Constitutive High Expression Level of a Synthetic Deleted Encoding Gene of Talaromyces minioluteus Endodextranase Variant (r–TmDEX49A–ΔSP–ΔN30) in Komagataella phaffii (Pichia pastoris)
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Vectors, and Media
2.2. Chemicals and Reagents
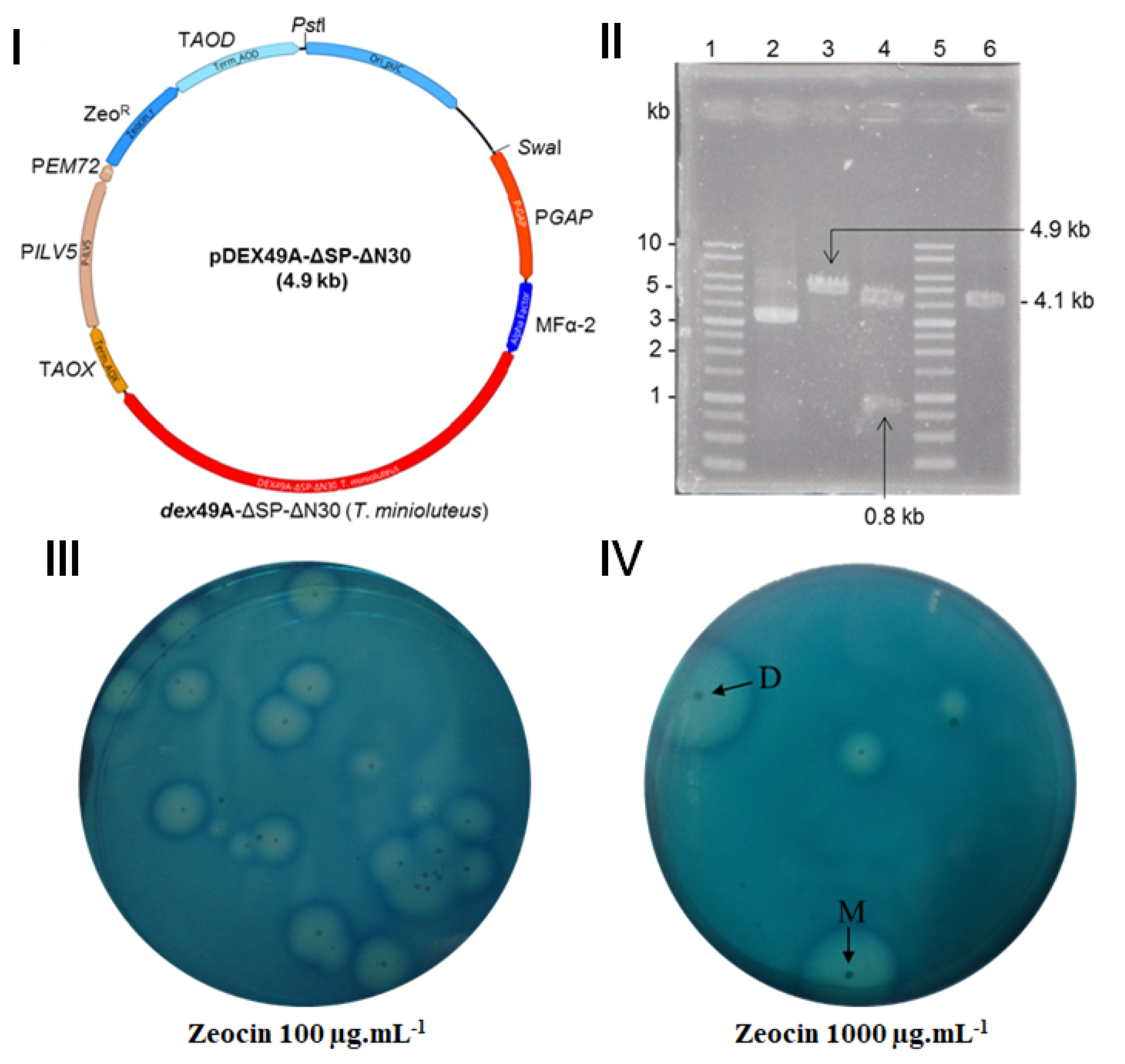
2.3. Vector Construction and Transformation
2.4. Screening of Recombinant K. phaffii Transformants and Production of the r–TmDEX49A–ΔSP–ΔN30
2.5. Production of the r–TmDEX49A–ΔSP–ΔN30 in Shake Flasks
2.6. Production of the r–TmDEX49A–ΔSP–ΔN30 in a 5 L Bioreactor (Fed-Batch Cultures)
2.7. Enzyme Assay and Protein Analysis
2.8. Statistical Analysis of the Data
3. Results and Discussion
3.1. r–TmDEX49A–ΔSP–ΔN30 Variant and Histochemical Screening of Dextranolytic Activity in K. phaffii Clones
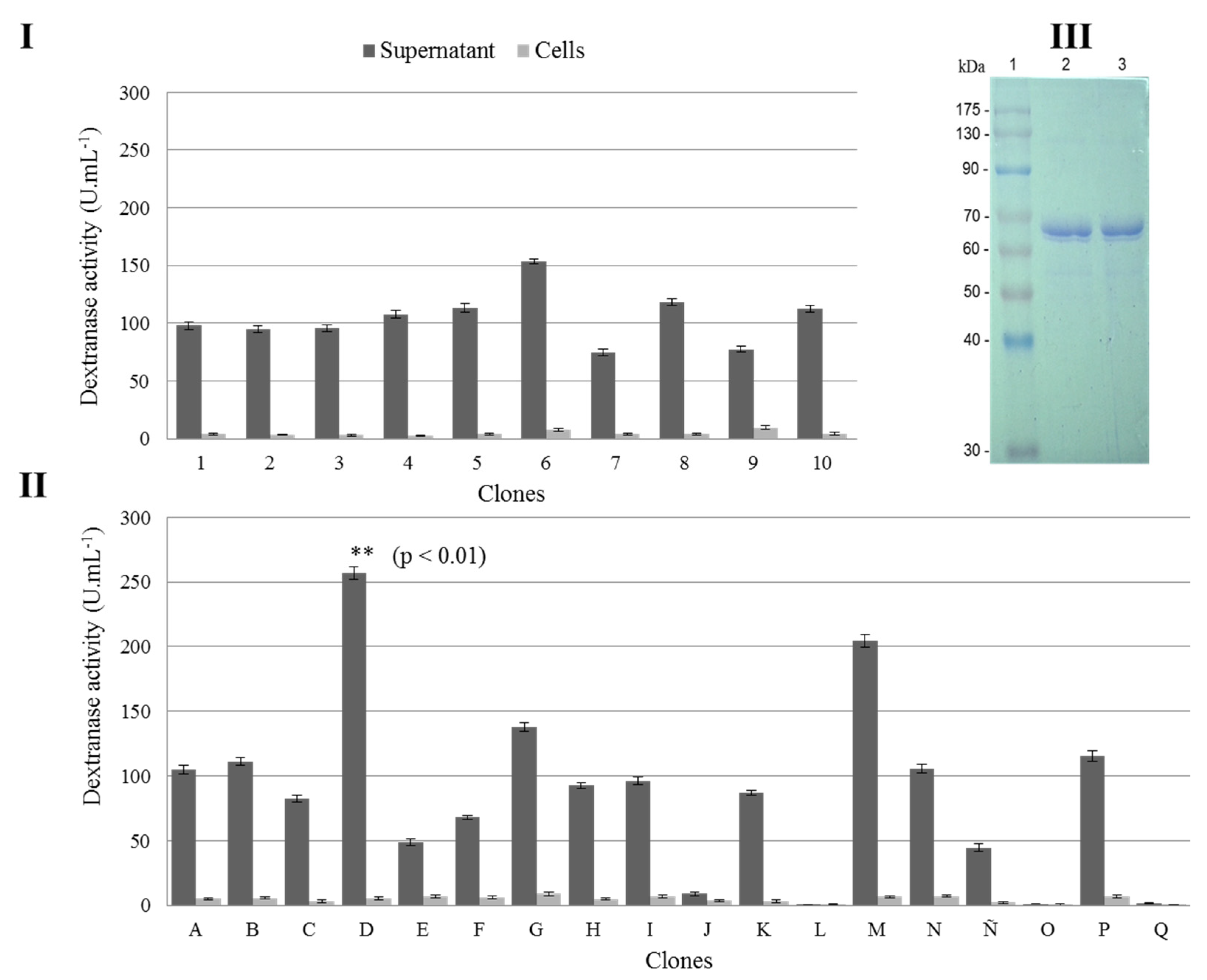
3.2. Selection of the Best K. phaffii Clone Producer of r–TmDEX49A–ΔSP–ΔN30
3.3. Dextranase Production at Shaker Scale by K. phaffii DEX49A–ΔSP–ΔN30
3.4. Detection in Polyacrylamide Gels of the Shaker Produced r–TmDEX49A–ΔSP–ΔN30
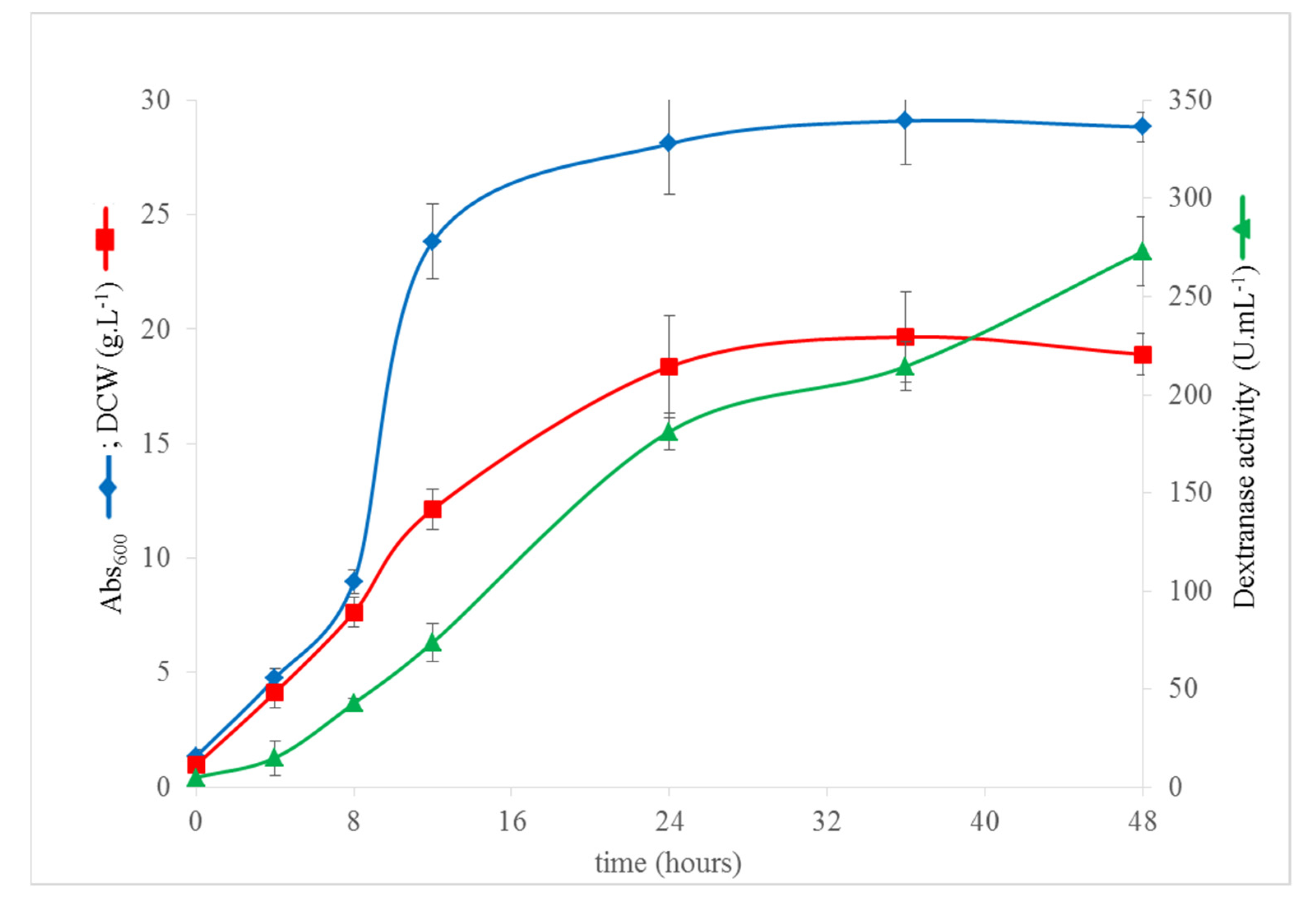
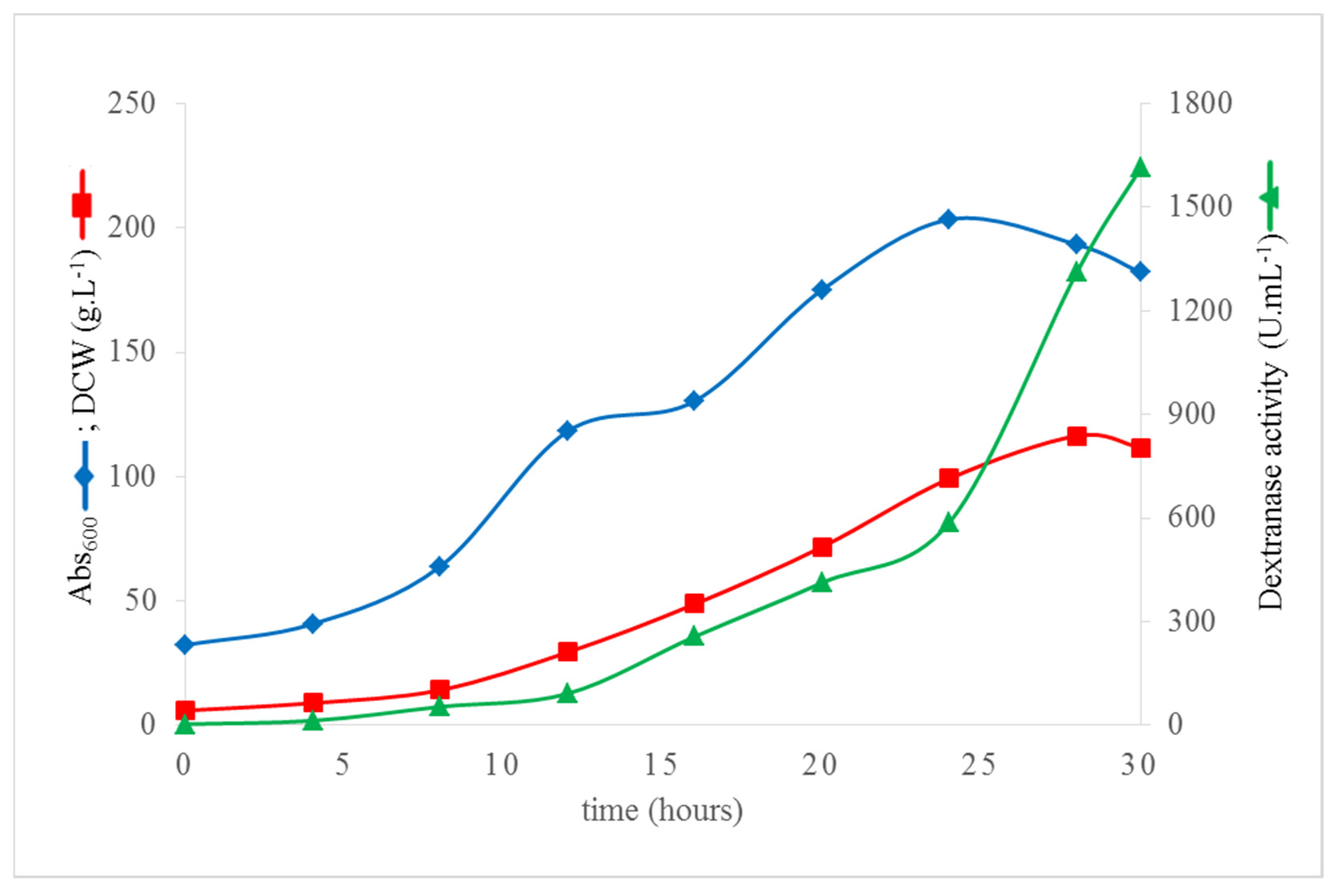
3.5. High Cell Density Fermentation of K. phaffii DEX49A–ΔSP–ΔN30 for High-Level Extracellular Production of r–TmDEX49A–ΔSP–ΔN30
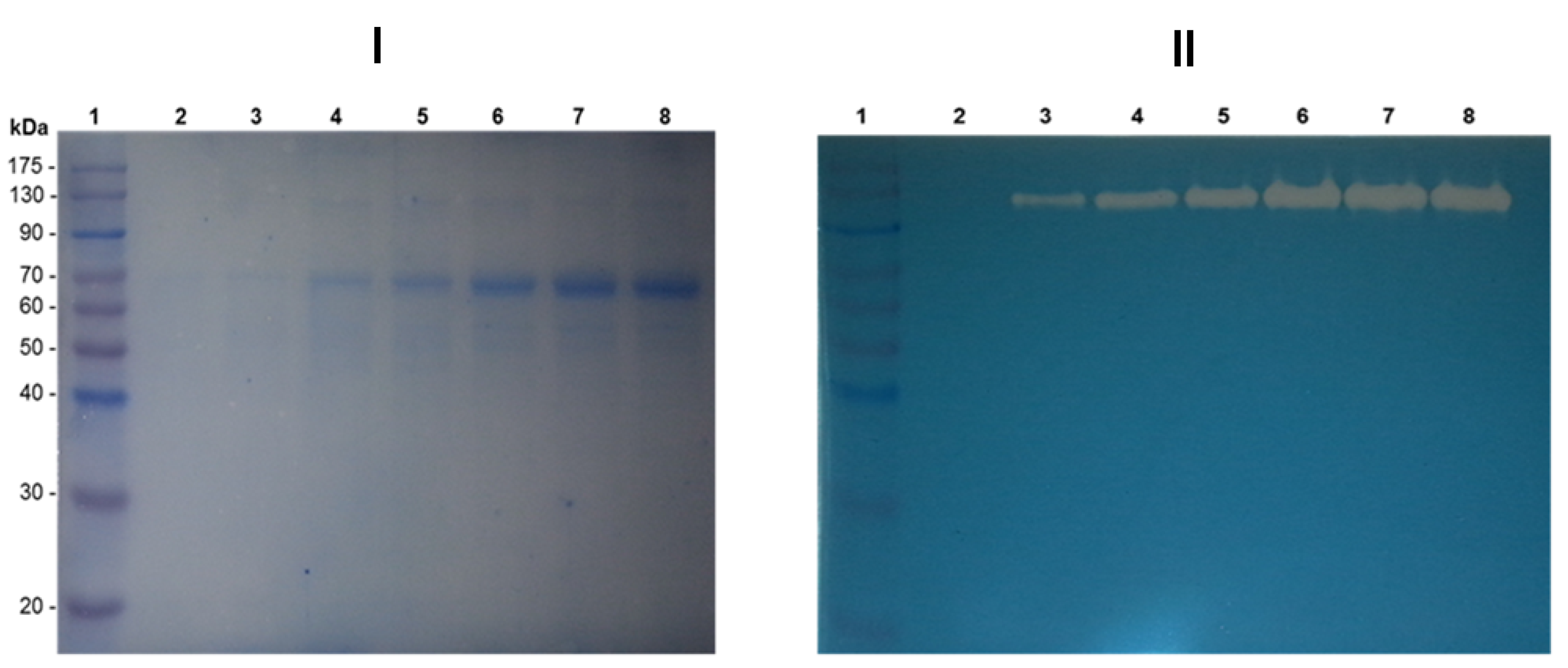
3.6. Detection of the Proteins Secreted to the Culture Supernatant Using the Synthetic Medium FM21–PTM1
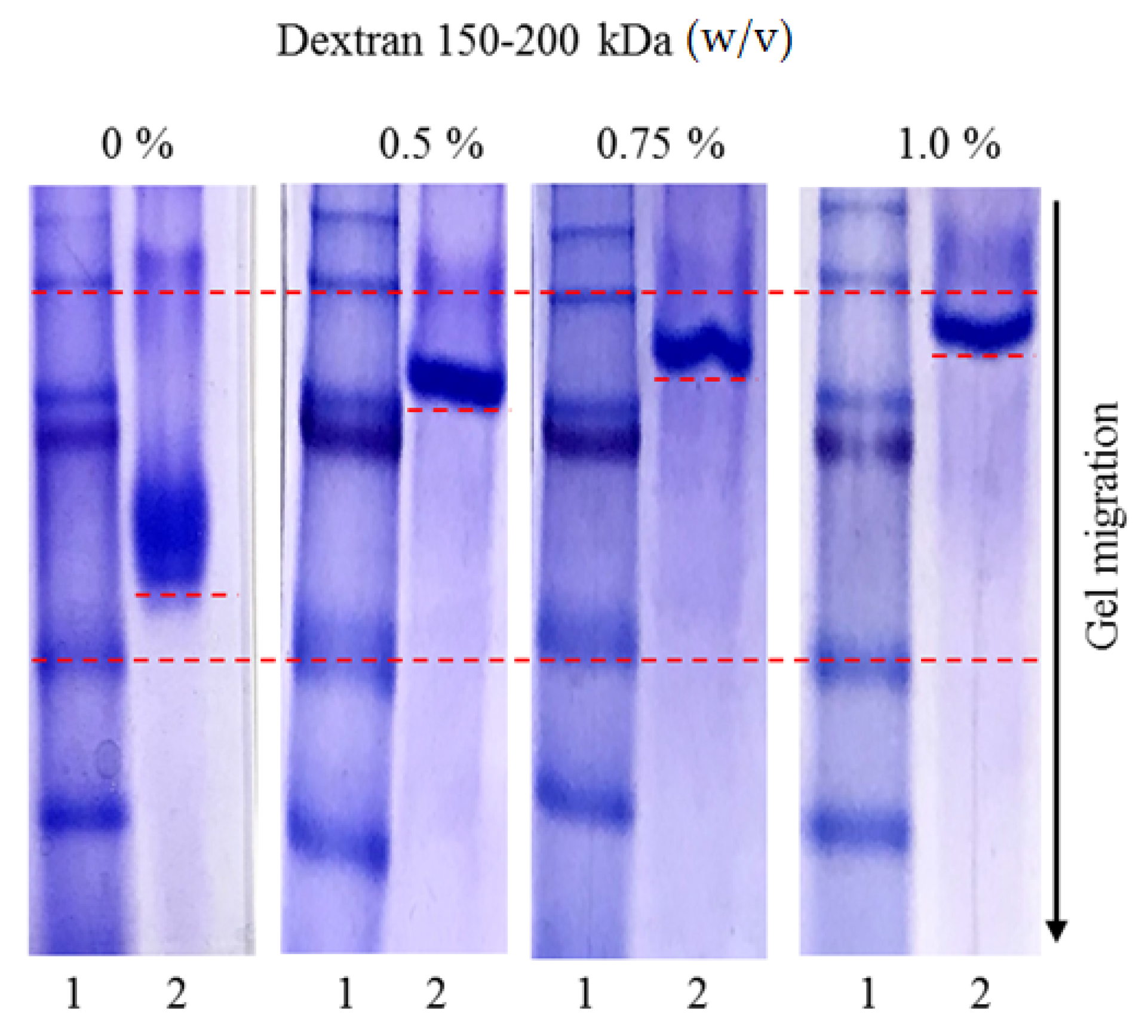
3.7. Affinity Study in Acrylamide Gels under Native Conditions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robyt, J.F.; Eklund, S.H. Stereochemistry involved in the mechanism of action of dextransucrase in the synthesis of dextran and the formation of acceptor products. Bioorganic Chem. 1982, 11, 115–132. [Google Scholar] [CrossRef]
