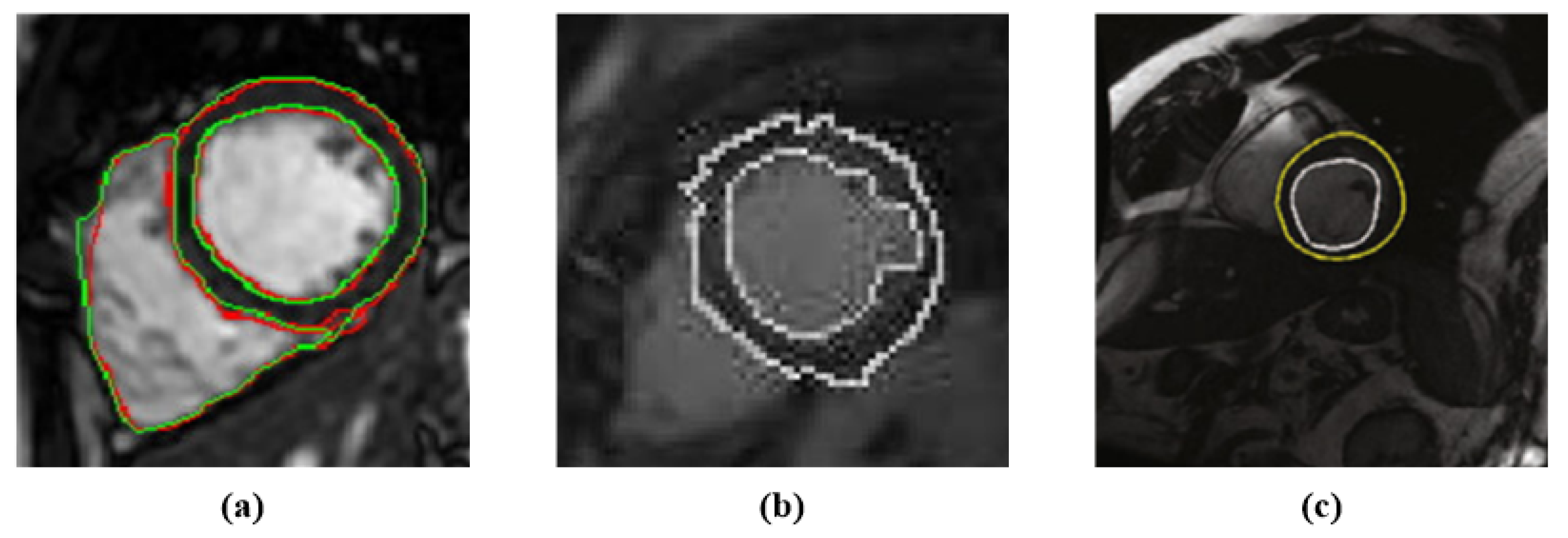
2.2. Loss Function
3. Experiments
3.1. Datasets
3.2. Implementation Details
3.3. Evaluation Metric
4. Experimental Results
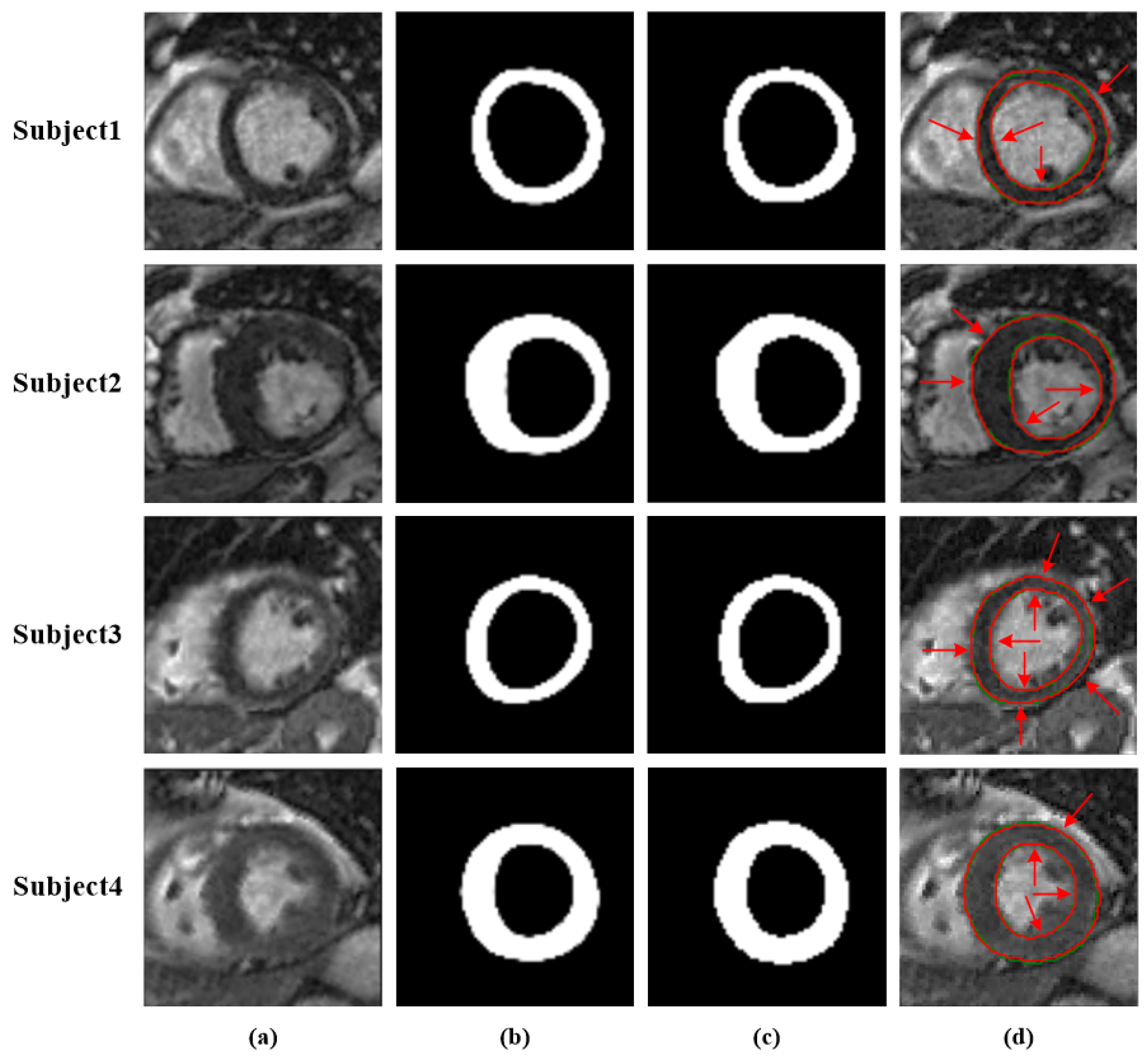
4.1. Performance of the Network
4.2. Performance Comparison
4.3. Ablation Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Fact-Sheets Cardiovascular Diseases (CVDs); WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Shaaf, Z.F.; Jamil, M.M.A.; Ambar, R.; Alattab, A.A.; Yahya, A.A.; Asiri, Y. Automatic Left Ventricle Segmentation from Short-Axis Cardiac MRI Images Based on Fully Convolutional Neural Network. Diagnostics 2022, 12, 414. [Google Scholar] [CrossRef] [PubMed]
- Gessert, N.; Schlaefer, A. Left Ventricle Quantification Using Direct Regression with Segmentation Regularization and Ensembles of Pretrained 2D and 3D CNNs. arXiv 2019, arXiv:1908.04181. [Google Scholar]
- Tavakoli, V.; Amini, A.A. A survey of shaped-based registration and segmentation techniques for cardiac images. Comput. Vis. Image Underst. 2013, 117, 966–989. [Google Scholar] [CrossRef]
- Petitjean, C.; Dacher, J.N. A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 2011, 15, 169–184. [Google Scholar] [CrossRef]
- Dakua, S.P. Towards Left Ventricle Segmentation From Magnetic Resonance Images. IEEE Sens. J. 2017, 17, 5971–5981. [Google Scholar] [CrossRef]
- Tran, P.V. A Fully Convolutional Neural Network for Cardiac Segmentation in Short-Axis MRI. arXiv 2016, arXiv:1604.00494. [Google Scholar]
- Xue, W.; Lum, A.; Mercado, A.; Landis, M.; Warrington, J.; Li, S. Full Quantification of Left Ventricle via Deep Multitask Learning Network Respecting Intra- and Inter-Task Relatedness. In Lecture Notes in Computer Science, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Wernick, M.N.; Yang, Y.; Brankov, J.G.; Yourganov, G.; Strother, S.C. Machine Learning in Medical Imaging. IEEE Signal Process. Mag. 2010, 27, 25–38. [Google Scholar] [CrossRef]
