A Progressively Expanded Database for Automated Lung Sound Analysis: An Update
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
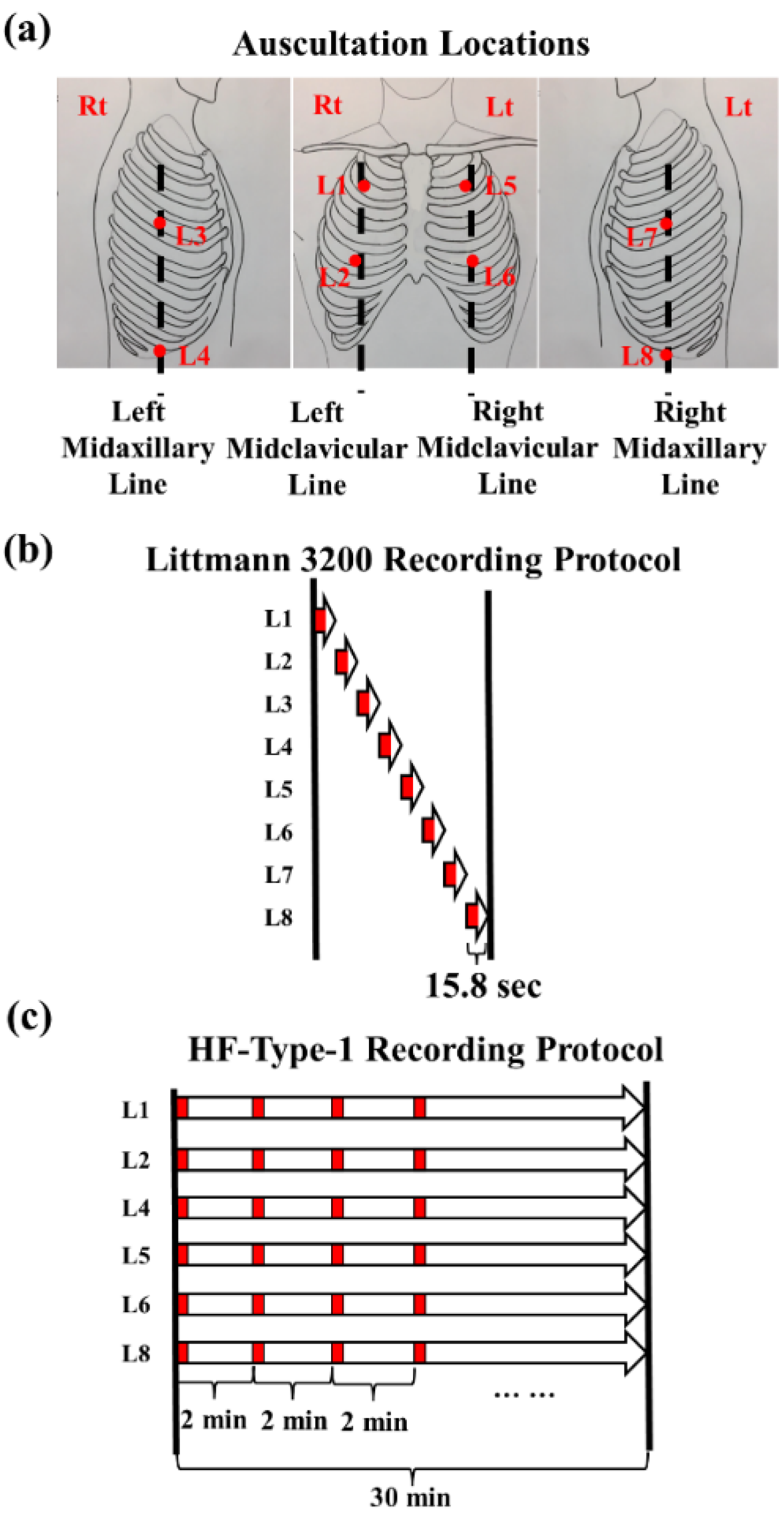
2.2. Lung Sound Recordings
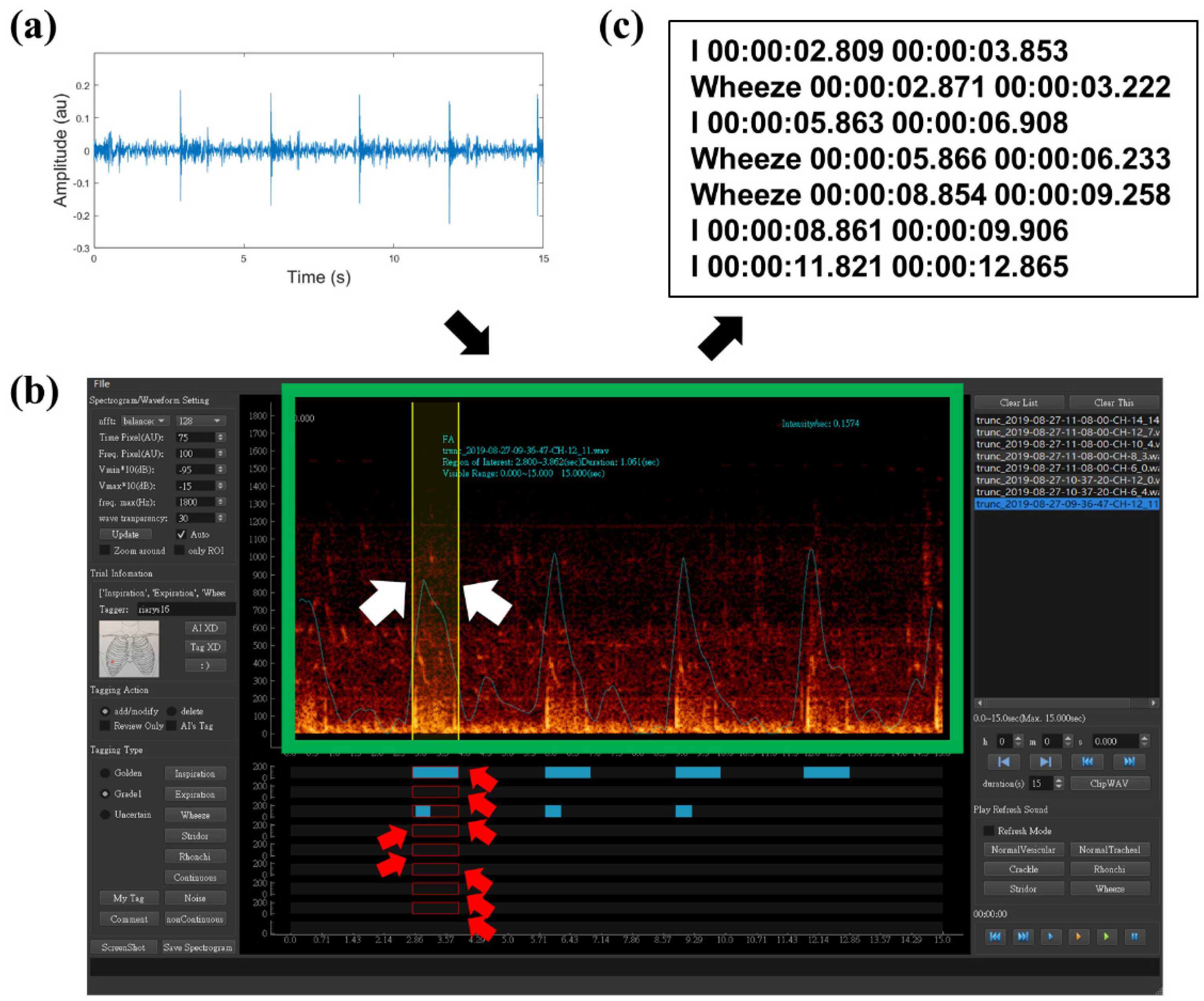
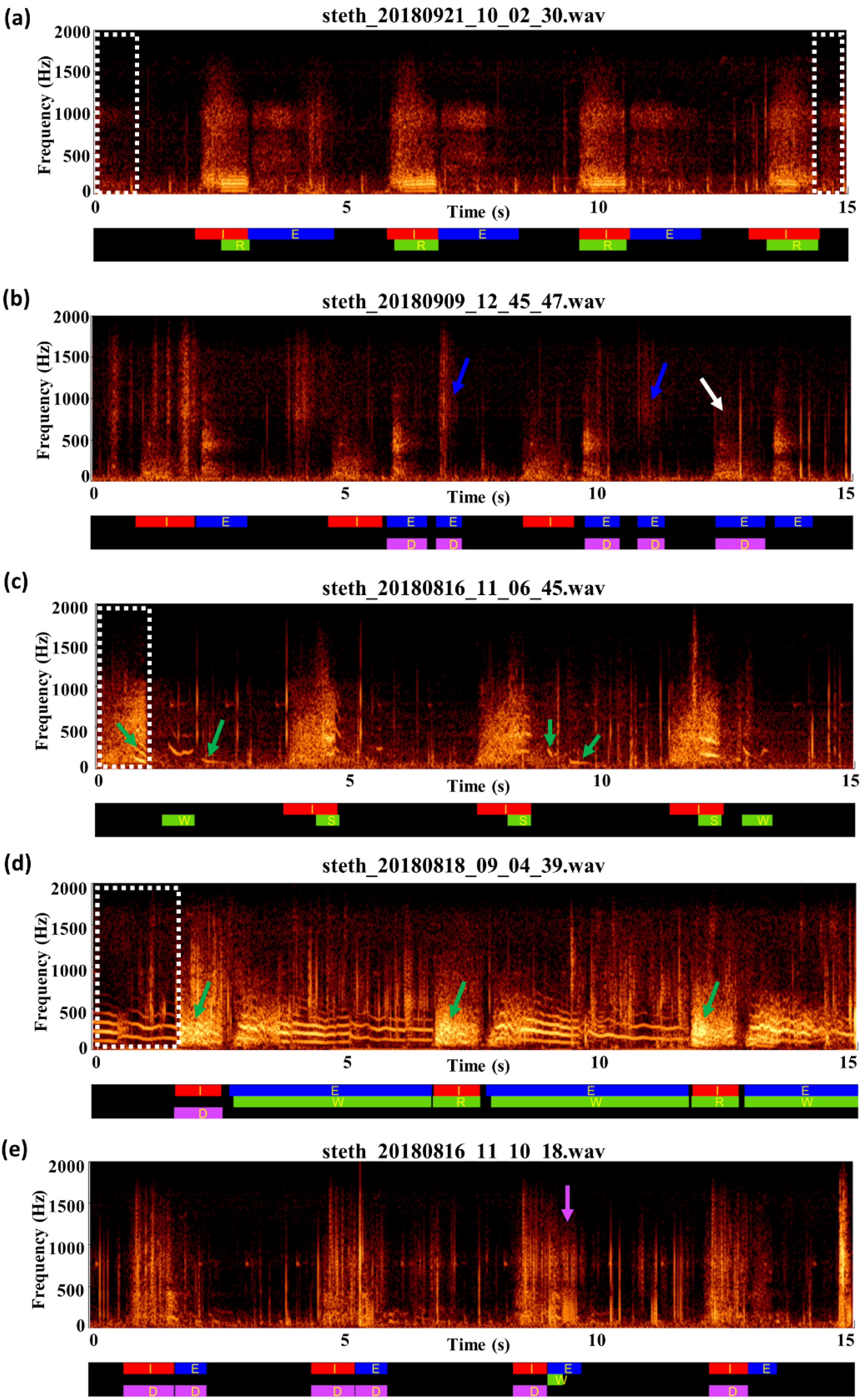
2.3. Data Preparation and Labeling
2.4. Dataset Arrangement
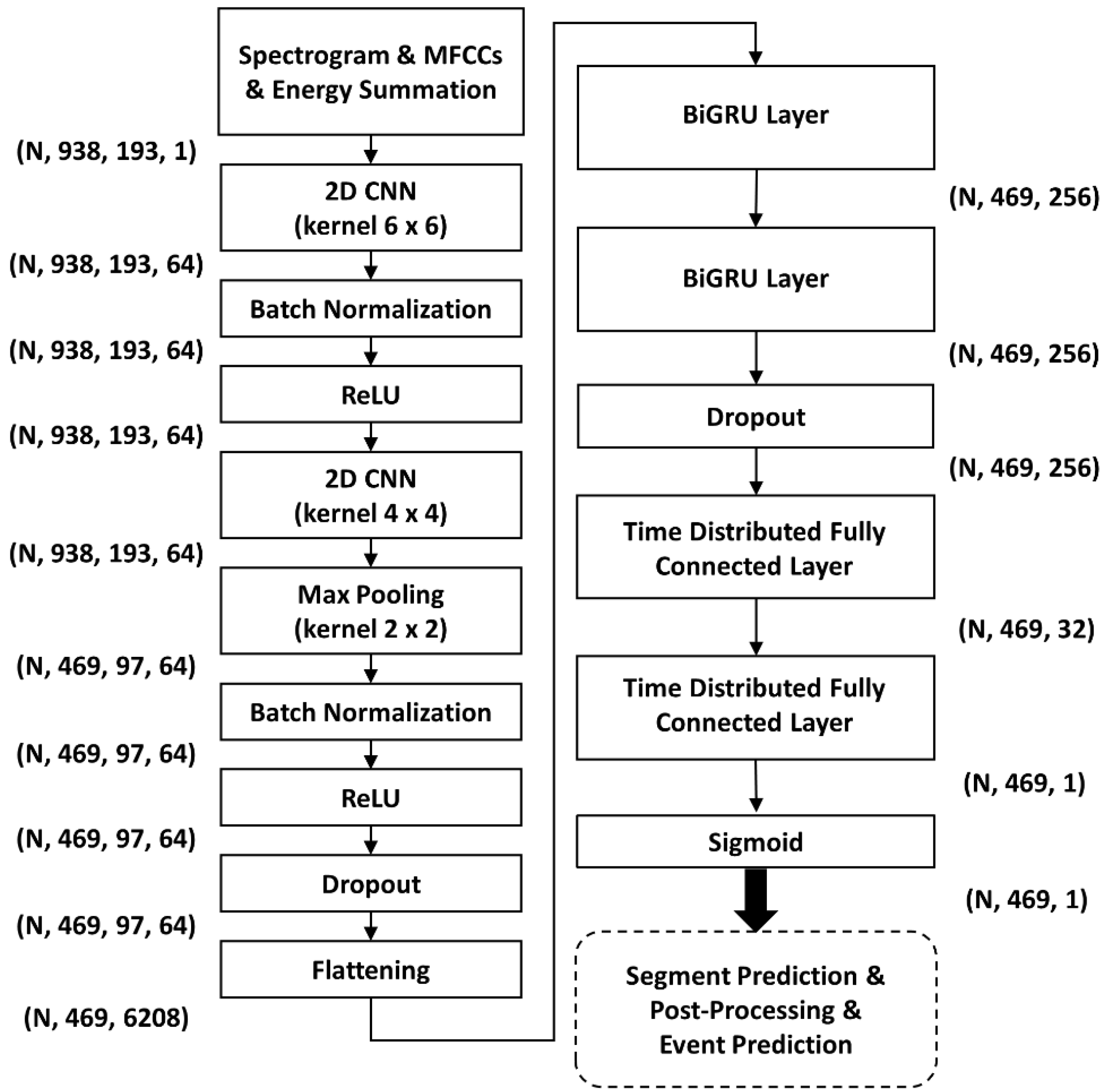
2.5. Benchmarking of HF_Lung_V2
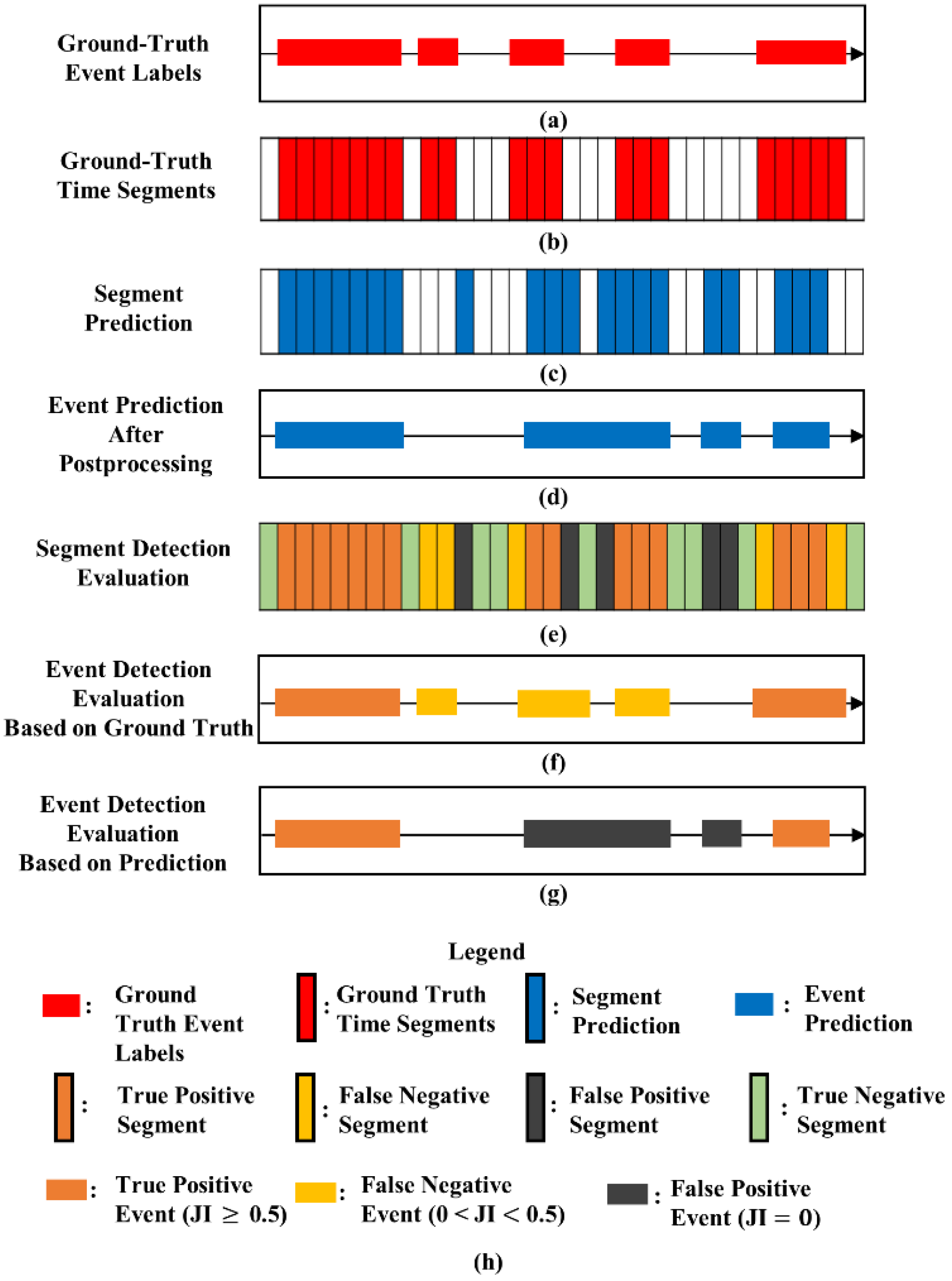
2.6. Performance Evaluation
2.7. Investigation of Label Quality and Sound Overlapping
3. Results
3.1. Summary of HF_Lung_V1 and HF_Lung_V2 Databases
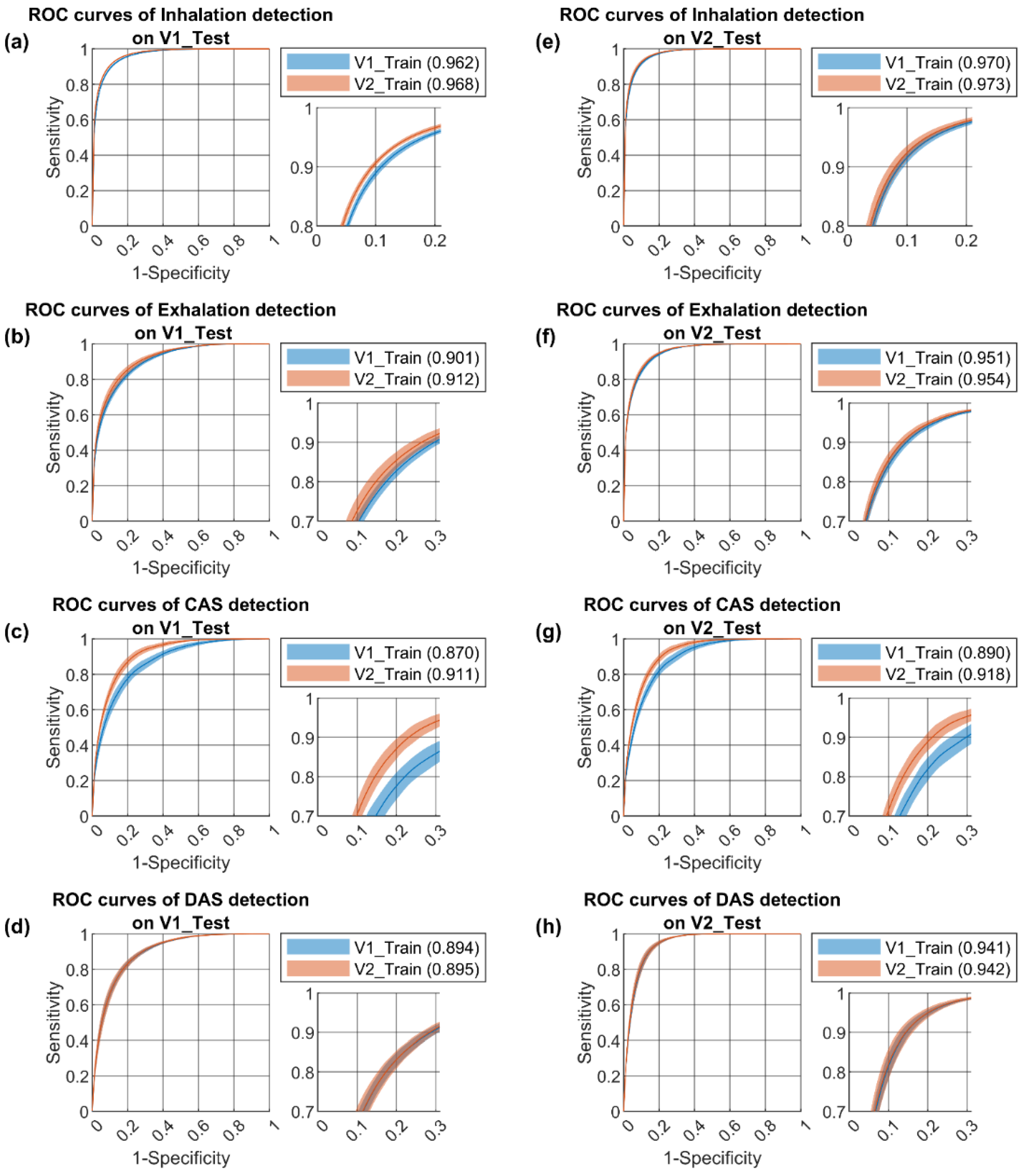
3.2. Performance Benchmark
3.3. Review of Label Quality
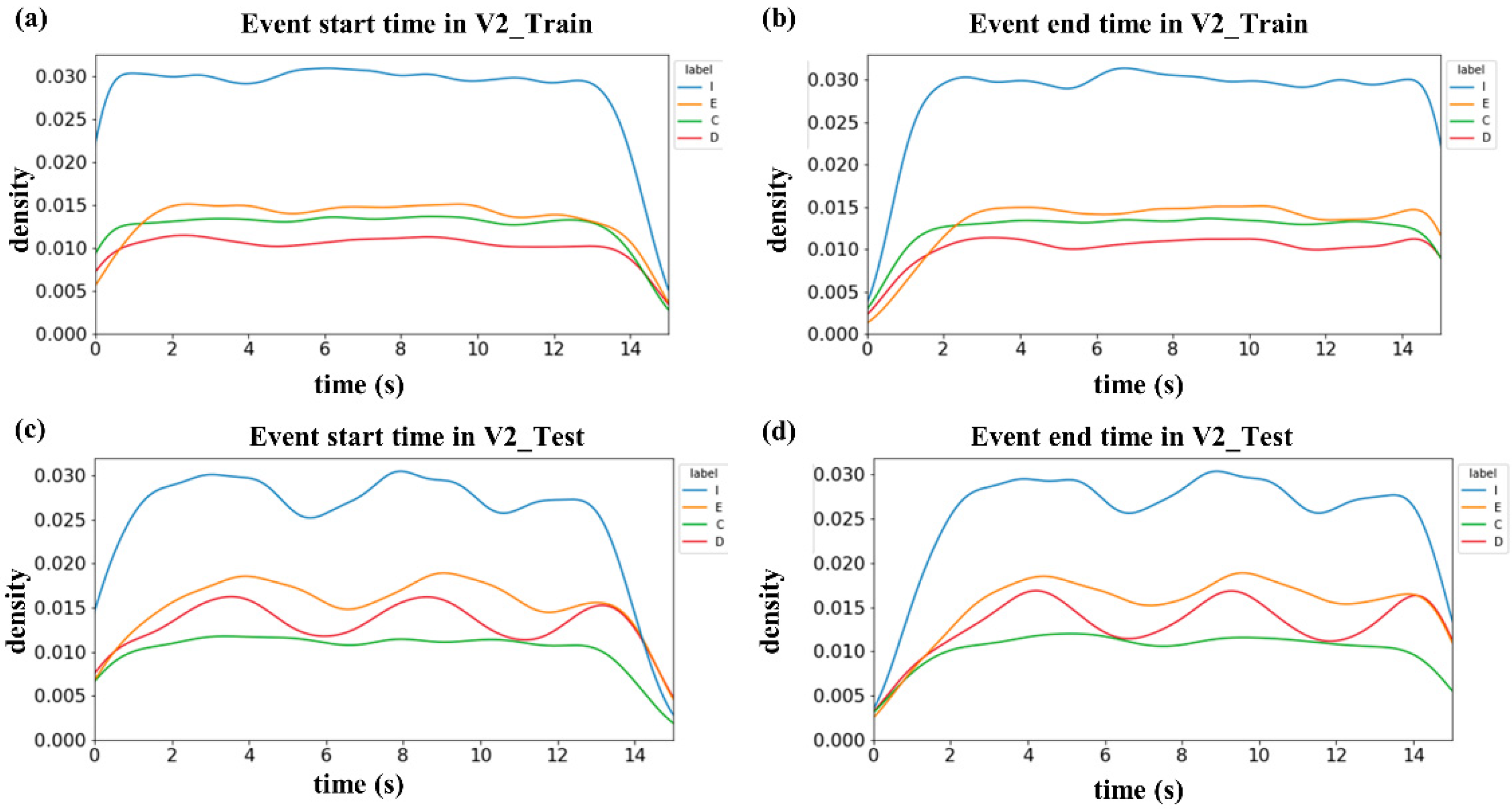
3.4. Overlap Ratios between Labels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bohadana, A.; Izbicki, G.; Kraman, S.S. Fundamentals of lung auscultation. N. Engl. J. Med. 2014, 370, 744–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouzas, S.; Anthracopoulos, M.B.; Bohadana, A. Clinical Usefulness of Breath Sounds. In Breath Sounds; Priftis, K.N., Hadjileontiadis, L.J., Everard, M.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 33–52. [Google Scholar]
- Kim, Y.; Hyon, Y.; Lee, S.; Woo, S.-D.; Ha, T.; Chung, C. The coming era of a new auscultation system for analyzing respiratory sounds. BMC Pulm. Med. 2022, 22, 119. [Google Scholar] [CrossRef]
- Gurung, A.; Scrafford, C.G.; Tielsch, J.M.; Levine, O.S.; Checkley, W. Computerized lung sound analysis as diagnostic aid for the detection of abnormal lung sounds: A systematic review and meta-analysis. Respir. Med. 2011, 105, 1396–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pramono, R.X.A.; Bowyer, S.; Rodriguez-Villegas, E. Automatic adventitious respiratory sound analysis: A systematic review. PLoS ONE 2017, 12, e0177926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthusamy, P.D.; Sundaraj, K.; Abd Manap, N. Computerized acoustical techniques for respiratory flow-sound analysis: A systematic review. Artif. Intell. Rev. 2020, 53, 3501–3574. [Google Scholar] [CrossRef]