- Bashari, M.; Tounkara, F.; Abdelhai, M.H.; Lagnika, C.; Xu, X.; Jin, Z. Impact of Dextranase on Sugar Manufacturing and its Kinetic on the Molecular Weights of Remaining Dextran. Sugar Tech 2013, 15, 84–93. [Google Scholar] [CrossRef]
- Monchois, V.; Willemot, R.M.; Monsan, P. Glucansucrases: Mechanism of action and structure-function relationships. FEMS Microbiol. Rev. 1999, 23, 131–151. [Google Scholar] [CrossRef]
- Bounaix, M.S.; Robert, H.; Gabriel, V.; Morel, S.; Remaud-Siméon, M.; Gabriel, B.; Fontagné-Faucher, C. Characterization of dextran-producing Weissella strains isolated from sourdoughs and evidence of constitutive dextransucrase expression. FEMS Microbiol. Lett. 2010, 311, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Hector, S.; Willard, K.; Bauer, R.; Mulako, I.; Slabbert, E.; Kossmann, J.; George, G.M. Diverse exopolysaccharide producing bacteria isolated from milled sugarcane: Implications for cane spoilage and sucrose yield. PLoS ONE 2015, 10, e0145487. [Google Scholar] [CrossRef] [PubMed]
- Leemhuis, H.; Pijning, T.; Dobruchowskaa, J.M.; van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, alpha-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272. [Google Scholar] [CrossRef]
- Ninchan, B.; Vanichsriratana, W.; Sriroth, K. Investigation of the optimized dextran-degrading enzyme conditions on the decomposition of different molecular weights of pure dextran using response surface methodology. Arch. Ind. Biotechnol. 2017, 1, 8–19. [Google Scholar]
- Eggleston, G.; Monge, A.; Montes, B.; Stewart, D. Application of dextranases in sugarcane factory: Overcoming practical problems. Sugar Tech 2009, 11, 135–141. [Google Scholar] [CrossRef]