- Yabrin, A.; Amin, B.S.; Mir, A.H. A Comparative Study on Left and Right Endocardium Segmentation using Gradient Vector Field and Adaptive Diffusion Flow Algorithms. Int. J. Bio-Sci. Bio-Technol. 2016, 8, 105–120. [Google Scholar]
- Li, W.; Ma, Y.; Zhan, K.; Ma, Y. Automatic Left Ventricle Segmentation in Cardiac MRI via Level Set and Fuzzy C-Means. In Proceedings of the International Conference on Recent Advances in Engineering & Computational Sciences, Chandigarh, India, 21–22 December 2015. [Google Scholar]
- Katouzian, A.; Prakash, A.; Konofagou, E. A new automated technique for left-and right-ventricular segmentation in magnetic resonance imaging. In Proceedings of the International Conference of the IEEE Engineering in Medicine and Biology Society, New York, NY, USA, 30 August–3 September 2006; Volume 2006, pp. 3074–3077. [Google Scholar] [CrossRef]
- Lynch, M.; Ghita, O.; Whelan, P.F. Automatic segmentation of the left ventricle cavity and myocardium in MRI data. Comput. Biol. Med. 2006, 36, 389–407. [Google Scholar] [CrossRef]
- Zhang, Z.; Duan, C.; Lin, T.; Zhou, S.; Wang, Y.; Gao, X. GVFOM: A novel external force for active contour based image segmentation. Inf. Sci. 2020, 506, 1–18. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Jia, Y. Segmentation of the left ventricle in cardiac cine MRI using a shape-constrained snake model. Comput. Vis. Image Underst. 2013, 117, 990–1003. [Google Scholar] [CrossRef]
- Chakraborty, A.; Staib, L.H.; Duncan, J.S. Deformable boundary finding in medical images by integrating gradient and region information. IEEE Trans. Med. Imaging 1996, 15, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Ghita, O.; Whelan, P.F. Segmentation of the left ventricle of the heart in 3-D+t MRI data using an optimized nonrigid temporal model. IEEE Trans. Med. Imaging 2008, 27, 195–203. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Wen, Z.; Tian, B.; Kao, E.; Liu, X.; Xuan, W.; Ordovas, K.; Saloner, D.; Liu, J. Deep learning based fully automatic segmentation of the left ventricular endocardium and epicardium from cardiac cine MRI. Quant. Imaging Med. Surg. 2021, 11, 1600–1612. [Google Scholar] [CrossRef] [PubMed]