- Rocha, B.M.; Pessoa, D.; Marques, A.; Carvalho, P.; Paiva, R.P. Automatic classification of adventitious respiratory sounds: A (un) solved problem? Sensors 2020, 21, 57. [Google Scholar] [CrossRef] [PubMed]
- Demir, F.; Sengur, A.; Bajaj, V. Convolutional neural networks based efficient approach for classification of lung diseases. Health Inf. Sci. Syst. 2020, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Fraiwan, M.; Fraiwan, L.; Alkhodari, M.; Hassanin, O. Recognition of pulmonary diseases from lung sounds using convolutional neural networks and long short-term memory. J. Ambient Intell. Humaniz. Comput. 2021, 1–13. [Google Scholar] [CrossRef]
- Aykanat, M.; Kılıç, Ö.; Kurt, B.; Saryal, S. Classification of lung sounds using convolutional neural networks. EURASIP J. Image Video Process. 2017, 2017, 65. [Google Scholar] [CrossRef]
- Bardou, D.; Zhang, K.; Ahmad, S.M. Lung sounds classification using convolutional neural networks. Artif. Intell. Med. 2018, 88, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-Y.; Liao, C.-H.; Wu, Y.-S.; Yuan, S.-M.; Sun, C.-T. Efficiently classifying lung sounds through depthwise separable CNN models with fused STFT and MFCC features. Diagnostics 2021, 11, 732. [Google Scholar] [CrossRef] [PubMed]
- Petmezas, G.; Cheimariotis, G.-A.; Stefanopoulos, L.; Rocha, B.; Paiva, R.P.; Katsaggelos, A.K.; Maglaveras, N. Automated Lung Sound Classification Using a Hybrid CNN-LSTM Network and Focal Loss Function. Sensors 2022, 22, 1232. [Google Scholar] [CrossRef] [PubMed]
- García-Ordás, M.T.; Benítez-Andrades, J.A.; García-Rodríguez, I.; Benavides, C.; Alaiz-Moretón, H. Detecting respiratory pathologies using convolutional neural networks and variational autoencoders for unbalancing data. Sensors 2020, 20, 1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gairola, S.; Tom, F.; Kwatra, N.; Jain, M. Respirenet: A deep neural network for accurately detecting abnormal lung sounds in limited data setting. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 527–530. [Google Scholar]