- Rodríguez, E. Dextranase in sugar industry: A review. Sugar Tech 2009, 11, 124–134. [Google Scholar] [CrossRef]
- Xiao, Z.; Liao, X.; Guo, S. Analysis of Sugarcane Juice Quality Indexes. J. Food Qual. 2017, 2017, 1746982. [Google Scholar] [CrossRef]
- Khalikova, E.; Susi, P.; Korpela, T.; Khalikova, E.; Susi, P.; Korpela, T. Microbial Dextran-Hydrolyzing Enzymes: Fundamentals and Applications. MMBR 2005, 69, 306–325. [Google Scholar] [CrossRef]
- Jiao, Y.L.; Wang, S.J.; Lv, M.S.; Jiao, B.H.; Li, W.J.; Fang, Y.W.; Liu, S. Characterization of a marine-derived dextranase and its application to the prevention of dental caries. Ind. Microbiol. Biotechnol. 2014, 41, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Martínez, D.; Menéndez, C.; Chacón, O.; Fuentes, A.D.; Borges, D.; Sobrino, A.; Ramírez, R.; Pérez, E.R.; Hernández, L. Removal of bacterial dextran in sugarcane juice by Talaromyces minioluteus dextranase expressed constitutively in Pichia pastoris. J. Biotechnol. 2021, 333, 10–20. [Google Scholar] [CrossRef]
- Kim, D.; Robyt, J.F.; Lee, S.Y.; Lee, J.H.; Kim, Y.M. Dextran molecular size and degree of branching as a function of sucrose concentration, pH, and temperature of reaction of Leuconostoc mesenteroides B-512FMCM dextransucrase. Carbohydr. Res. 2003, 338, 1183–1189. [Google Scholar] [CrossRef]
- Roca, H.; García, B.; Rodríguez, E.; Mateu, D.; Coroas, L.; Cremata, J.; García, R.; Pons, T.; Delgado, J. Cloning of the Penicillium minioluteum gene encoding dextranase and its expression in Pichia pastoris. Yeast 1996, 12, 1187–1200. [Google Scholar] [CrossRef]
- Raices, M.; Molerio, M.C.; Li, I.M.; Morera, V.; Roca, H.; Delgado, J.; Cremata, J.; Herrera, L. Purificación parcial de una enzima dextranasa a partir de una cepa de hongo del género Penicillium. Biotecnol. Apl. 1991, 8, 248–255. [Google Scholar]
