Deep-Learning-Based Framework for PET Image Reconstruction from Sinogram Domain
Abstract
:1. Introduction
2. Methodology
2.1. Problem Definition
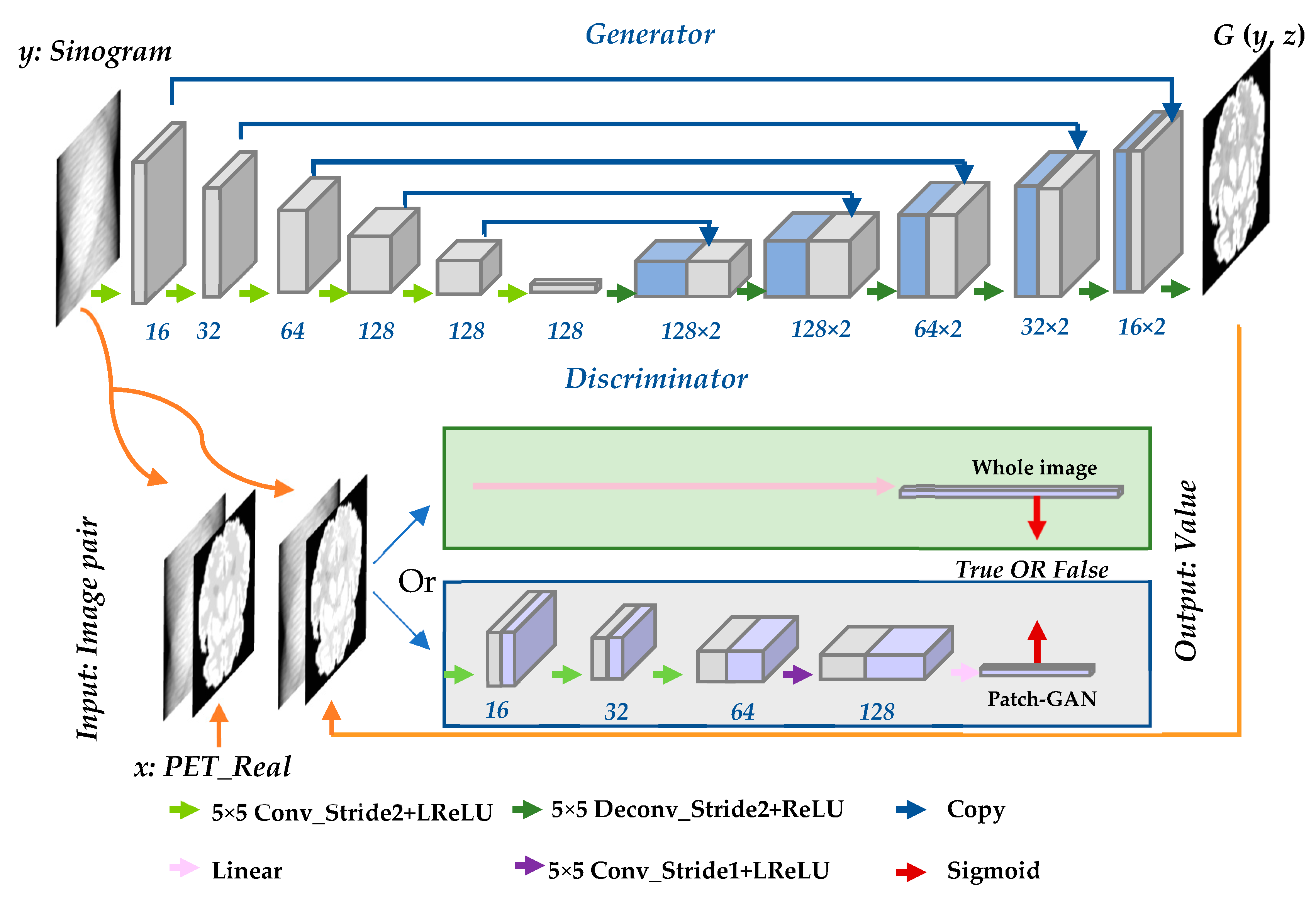
2.2. Framework Based on Conditional Generative Adversarial Network
2.2.1. Network Design
2.2.2. Objective Function
3. Experiments
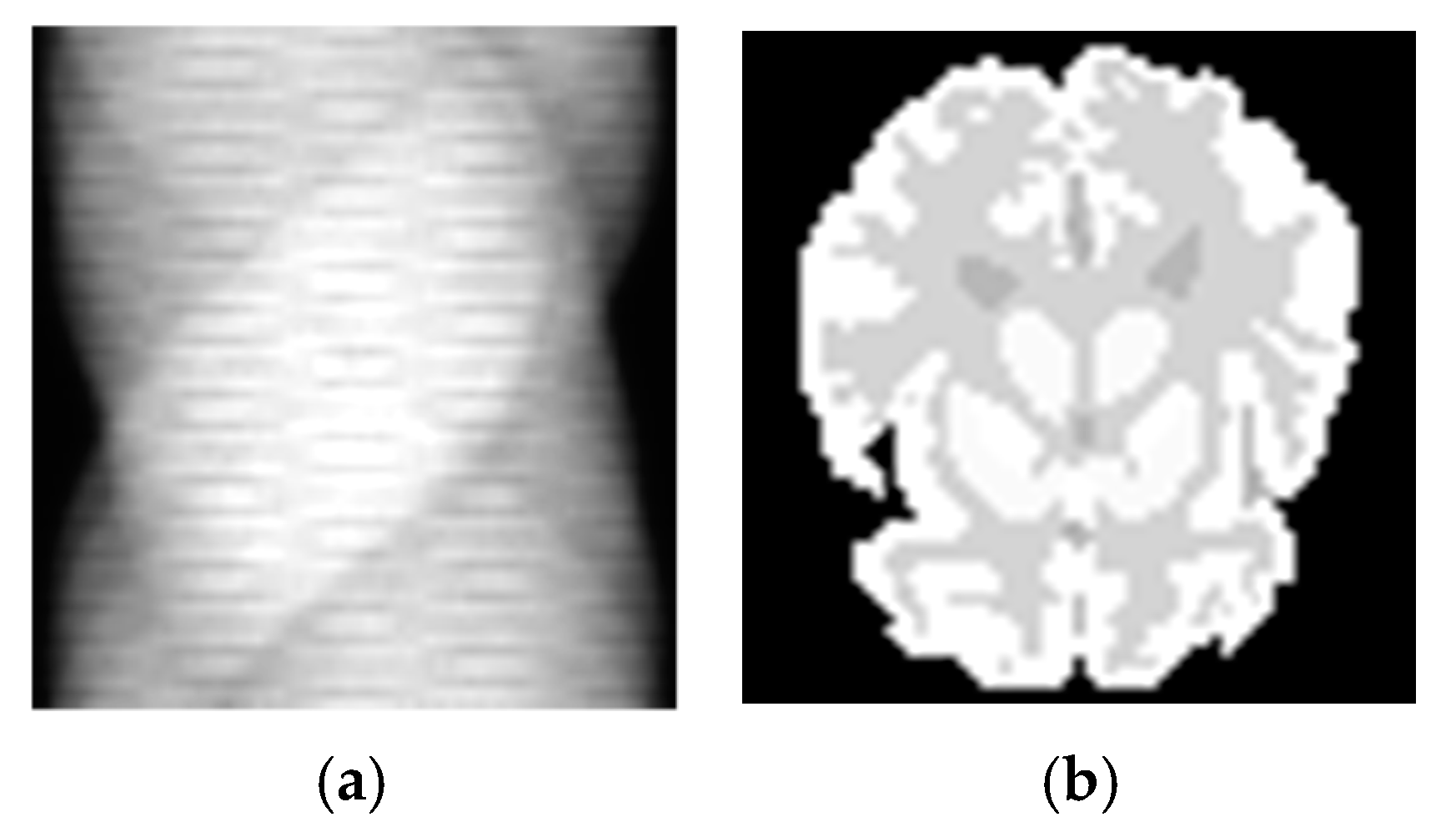
3.1. Simulation Experiments
3.2. SD Rat Experiments
3.3. Real Patient Experiments
4. Results
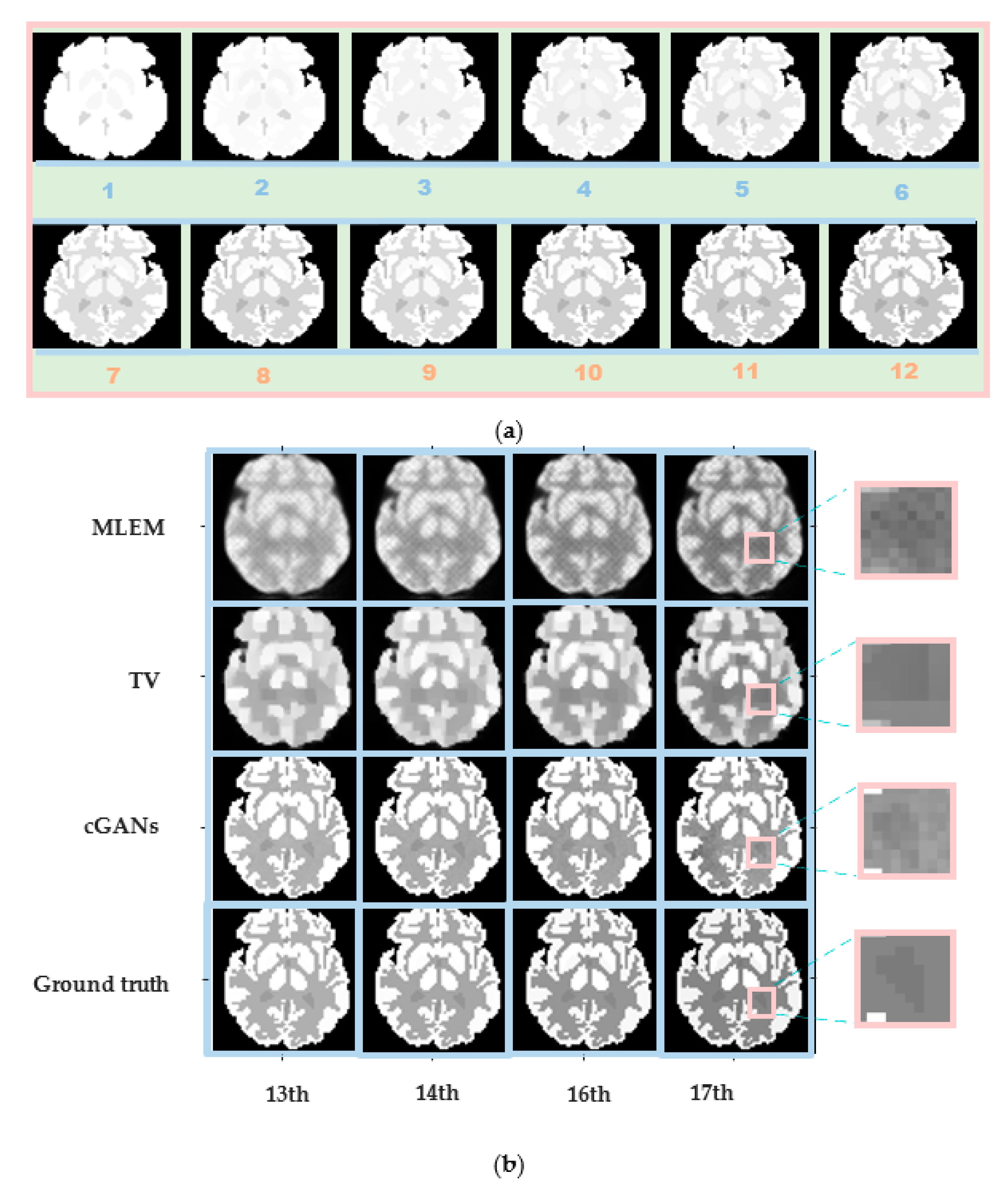
4.1. Simulation Experiments
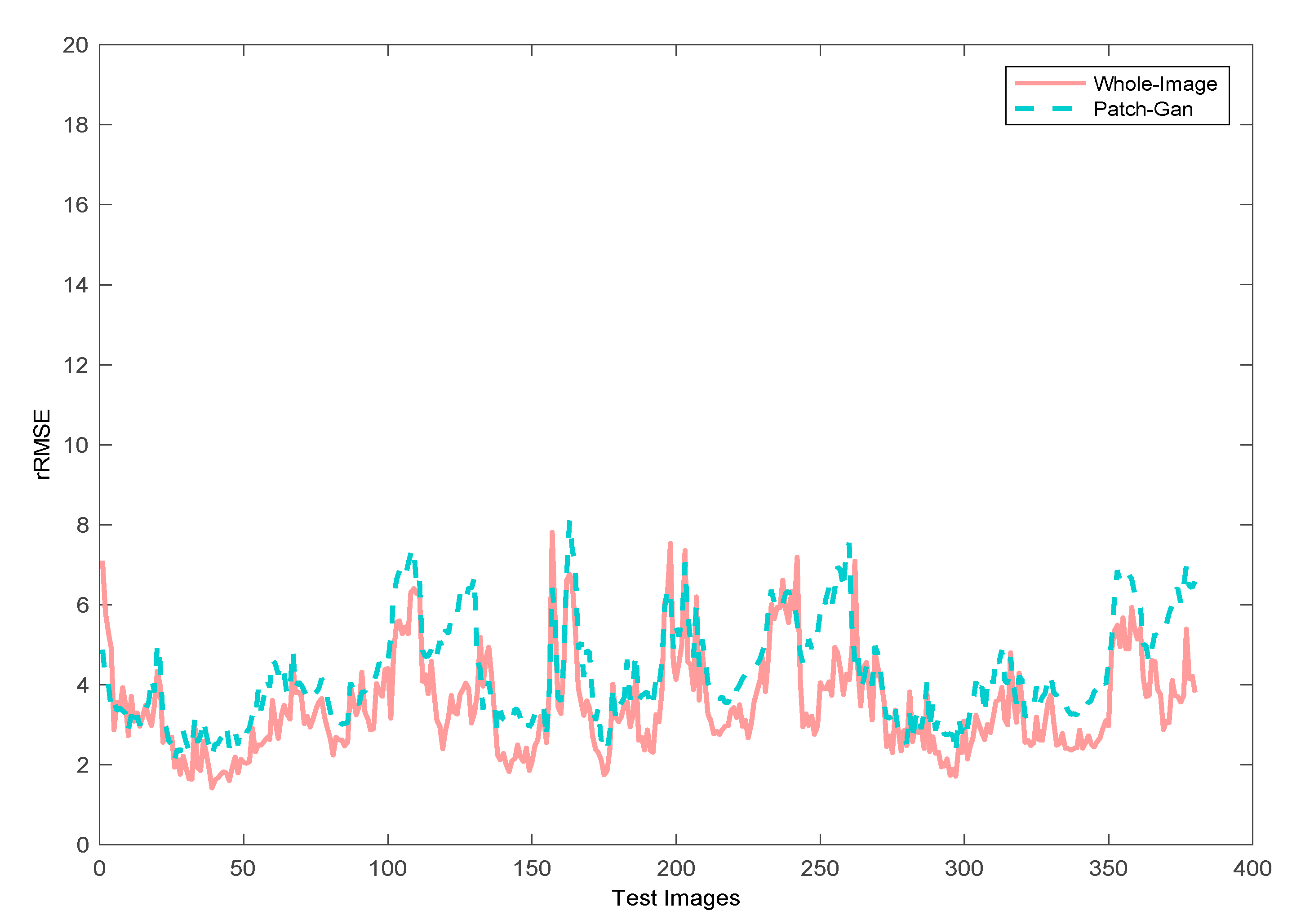
4.1.1. Discriminator Comparison Experiments
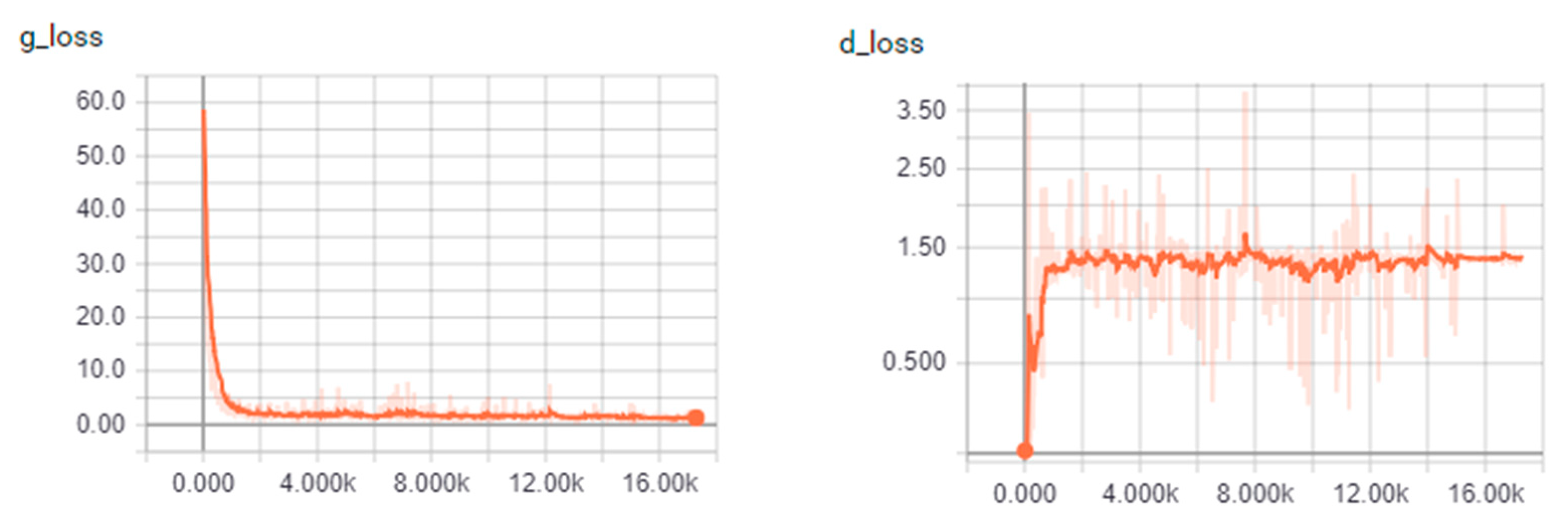
4.1.2. Convergence of the Algorithm
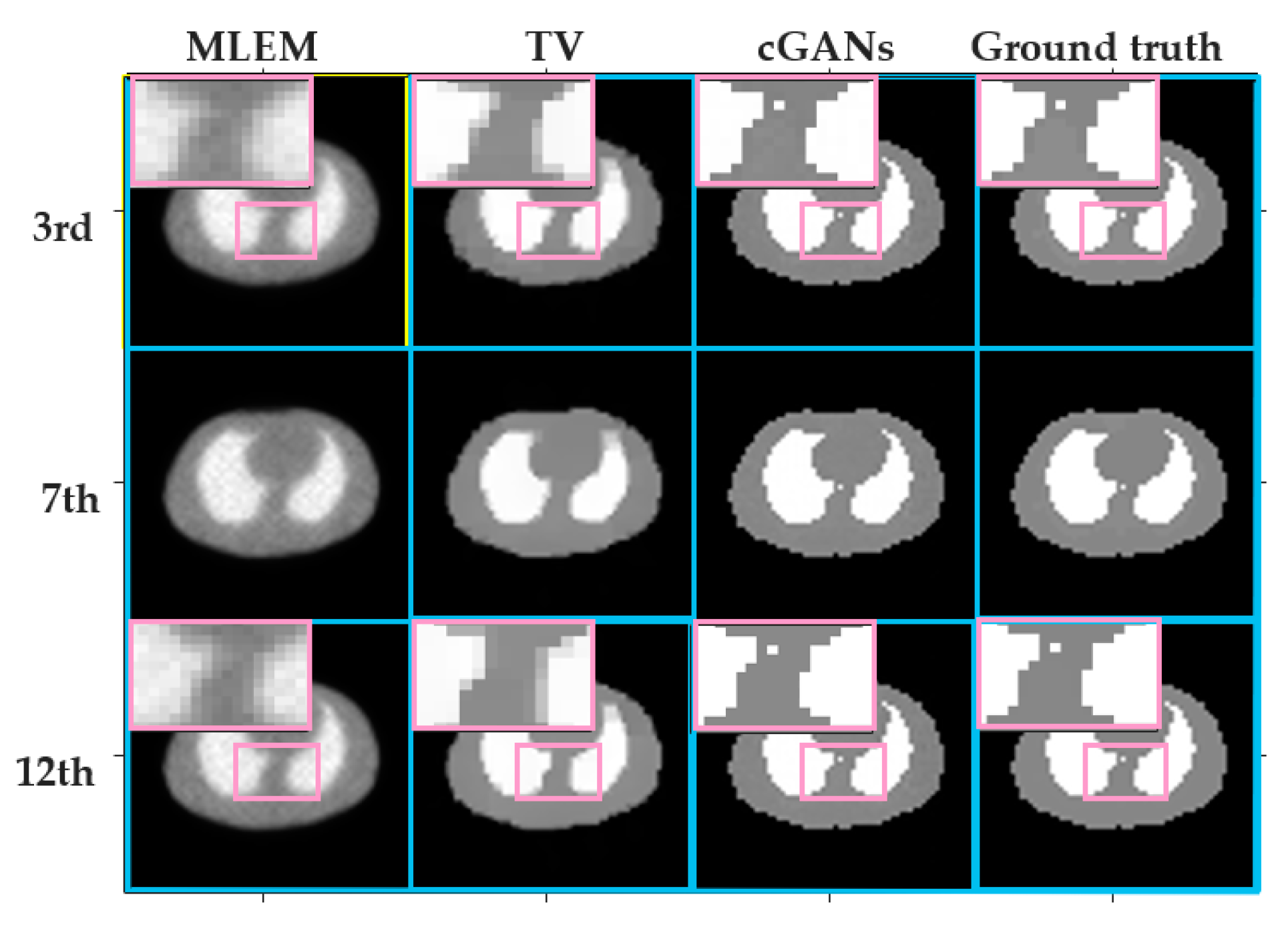
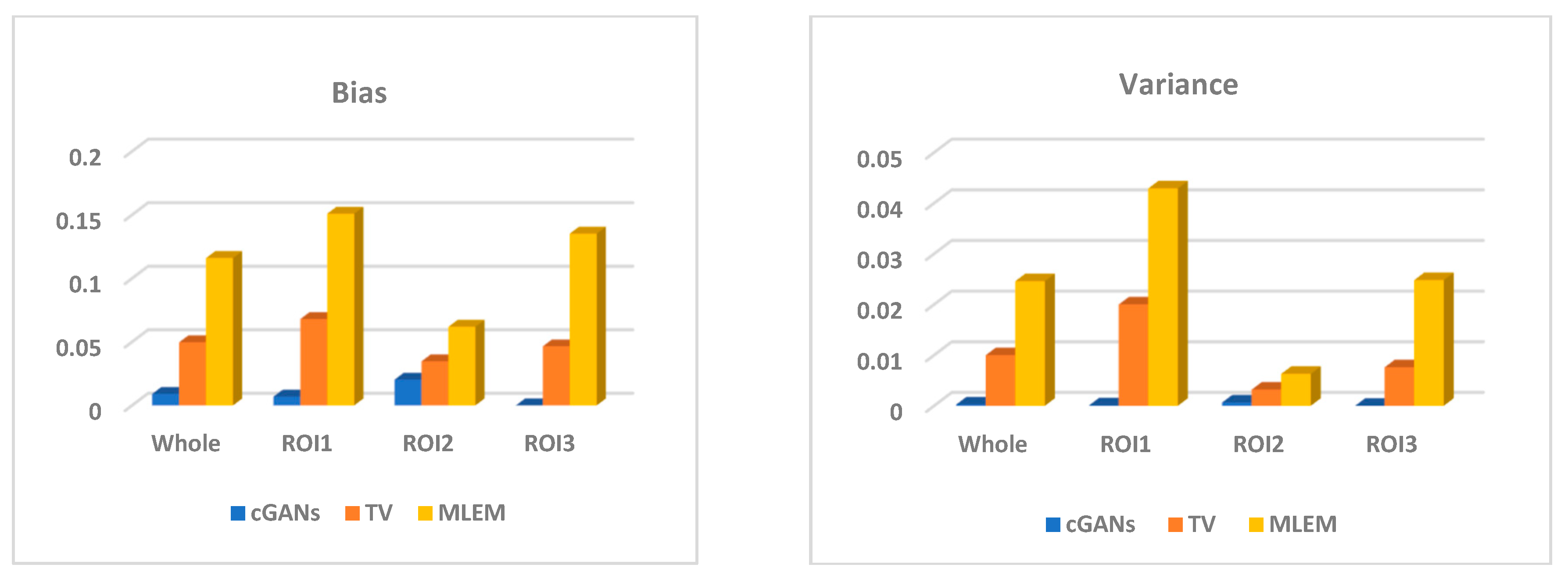
4.1.3. Accuracy
4.1.4. Robustness and Runtime Analysis
4.2. SD Rat Experiments
4.3. Real Patient Experiments
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kak, A.C.; Slaney, M.; Wang, G. Principles of Computerized Tomographic Imaging. Am. Assoc. Phys. Med. 2002, 29, 107. [Google Scholar] [CrossRef]
- Shepp, L.A.; Vardi, Y. Maximum Likelihood Reconstruction for Emission Tomography. IEEE Trans. Med. Imaging 1982, 1, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Levitan, E.; Herman, G.T. A Maximum a Posteriori Probability Expectation Maximization Algorithm for Image Reconstruction in Emission Tomography. IEEE Trans. Med. Imaging 1987, 6, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Luo, L. Sequential weighted least squares algorithm for PET image reconstruction. Digit. Signal Process. 2006, 16, 735–745. [Google Scholar] [CrossRef]