- Xijing, Z.; Qian, W.; Ting, L. A novel approach for left ventricle segmentation in tagged MRI. Comput. Electr. Eng. 2021, 95, 107416. [Google Scholar]
- Haque, I.R.I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 100297. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Baloglu, U.B.; Talo, M.; Yildirim, O.; Tan, R.S.; Acharya, U.R. Classification of myocardial infarction with multi-lead ECG signals and deep CNN. Pattern Recognit. Lett. 2019, 122, 23–30. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Laurens, V.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Computer Society, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in Medical Imaging: A Survey. arXiv 2022, arXiv:2201.09873. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020. [Google Scholar]
- Goshtasby, A.; Turner, D.A. Segmentation of cardiac cine MR images for extraction of right and left ventricular chambers. IEEE Trans. Med. Imaging 1995, 14, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Peng, Y.; Li, D.; Guo, Y.; Zhang, B. MMNet: A multi-scale deep learning network for the left ventricular segmentation of cardiac MRI images. Appl. Intell. 2021, 52, 5225–5240. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H.S. Rethinking Semantic Segmentation from a Sequence-to-Sequence Perspective with Transformers. In Proceedings of the Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. arXiv 2020, arXiv:2005.12872. [Google Scholar]
- Parmar, N.; Vaswani, A.; Uszkoreit, J.; Kaiser, L.; Shazeer, N.; Ku, A.; Tran, D. Image Transformer. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; Volume 80, pp. 4055–4064. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. TransFuse: Fusing Transformers and CNNs for Medical Image Segmentation. In Lecture Notes in Computer Science, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Xue, W.; Islam, A.; Bhaduri, M.; Li, S. Direct Multitype Cardiac Indices Estimation via Joint Representation and Regression Learning. IEEE Trans. Med. Imaging 2017, 36, 2057–2067. [Google Scholar] [CrossRef] [Green Version]