- Betancourt, L.H.; García, R.; González, J.; Montesino, R.; Quintero, O.; Takao, T.; Shimonishi, Y.; Cremata, J.A. Dextranase (α-1,6 glucan-6-glucanohydrolase) from Penicillium minioluteum expressed in Pichia pastoris: Two host cells with minor differences in N-glycosylation. FEMS Yeast Res. 2001, 1, 151–160. [Google Scholar] [CrossRef] [PubMed]
- García, B.; Margolles, E.; Roca, H.; Mateu, D.; Raices, M.; Gonzáles, M.E.; Herrera, L.; Delgado, J. Cloning and sequencing of a dextranase-encoding cDNA from Penicillium minioluteum. FEMS Microbiol. Lett. 1996, 143, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.M.; Andersson, R.; Stahlberg, J.; Kenne, L.; Jones, T.A. Dextranase from Penicillium minioluteum: Reaction course, crystal structure, and product complex. Structure 2003, 11, 1111–1121. [Google Scholar] [CrossRef]
- Yoder, M.D.; Lietzke, S.E.; Jurnak, F. Unusual structural features in the parallel beta-helix in pectate lyases. Structure 1993, 1, 241–251. [Google Scholar] [CrossRef]
- Marín, A.M.; Jáuregui, J.; Carreón, L.; Chávez, N.A. Dextranase production by recombinant Pichia pastoris under operational volumetric mass transfer coefficient (kLa) and volumetric gassed power input (Pg/V) attainable at commercial large scale. Prep. Biochem. Biotechnol. 2019, 49, 606–615. [Google Scholar] [CrossRef]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317. [Google Scholar] [CrossRef] [PubMed]
- Cos, O.; Ramon, R.; Montesinos, J.L.; Valero, F. Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: A review. Microb. Cell Factories 2006, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Emr, S.; Schekman, R.; Flessel, M.; Thorner, J. An MFαl-SUC2 (α-factor-invertase) gene fusion for study of protein localization and gene expression in yeast. Proc. Natl. Acad. Sci. USA 1983, 80, 7080–7084. [Google Scholar] [CrossRef] [PubMed]