- Cabello, J.; Torres-Espallardo, I.; Gillam, J.E.; Rafecas, M. PET Reconstruction From Truncated Projections Using Total-Variation Regularization for Hadron Therapy Monitoring. IEEE Trans. Nucl. Sci. 2013, 60, 3364–3372. [Google Scholar] [CrossRef]
- Verhaeghe, J.; D’Asseler, Y.; Vandenberghe, S.; Staelens, S.; Lemahieu, I. An investigation of temporal regularization techniques for dynamic PET reconstructions using temporal splines. Med. Phys. 2007, 34, 1766–1778. [Google Scholar] [CrossRef]
- Marin, T.; Djebra, Y.; Han, P.K.; Chemli, Y.; Bloch, I.; El Fakhri, G.; Ouyang, J.; Petibon, Y.; Ma, C. Motion correction for PET data using subspace-based real-time MR imaging in simultaneous PET/MR. Phys. Med. Biol. 2020, 65, 235022. [Google Scholar] [CrossRef]
- Higaki, T.; Nakamura, Y.; Tatsugami, F.; Nakaura, T.; Awai, K. Improvement of image quality at CT and MRI using deep learning. Jpn. J. Radiol. 2018, 37, 73–80. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, B.; Wang, L.; Zu, C.; Lalush, D.S.; Lin, W.; Wu, X.; Zhou, J.; Shen, D.; Zhou, L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. NeuroImage 2018, 174, 550–562. [Google Scholar] [CrossRef]
- Liu, H.; Xu, J.; Wu, Y.; Guo, Q.; Ibragimov, B.; Xing, L. Learning deconvolutional deep neural network for high resolution medical image reconstruction. Inform. Sci. 2018, 468, 142–154. [Google Scholar] [CrossRef]
- Tezcan, K.C.; Baumgartner, C.F.; Luechinger, R.; Pruessmann, K.P.; Konukoglu, E. MR Image Reconstruction Using Deep Density Priors. IEEE Trans. Med. Imaging 2018, 38, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Luis, C.O.D.; Reader, A.J. Deep learning for suppression of resolution-recovery artefacts in MLEM PET image reconstruction. In Proceedings of the IEEE Nuclear Science Symposium and Medical Imaging Conference, NSS/MIC 2017, Atlanta, GA, USA, 21–28 October 2017. [Google Scholar]