- Pham, L.; Phan, H.; Palaniappan, R.; Mertins, A.; McLoughlin, I. Cnn-moe based framework for classification of respiratory anomalies and lung disease detection. IEEE J. Biomed. Health Inform. 2021, 25, 2938–2947. [Google Scholar] [CrossRef]
- Minami, K.; Lu, H.; Kim, H.; Mabu, S.; Hirano, Y.; Kido, S. Automatic classification of large-scale respiratory sound dataset based on convolutional neural network. In Proceedings of the 2019 19th International Conference on Control, Automation and Systems (ICCAS), Jeju, Korea, 15–18 October 2019; pp. 804–807. [Google Scholar]
- Hsu, F.-S.; Huang, S.-R.; Huang, C.-W.; Huang, C.-J.; Cheng, Y.-R.; Chen, C.-C.; Hsiao, J.; Chen, C.-W.; Chen, L.-C.; Lai, Y.-C.; et al. Benchmarking of eight recurrent neural network variants for breath phase and adventitious sound detection on a self-developed open-access lung sound database-HF_Lung_V1. PLoS ONE 2021, 16, e0254134. [Google Scholar] [CrossRef]
- Messner, E.; Fediuk, M.; Swatek, P.; Scheidl, S.; Smolle-Juttner, F.-M.; Olschewski, H.; Pernkopf, F. Crackle and breathing phase detection in lung sounds with deep bidirectional gated recurrent neural networks. In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 356–359. [Google Scholar]
- Perna, D.; Tagarelli, A. Deep auscultation: Predicting respiratory anomalies and diseases via recurrent neural networks. In Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), Cordoba, Spain, 5–7 June 2019; pp. 50–55. [Google Scholar]
- Kochetov, K.; Putin, E.; Balashov, M.; Filchenkov, A.; Shalyto, A. Noise masking recurrent neural network for respiratory sound classification. In Proceedings of the International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; pp. 208–217. [Google Scholar]
- Chen, H.; Yuan, X.; Pei, Z.; Li, M.; Li, J. Triple-classification of respiratory sounds using optimized s-transform and deep residual networks. IEEE Access 2019, 7, 32845–32852. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, X.; Yu, Q.; Zhang, Y.; Li, Y.; Zhao, J.; Wang, G. LungBRN: A smart digital stethoscope for detecting respiratory disease using bi-resnet deep learning algorithm. In Proceedings of the 2019 IEEE Biomedical Circuits and Systems Conference (BioCAS), Nara, Japan, 17–19 October 2019; pp. 1–4. [Google Scholar]