- Lin-Cereghino, J.; Wong, W.W.; Xiong, S.; Giang, W.; Luong, L.T.; Vu, J.; Johnson, S.D.; Lin-Cereghino, G.P. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. BioTechniques 2005, 38, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Wegner, E.H. Biochemical Conversions by Yeast Fermentation at High Cell Densities. U.S. Patent 4,414,329, 8 November 1983. [Google Scholar]
- Kosaric, N.; Yu, K.; Zajic, J.E.; Rozanis, J. Dextranase production from Penicillium funiculosum. Biotech. Bioeng. 1973, 15, 729–741. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T-4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Shin, S.Y.; Moon, J.S.; Li, L.; Cho, S.K.; Kim, T.J.; Han, N.S. Isolation of dextran-hydrolyzing intestinal bacteria and characterization of their dextranolytic activities. Biopolymers 2015, 103, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Brison, Y.; Malbert, Y.; Czaplicki, G.; Mourey, L.; Remaud-Simeón, M.; Tranier, S. Structural Insights into the Carbohydrate Binding Ability of an α-(1→2) Branching Sucrase from Glycoside Hydrolase Family 70. J. Biol. Chem. 2016, 291, 7527–7540. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.K.; Kim, S.H.; Park, J.Y.; Jin, X.J.; Oh, D.K.; Kang, S.S.; Kim, D. Cloning and characterization of a dextranase gene from Lipomyces starkeyi and its expression in Saccharomyces cerevisiae. Yeast 2005, 22, 1239–1248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kang, H.K.; Park, J.Y.; Ahn, J.S.; Kim, S.H.; Kim, D. Cloning of a gene encoding dextranase from Lipomyces starkeyi and its expression in Pichia pastoris. J. Microbiol. Biotechnol. 