- Xie, S.; Zheng, X.; Chen, Y.; Xie, L.; Liu, J.; Zhang, Y.; Yan, J.; Zhu, H.; Hu, Y. Artifact Removal using Improved GoogleNet for Sparse-view CT Reconstruction. Sci. Rep. 2018, 8, 6700–6709. [Google Scholar] [CrossRef]
- Kim, K.; Wu, D.; Gong, K.; Dutta, J.; Kim, J.H.; Son, Y.D.; Kim, H.K.; El Fakhri, G.; Li, Q. Penalized PET Reconstruction Using Deep Learning Prior and Local Linear Fitting. IEEE Trans. Med. Imaging 2018, 37, 1478–1487. [Google Scholar] [CrossRef]
- Hong, X.; Zan, Y.; Weng, F.; Tao, W.; Peng, Q.; Huang, Q. Enhancing the Image Quality via Transferred Deep Residual Learning of Coarse PET Sinograms. IEEE Trans. Med. Imaging 2018, 37, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yoo, J.; Kim, H.H.; Shin, H.J.; Sung, K.; Ye, J.C. Deep learning with domain adaptation for accelerated projection-reconstruction MR. Magn. Reson. Med. 2018, 80, 1189–1205. [Google Scholar] [CrossRef] [PubMed]
- Hammernik, K.; Klatzer, T.; Kobler, E.; Recht, M.P.; Sodickson, D.; Pock, T.; Knoll, F. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 2017, 79, 3055–3071. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Su, Z.; Ying, L.; Peng, X.; Zhu, S.; Liang, F.; Feng, D.; Liang, D. Accelerating magnetic resonance imaging via deep learning. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016. [Google Scholar]
- Kang, E.; Min, J.; Ye, J.C. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med. Phys. 2017, 44, e360–e375. [Google Scholar] [CrossRef]