- Fernando, T.; Sridharan, S.; Denman, S.; Ghaemmaghami, H.; Fookes, C. Robust and Interpretable Temporal Convolution Network for Event Detection in Lung Sound Recordings. IEEE J. Biomed. Health Inform. 2022, 26, 2898–2908. [Google Scholar] [CrossRef]
- Jácome, C.; Ravn, J.; Holsbø, E.; Aviles-Solis, J.C.; Melbye, H.; Ailo Bongo, L. Convolutional neural network for breathing phase detection in lung sounds. Sensors 2019, 19, 1798. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Liu, S.; Song, M.; Parada-Cabaleiro, E.; Schuller, B.W. Adventitious respiratory classification using attentive residual neural networks. In Proceedings of the 21st Annual Conference of the International Speech Communication Association (Interspeech 2020), Shanghai, China, 25–29 October 2020; pp. 2912–2916. [Google Scholar]
- Zhao, X.; Shao, Y.; Mai, J.; Yin, A.; Xu, S. Respiratory Sound Classification Based on BiGRU-Attention Network with XGBoost. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020; pp. 915–920. [Google Scholar]
- Rocha, B.M.; Filos, D.; Mendes, L.; Serbes, G.; Ulukaya, S.; Kahya, Y.P.; Jakovljevic, N.; Turukalo, T.L.; Vogiatzis, I.M.; Perantoni, E. An open access database for the evaluation of respiratory sound classification algorithms. Physiol. Meas. 2019, 40, 035001. [Google Scholar] [CrossRef]
- Fraiwan, M.; Fraiwan, L.; Khassawneh, B.; Ibnian, A. A dataset of lung sounds recorded from the chest wall using an electronic stethoscope. Data Brief 2021, 35, 106913. [Google Scholar] [CrossRef] [PubMed]
- Hestness, J.; Narang, S.; Ardalani, N.; Diamos, G.; Jun, H.; Kianinejad, H.; Patwary, M.; Ali, M.; Yang, Y.; Zhou, Y. Deep learning scaling is predictable, empirically. arXiv 2017, arXiv:1712.00409. [Google Scholar]
- Hsu, F.-S.; Huang, C.-J.; Kuo, C.-Y.; Huang, S.-R.; Cheng, Y.-R.; Wang, J.-H.; Wu, Y.-L.; Tzeng, T.-L.; Lai, F. Development of a Respiratory Sound Labeling Software for Training a Deep Learning-Based Respiratory Sound Analysis Model. arXiv 2021, arXiv:2101.01352. [Google Scholar]