2009, 19, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Liu, X.; Liu, X.; Deng, T.; Feng, Y.; Tian, X.; Lyu, M. The Marine Catenovulum agarivorans MNH15 and Dextranase: Removing Dental Plaque. Mar. Drugs 2019, 17, 592. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, J.; Valdés, I.; Cabrera, N. The ICL1 gene of Pichia pastoris, transcriptional regulation and use of its promoter. Yeast 2003, 20, 1097–1108. [Google Scholar] [CrossRef]
- Clare, J.J.; Rayment, F.B.; Ballantine, S.P.; Sreekrishna, K.; Romanos, M.A. High-level expression of tetanus toxin fragment C in Pichia pastoris strains containing multiple tandem integrations of the gene. Bio/Technology 1991, 9, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Parashar, D.; Satyanarayana, T. Enhancing the production of recombinant acidic α-amylase and phytase in Pichia pastoris under dual promoters [constitutive (GAP) and inducible (AOX)] in mixed fed batch high cell density cultivation. Process. Biochem. 2016, 51, 1315–1322. [Google Scholar] [CrossRef]
- García, B.M.; Rodríguez, E.; Rivero, T.; Hidalgo, Y.; Menéndez, J.J. Dextranase Expression in two Different Host-vector Systems of the Methylotrophic Yeast Pichia pastoris. Biotecnol. Apl. 2000, 17, 11–15. [Google Scholar]
- Safder, I.; Khan, S.; Islam, I.; Kazim, M.; Bibi, Z.; Waqas, M. Pichia pastoris expression system: A potential candidate to express protein in industrial and biopharmaceutical domains. Biomed. Lett. 2018, 4, 1–14. [Google Scholar]
- Rerngsamran, P.; Temjitpukdee, P.; Assavasirijinda, N.; Chareonpornwattana, S.; Thaniyavarn, S. Cloning, characterization, and heterologous expression of a dextranase gene from Penicillium pinophilum SMCU3-14. ScienceAsia 2014, 40, 405–413. [Google Scholar] [CrossRef]