- Wu, D.; Kim, K.; Fakhri, G.E.; Li, Q. A Cascaded Convolutional Neural Network for X-ray Low-dose CT Image Denoising. arXiv 2017, arXiv:1705.04267. [Google Scholar]
- Wang, T.; Lei, Y.; Fu, Y.; Wynne, J.F.; Curran, W.J.; Liu, T.; Yang, X. A review on medical imaging synthesis using deep learning and its clinical applications. J. Appl. Clin. Med. Phys. 2021, 22, 11–36. [Google Scholar] [CrossRef]
- Cheng, Z.; Wen, J.; Huang, G.; Yan, J. Applications of artificial intelligence in nuclear medicine image generation. Quant. Imaging Med. Surg. 2021, 11, 2792–2822. [Google Scholar] [CrossRef]
- Kawauchi, K.; Furuya, S.; Hirata, K.; Katoh, C.; Manabe, O.; Kobayashi, K.; Watanabe, S.; Shiga, T. A convolutional neural network-based system to classify patients using FDG PET/CT examinations. BMC Cancer 2020, 20, 227. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Fulham, M.; Feng, D.; Kim, J. Co-Learning Feature Fusion Maps From PET-CT Images of Lung Cancer. IEEE Trans. Med. Imaging 2019, 39, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Protonotarios, N.E.; Katsamenis, I.; Sykiotis, S.; Dikaios, N.; Kastis, G.A.; Chatziioannou, S.N.; Metaxas, M.; Doulamis, N.; Doulamis, A. A few-shot U-Net deep learning model for lung cancer lesion segmentation via PET/CT imaging. Biomed. Phys. Eng. Express 2022, 8, 025019. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kim, J.; Wen, L.; Fulham, M.; Feng, D. A graph-based approach for the retrieval of multi-modality medical images. Med Image Anal. 2014, 18, 330–342. [Google Scholar] [CrossRef]