- Rasamoelina, A.D.; Adjailia, F.; Sinčák, P. A review of activation function for artificial neural network. In Proceedings of the 2020 IEEE 18th World Symposium on Applied Machine Intelligence and Informatics (SAMI), Herlany, Slovakia, 23–25 January 2020; pp. 281–286. [Google Scholar]
- Das, P.P.; Allayear, S.M.; Amin, R.; Rahman, Z. Bangladeshi dialect recognition using Mel frequency cepstral coefficient, delta, delta-delta and Gaussian mixture model. In Proceedings of the 2016 Eighth International Conference on Advanced Computational Intelligence (ICACI), Chiang Mai, Thailand, 14–16 February 2016; pp. 359–364. [Google Scholar]
- Nakano, H.; Furukawa, T.; Tanigawa, T. Tracheal sound analysis using a deep neural network to detect sleep apnea. J. Clin. Sleep Med. 2019, 15, 1125–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Subjects from RCC/RCW | Subjects from FEMH | Subjects in TSECCM Database | |
|---|---|---|---|
| Number (n) | 25 | 32 | 243 |
| Sex (M/F/NA) | 16/8/1 | 19/13 | NA |
| Age | 64.5 ± 18.5 | 68.2 ± 11.7 | NA |
| Height (cm) | 162.3 ± 9.1 * | 160.0 ± 8.8 | NA |
| Weight (kg) | 63.4 ± 12.7 * | 59.7 ± 12.3 | NA |
| BMI (kg/m2) | 24.0 ± 4.3 * | 23.6 ± 4.6 | NA |
| Recording device | HF-Type-1 & Littmann 3200 | Littmann 3200 | Littmann 3200 |
| Respiratory disease | |||
| Acute exacerbation of chronic obstructive pulmonary disease | 1 (4%) | 1 (3%) | NA |
| Acute respiratory distress syndrome | 1 (4%) | 0 (0%) | NA |
| Acute respiratory failure | 4 (16%) | 0 (0%) | NA |
| Asthma | 0 (0%) | 1 (3%) | NA |
| Bronchitis | 1 (4%) | 0 (0%) | NA |
| Chronic respiratory failure | 12 (48%) | 2 (6%) | NA |
| Chronic obstructive pulmonary disease | 2 (8%) | 5 (16%) | NA |
| Emphysema | 1 (4%) | 0 (0%) | NA |
| Pleural effusion | 0 (0%) | 1 (3%) | NA |
| Pneumoconiosis | 0 (0%) | 1 (3%) | NA |
| Pneumonia | 6 (24%) | 7 (22%) | NA |
| Pulmonary embolism | 0 (0%) | 1 (3%) | NA |
| Database | HF_Lung_V1 | HF_Lung_V2 | |
|---|---|---|---|
| Subjects | 261 | 300 | |