- Netsopa, S.; Niamsanit, S.; Araki, T.; Kongkeitkajorn, M.B.; Milintawisamai, N. Purification and Characterization Including Dextran Hydrolysis of Dextranase from Aspergillus allahabadii X26. Sugar Tech 2019, 21, 329–340. [Google Scholar] [CrossRef]
- Matthews, C.B.; Kuo, A.; Love, K.R.; Love, J.C. Development of a general defined medium for Pichia pastoris. Biotechnol. Bioeng. 2017, 115, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Barrero, J.J.; Casler, J.C.; Valero, F.; Ferrer, P.; Glick, B.S. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb. Cell Factories 2018, 17, 161. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, E.; Sánchez, K.; Roca, H.; Delgado, J.M. Different methanol feeding strategies to recombinant Pichia pastoris cultures producing high level of dextranase. Biotechnol. Tech. 1997, 11, 461–466. [Google Scholar] [CrossRef]
- Goodrick, J.C.; Xu, M.; Finnegan, R.; Schilling, B.M.; Schiavi, S.; Hoppe, H.; Wan, N.C. High- level expression and stabilization of recombinant human chitinase produced in a continuous constitutive Pichia pastoris expression system. Biotechnol. Bioeng. 2001, 74, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Piva, L.C.; Bentacur, M.O.; Castelo, V.; Reis, B.; De Marco, J.L.; de Moraes, L.M.P.; Gonçalves, F.A. Molecular strategies to increase the levels of heterologous transcripts in Komagataella phaffii for protein production. Bioengineered 2017, 8, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.; Mall, A.K.; Pathak, A.D.; Solomon, S.; Kishor, R. Microorganisms affecting Post-Harvest Sucrose Losses in Sugarcane. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2554–2566. [Google Scholar] [CrossRef][Green Version]