- Gong, K.; Guan, J.; Kim, K.; Zhang, X.; Yang, J.; Seo, Y.; El Fakhri, G.; Qi, J.; Li, Q. Iterative PET Image Reconstruction Using Convolutional Neural Network Representation. IEEE Trans. Med. Imaging 2018, 38, 675–685. [Google Scholar] [CrossRef]
- Gong, K.; Catana, C.; Qi, J.; Li, Q. Direct Reconstruction of Linear Parametric Images From Dynamic PET Using Nonlocal Deep Image Prior. IEEE Trans. Med. Imaging 2022, 41, 680–689. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, H.; Duan, X.; Hong, X.; Sun, H.; Lv, W.; Lu, L.; Feng, Q. GapFill-recon net: A cascade network for simultaneously PET gap filling and image reconstruction. Comput. Methods Programs Biomed. 2021, 208, 106271. [Google Scholar] [CrossRef]
- Lempitsky, V.; Vedaldi, A.; Ulyanov, D. Deep image prior. In Proceedings of the 2018 IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–13 June 2018. [Google Scholar]
- Gong, K.; Catana, C.; Qi, J.; Li, Q. PET Image Reconstruction Using Deep Image Prior. IEEE Trans. Med. Imaging 2018, 38, 1655–1665. [Google Scholar] [CrossRef]
- Song, T.-A.; Chowdhury, S.R.; Yang, F.; Dutta, J. Super-Resolution PET Imaging Using Convolutional Neural Networks. IEEE Trans. Comput. Imaging 2020, 6, 518–528. [Google Scholar] [CrossRef]
- Häggström, I.; Schmidtlein, C.R.; Campanella, G.; Fuchs, T.J. DeepPET: A deep encoder–decoder network for directly solving the PET image reconstruction inverse problem. Med. Image Anal. 2019, 54, 253–262. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Isola, P.; Zhu, J.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar]