| No. of 15-s recordings | 9765 | 13,957 | |
| Total duration (min) | 2441.25 | 3489.25 | |
| Inhalation | No. | 34,095 | 49,373 |
| Duration (min) | 528.14 | 785.48 | |
| Mean (s) | 0.93 | 0.95 | |
| Exhalation | No. | 18,349 | 24,552 |
| Duration (min) | 292.85 | 374.24 | |
| Mean (s) | 0.96 | 0.91 | |
| CAS | No. C/W/S/R | 13,883/8457/686/4740 | 21,558/13,139/914/7505 |
| Duration (min) C/W/S/R | 191.16/119.73/9.46/61.98 | 292.85/186.97/12.82/93.06 | |
| Mean (s) C/W/S/R | 0.83/0.85/0.83/0.78 | 0.82/0.85/0.84/0.74 | |
| DAS | No. | 15,606 | 19,617 |
| Duration (min) | 230.87 | 281.55 | |
| Mean (s) | 0.89 | 0.86 | |
| Database | HF_Lung_V1 | HF_Lung_V2 | |||
|---|---|---|---|---|---|
| Data Set | Training | Test | Training | Test | |
| No. of 15-s recordings | 7809 | 1956 | 10,554 | 3403 | |
| Total duration (min) | 1952.25 | 489 | 2638.5 | 850.75 | |
| I | No. | 27,223 | 6872 | 39,057 | 10,316 |
| Duration (min) | 422.17 | 105.97 | 623.02 | 162.46 | |
| Mean (s) | 0.93 | 0.93 | 0.96 | 0.94 | |
| E | No. | 15,601 | 2748 | 18,334 | 6218 |
| Duration (min) | 248.05 | 44.81 | 292.88 | 81.37 | |
| Mean (s) | 0.95 | 0.98 | 0.96 | 0.79 | |
| C | No. C/W/S/R | 11,464/7027/657/3780 | 2419/1430/29/960 | 17,361/11,453/866/5042 | 4197/1686/48/2463 |
| Duration (min) C/W/S/R | 160.16/100.71/9.10/50.35 | 31.01/19.02/0.36/11.63 | 240.40/163.77/12.34/64.29 | 52.45/23.19/0.48/28.77 | |
| Mean (s) C/W/S/R | 0.84/0.86/0.83/0.80 | 0.77/0.80/0.74/0.73 | 0.83/0.86/0.85/0.77 | 0.75/0.83/0.60/0.70 | |
| D | No. | 13,794 | 1812 | 14,239 | 5378 |
| Duration (min) | 203.59 | 27.29 | 210.96 | 70.59 | |
| Mean (s) | 0.89 | 0.90 | 0.89 | 0.79 | |
| Segment Detection | Event Detection | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | PPV | Sensitivity | Specificity | F1 Score | AUC | PPV | Sensitivity | F1 Score | ||
| Inhalation | ||||||||||
| Train_V1 on Test_V1 | 0.915 ± 0.001 | 0.833 ± 0.006 | 0.781 ± 0.010 | 0.954 ± 0.003 | 0.806 ± 0.004 | 0.962 ± 0.001 | 0.818 ± 0.005 | 0.855 ± 0.009 | 0.836 ± 0.005 | |