- Noh, H.; Hong, S.; Han, B. Learning Deconvolution Network for Semantic Segmentation. arXiv 2015, arXiv:1505.04366. [Google Scholar]
- Du, X.; Tang, R.; Yin, S.; Zhang, Y.; Li, S. Direct Segmentation-Based Full Quantification for Left Ventricle via Deep Multi-Task Regression Learning Network. IEEE J. Biomed. Health Inform. 2019, 23, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Yin, S.; Tang, R.; Zhang, Y.; Li, S. Cardiac-DeepIED: Automatic Pixel-Level Deep Segmentation for Cardiac Bi-Ventricle Using Improved End-to-End Encoder-Decoder Network. IEEE J. Transl. Eng. Health Med. 2019, 7, 1900110. [Google Scholar] [CrossRef] [PubMed]
| Methods | DM | JC | ACC | PPV |
|---|---|---|---|---|
| FCN | 0.873 | 0.778 | 0.972 | 0.878 |
| SegNet | 0.843 | 0.728 | 0.964 | 0.845 |
| U-Net | 0.887 | 0.800 | 0.975 | 0.887 |
| Conv-Deconv | 0.846 | 0.733 | 0.965 | 0.840 |
| Indices-JSQ | 0.870 | − 1 | − | − |
| Cardiac-DeepIED [41] | 0.890 | 0.801 | 0.976 | 0.891 |
| Our Method | 0.908 | 0.834 | 0.979 | 0.903 |
| UNet 3+ | Transformer | DM | ||
|---|---|---|---|---|
| ✓ | × | ✓ | × | 0.896 |
| ✓ | ✓ | ✓ | × | 0.907 |
| ✓ | ✓ | ✓ | ✓ | 0.908 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; He, X.; Lu, Y. Combining UNet 3+ and Transformer for Left Ventricle Segmentation via Signed Distance and Focal Loss. Appl. Sci. 2022, 12, 9208. https://doi.org/10.3390/app12189208
Liu Z, He X, Lu Y. Combining UNet 3+ and Transformer for Left Ventricle Segmentation via Signed Distance and Focal Loss. Applied Sciences. 2022; 12(18):9208. https://doi.org/10.3390/app12189208
Chicago/Turabian StyleLiu, Zhi, Xuelin He, and Yunhua Lu. 2022. "Combining UNet 3+ and Transformer for Left Ventricle Segmentation via Signed Distance and Focal Loss" Applied Sciences 12, no. 18: 9208. https://doi.org/10.3390/app12189208
APA StyleLiu, Z., He, X., & Lu, Y. (2022). Combining UNet 3+ and Transformer for Left Ventricle Segmentation via Signed Distance and Focal Loss. Applied Sciences, 12(18), 9208. https://doi.org/10.3390/app12189208