- Liu, Z.; Chen, H.; Liu, H. Deep Learning Based Framework for Direct Reconstruction of PET Images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, 22nd International Conference, Shenzhen, China, 13–17 October 2019; pp. 48–56. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Bengio, Y.; Courville, A. Generative adversarial nets. Adv. Neural Inform. Processing Syst. 2014, 27, 1–9. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Fei, G.; Ryoko, Y.; Mitsuo, W.; Hua-Feng, L. An effective scatter correction method based on single scatter simulation for a 3D whole-body PET scanner. Chin. Phys. B 2009, 18, 3066–3072. [Google Scholar] [CrossRef]
- Koeppe, R.A.; Frey, K.A.; Borght, T.M.V.; Karlamangla, A.; Jewett, D.M.; Lee, L.C.; Kilbourn, M.R.; Kuhl, D.E. Kinetic Evaluation of [11C]Dihydrotetrabenazine by Dynamic PET: Measurement of Vesicular Monoamine Transporter. J. Cereb. Blood Flow Metab. 1996, 16, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Muzi, M.; Spence, A.M.; O’Sullivan, F.; Mankoff, D.A.; Wells, J.M.; Grierson, J.R.; Link, J.M.; Krohn, K.A. Kinetic analysis of 3′-deoxy-3′-18F-fluorothymidine in patients with gliomas. J. Nucl. Med. 2006, 47, 1612. [Google Scholar] [PubMed]
- Tong, S.; Shi, P. Tracer kinetics guided dynamic PET reconstruction. In Proceedings of the Biennial International Conference on Information Processing in Medical Imaging, Kerkrade, The Netherlands, 2–6 July 2007; Springer: Berlin/Heidelberg, Germany, 2007; p. 421. [Google Scholar]





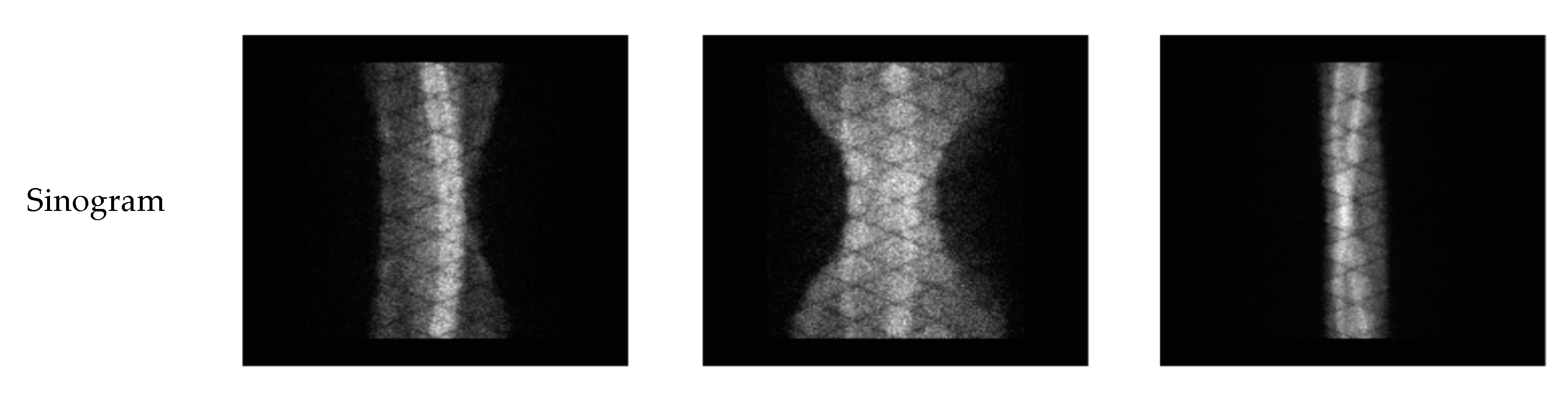



| Phantom | Sampling Time | Sampling Interval | Counts | Dataset |
|---|---|---|---|---|
| Zubal thorax | 20 min | 14 × 50, 2 × 100, 2 × 150 s | 5 × 106 | training |
| 1 × 107 | ||||
| 5 × 107 | ||||
| 30 min | 10 × 30, 2 × 150, 6 × 200 s | 5 × 106 | training | |