| Train_V2 on Test_V1 | 0.921 ± 0.001 *** | 0.849 ± 0.005 *** | 0.794 ± 0.010 *** | 0.959 ± 0.002 *** | 0.821 ± 0.004 *** | 0.968 ± 0.001 *** | 0.836 ± 0.005 *** | 0.864 ± 0.009 ** | 0.850 ± 0.005 *** | |
| Train_V1 on Test_V2 | 0.927 ± 0.002 | 0.834 ± 0.009 | 0.789 ± 0.010 | 0.961 ± 0.003 | 0.811 ± 0.005 | 0.970 ± 0.001 | 0.819 ± 0.007 | 0.864 ± 0.009 | 0.842 ± 0.005 | |
| Train_V2 on Test_V2 | 0.931 ± 0.003 ††† | 0.844 ± 0.011 † | 0.799 ± 0.012 † | 0.963 ± 0.003 | 0.821 ± 0.008 ††† | 0.973 ± 0.002 ††† | 0.832 ± 0.010 ††† | 0.869 ± 0.011 | 0.851 ± 0.009 †† | |
| Exhalation | ||||||||||
| Train_V1 on Test_V1 | 0.875 ± 0.003 | 0.750 ± 0.013 | 0.536 ± 0.020 | 0.957 ± 0.004 | 0.625 ± 0.013 | 0.901 ± 0.003 | 0.604 ± 0.017 | 0.629 ± 0.022 | 0.616 ± 0.018 | |
| Train_V2 on Test_V1 | 0.882 ± 0.004 *** | 0.763 ± 0.011 ** | 0.573 ± 0.026 *** | 0.957 ± 0.003 | 0.654 ± 0.018 *** | 0.912 ± 0.005 *** | 0.626 ± 0.026 * | 0.652 ± 0.025 * | 0.639 ± 0.025 ** | |
| Train_V1 on Test_V2 | 0.923 ± 0.002 | 0.782 ± 0.009 | 0.630 ± 0.018 | 0.971 ± 0.002 | 0.698 ± 0.011 | 0.951 ± 0.002 | 0.751 ± 0.011 | 0.752 ± 0.020 | 0.752 ± 0.015 | |
| Train_V2 on Test_V2 | 0.924 ± 0.003 | 0.783 ± 0.011 | 0.644 ± 0.019 | 0.970 ± 0.002 | 0.706 ± 0.012 | 0.954 ± 0.002 †† | 0.754 ± 0.017 | 0.749 ± 0.019 | 0.751 ± 0.017 | |
| CAS | ||||||||||
| Train_V1 on Test_V1 | 0.852 ± 0.004 | 0.683 ± 0.014 | 0.448 ± 0.031 | 0.950 ± 0.005 | 0.540 ± 0.022 | 0.870 ± 0.008 | 0.470 ± 0.025 | 0.423 ± 0.029 | 0.446 ± 0.026 | |
| Train_V2 on Test_V1 | 0.869 ± 0.003 *** | 0.702 ± 0.007 *** | 0.566 ± 0.030 *** | 0.942 ± 0.004 | 0.626 ± 0.018 *** | 0.911 ± 0.005 *** | 0.539 ± 0.025 *** | 0.489 ± 0.030 *** | 0.514 ± 0.027 *** | |
| Train_V1 on Test_V2 | 0.873 ± 0.002 | 0.659 ± 0.010 | 0.383 ± 0.032 | 0.963 ± 0.004 | 0.484 ± 0.024 | 0.890 ± 0.006 | 0.379 ± 0.028 | 0.339 ± 0.030 | 0.359 ± 0.028 | |
| Train_V2 on Test_V2 | 0.884 ± 0.003 ††† | 0.675 ± 0.013 ††† | 0.495 ± 0.028 ††† | 0.956 ± 0.004 | 0.571 ± 0.018 ††† | 0.918 ± 0.005 ††† | 0.438 ± 0.023 ††† | 0.403 ± 0.028 ††† | 0.420 ± 0.025 ††† | |
| DAS | ||||||||||