| 1 × 107 | ||||
| 5 × 107 | ||||
| 40 min | 10 × 30, 5 × 240, 3 × 300 s | 5 × 106 | testing | |
| 1 × 107 | ||||
| 5 × 107 | ||||
| Zubal head | 20 min | 12 × 50 s | 5 × 106 | training |
| 2 × 50, 2 × 100, 2 × 150 s | 1 × 107 | testing | ||
| 5 × 107 | ||||
| 30 min | 5 × 30, 7 × 100 s | 5 × 106 | training | |
| 1 × 100, 3 × 150, 2 × 200 s | 1 × 107 | testing | ||
| 5 × 107 | ||||
| 40 min | 5 × 30, 7 × 150 s | 5 × 106 | training | |
| 6 × 200 s | 1 × 107 | testing | ||
| 5 × 107 |
| Frame | Method | Bias | Variance | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | ROI1 | ROI2 | ROI3 | Total | ROI1 | ROI2 | ROI3 | ||
| 3rd | MLEM | 0.1225 | 0.1739 | 0.0654 | 0.1284 | 0.0271 | 0.0510 | 0.0070 | 0.0233 |
| TV | 0.0611 | 0.0909 | 0.0503 | 0.0420 | 0.0110 | 0.0227 | 0.0040 | 0.0063 | |
| cGANs | 0.0189 | 0.0112 | 0.0454 | 2.0 × 10−5 | 8.1 × 10−4 | 2.4 × 10−4 | 0.0022 | 7.8 × 10−8 | |
| 7th | MLEM | 0.1034 | 0.1335 | 0.0547 | 0.1221 | 0.0204 | 0.0356 | 0.0047 | 0.0210 |
| TV | 0.0407 | 0.0586 | 0.0189 | 0.0446 | 0.0086 | 0.0168 | 0.0014 | 0.0077 | |
| cGANs | 0.0106 | 0.0066 | 0.0252 | 2.0 × 10−5 | 2.7 × 10−4 | 1.0 × 10−4 | 7.2 × 10−4 | 7.8 × 10−8 | |
| 12th | MLEM | 0.1049 | 0.1325 | 0.0573 | 0.1249 | 0.0208 | 0.0355 | 0.0055 | 0.0215 |
| TV | 0.0457 | 0.0631 | 0.0342 | 0.0399 | 0.0089 | 0.0173 | 0.0025 | 0.0065 | |
| cGANs | 0.0042 | 0.0058 | 0.0068 | 1.0 × 10−5 | 6.7 × 10−5 | 8.9 × 10−5 | 1.1 × 10−4 | 3.9 × 10−8 | |
| 18th | MLEM | 0.1211 | 0.1545 | 0.0649 | 0.1439 | 0.0263 | 0.0444 | 0.0068 | 0.0277 |
| TV | 0.0531 | 0.0689 | 0.0320 | 0.0584 | 0.0109 | 0.0195 | 0.0031 | 0.0103 | |
| cGANs | 0.0055 | 0.0067 | 0.0098 | 2.0 × 10−5 | 1.0 × 10−4 | 1.1 × 10−5 | 2.0 × 10−4 | 7.8 × 10−8 | |
| Frame | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | Mean | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | |||||||||||
| MLEM | 0.213 s | 0.211 s | 0.204 s | 0.215 s | 0.218 s | 0.210 s | 0.207 s | 0.217 s | 0.202 s | 0.211 s | |
| TV | 0.508 s | 0.449 s | 0.504 s | 0.542 s | 0.522 s | 0.525 s | 0.527 s | 0.519 s | 0.511 s | 0.512 s | |
| CGANs | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | |
| Frame | 13 | 14 | 15 | 16 | 17 | 18 | Mean | |
|---|---|---|---|---|---|---|---|---|
| Method | ||||||||
| MLEM | 0.219 s | 0.268 s | 0.226 s | 0.233 s | 0.260 s | 0.260 s | 0.244 s | |
| TV | 1.005 s | 1.120 s | 1.088 s | 1.176 s | 1.153 s | 1.216 s | 1.126 s | |
| CGANs | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | 0.007 s | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Ye, H.; Liu, H. Deep-Learning-Based Framework for PET Image Reconstruction from Sinogram Domain. Appl. Sci. 2022, 12, 8118. https://doi.org/10.3390/app12168118
Liu Z, Ye H, Liu H. Deep-Learning-Based Framework for PET Image Reconstruction from Sinogram Domain. Applied Sciences. 2022; 12(16):8118. https://doi.org/10.3390/app12168118
Chicago/Turabian StyleLiu, Zhiyuan, Huihui Ye, and Huafeng Liu. 2022. "Deep-Learning-Based Framework for PET Image Reconstruction from Sinogram Domain" Applied Sciences 12, no. 16: 8118. https://doi.org/10.3390/app12168118
APA StyleLiu, Z., Ye, H., & Liu, H. (2022). Deep-Learning-Based Framework for PET Image Reconstruction from Sinogram Domain. Applied Sciences, 12(16), 8118. https://doi.org/10.3390/app12168118