| Train_V1 on Test_V1 | 0.828 ± 0.005 | 0.733 ± 0.016 | 0.694 ± 0.022 | 0.888 ± 0.012 | 0.713 ± 0.009 | 0.894 ± 0.004 | 0.624 ± 0.017 | 0.593 ± 0.016 | 0.608 ± 0.014 | |
| Train_V2 on Test_V1 | 0.829 ± 0.006 | 0.728 ± 0.016 | 0.706 ± 0.017 | 0.883 ± 0.011 | 0.717 ± 0.010 | 0.895 ± 0.006 | 0.635 ± 0.017 | 0.605 ± 0.011 * | 0.620 ± 0.011 * | |
| Train_V1 on Test_V2 | 0.877 ± 0.004 | 0.758 ± 0.012 | 0.811 ± 0.019 | 0.901 ± 0.008 | 0.784 ± 0.008 | 0.941 ± 0.003 | 0.691 ± 0.014 | 0.481 ± 0.012 | 0.586 ± 0.011 | |
| Train_V2 on Test_V2 | 0.878 ± 0.006 | 0.759 ± 0.014 | 0.819 ± 0.016 | 0.901 ± 0.008 | 0.787 ± 0.010 | 0.942 ± 0.004 | 0.698 ± 0.024 | 0.485 ± 0.017 | 0.591 ± 0.019 | |
| Label | Overlapped with | Overlap Ratio | |
|---|---|---|---|
| Train | Test | ||
| I | E | 0.001 | 0.001 |
| C | 0.179 | 0.167 | |
| D | 0.198 | 0.287 | |
| E | I | 0.003 | 0.002 |
| C | 0.234 | 0.212 | |
| D | 0.283 | 0.266 | |
| C | I | 0.463 | 0.517 |
| E | 0.286 | 0.329 | |
| D | 0.006 | 0.009 | |
| D | I | 0.584 | 0.661 |
| E | 0.393 | 0.306 | |
| C | 0.007 | 0.006 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, F.-S.; Huang, S.-R.; Huang, C.-W.; Cheng, Y.-R.; Chen, C.-C.; Hsiao, J.; Chen, C.-W.; Lai, F. A Progressively Expanded Database for Automated Lung Sound Analysis: An Update. Appl. Sci. 2022, 12, 7623. https://doi.org/10.3390/app12157623
Hsu F-S, Huang S-R, Huang C-W, Cheng Y-R, Chen C-C, Hsiao J, Chen C-W, Lai F. A Progressively Expanded Database for Automated Lung Sound Analysis: An Update. Applied Sciences. 2022; 12(15):7623. https://doi.org/10.3390/app12157623
Chicago/Turabian StyleHsu, Fu-Shun, Shang-Ran Huang, Chien-Wen Huang, Yuan-Ren Cheng, Chun-Chieh Chen, Jack Hsiao, Chung-Wei Chen, and Feipei Lai. 2022. "A Progressively Expanded Database for Automated Lung Sound Analysis: An Update" Applied Sciences 12, no. 15: 7623. https://doi.org/10.3390/app12157623
APA StyleHsu, F.-S., Huang, S.-R., Huang, C.-W., Cheng, Y.-R., Chen, C.-C., Hsiao, J., Chen, C.-W., & Lai, F. (2022). A Progressively Expanded Database for Automated Lung Sound Analysis: An Update. Applied Sciences, 12(15), 7623. https://doi.org/10.3390/app12157623