- Zepeda, A.B.; Pessoa, A.; Farías, J.G. Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Braz. J. Microbiol. 2018, 49, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Potvin, G.; Ahmad, A.; Zhang, Z. Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: A review. Biochem. Eng. J. 2012, 64, 91–105. [Google Scholar] [CrossRef]
- Mattanovich, D.; Graf, A.; Stadlmann, J.; Dragosits, M.; Redl, A.; Maurer, M.; Kleinheinz, M.; Sauer, M.; Altmann, F.; Gasser, B. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb. Cell Factories 2009, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.A.; Akkermans, S.; Nimmegeers, P.; Van Impe, J.F.M. Bioproduction of the Recombinant Sweet Protein Thaumatin: Current State of the Art and Perspectives. Front. Microbiol. 2019, 10, 695. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.; Moulis, C.; Monties, N.; Guieysse, D.; Morel, S.; Cioci, G.; Remaud-Siméon, M. A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation. J. Biol. Chem. 2020, 295, 9474–9489. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arísticas Ribalta, R.C.; Valdés, L.M.; Lafargue Gámez, M.; Rodríguez Davydenko, S.; Dubreucq, E.; Perrier, V.; Moreau, B.; Vidal, R.F. Constitutive High Expression Level of a Synthetic Deleted Encoding Gene of Talaromyces minioluteus Endodextranase Variant (r–TmDEX49A–ΔSP–ΔN30) in Komagataella phaffii (Pichia pastoris). Appl. Sci. 2022, 12, 7562. https://doi.org/10.3390/app12157562
Arísticas Ribalta RC, Valdés LM, Lafargue Gámez M, Rodríguez Davydenko S, Dubreucq E, Perrier V, Moreau B, Vidal RF. Constitutive High Expression Level of a Synthetic Deleted Encoding Gene of Talaromyces minioluteus Endodextranase Variant (r–TmDEX49A–ΔSP–ΔN30) in Komagataella phaffii (Pichia pastoris). Applied Sciences. 2022; 12(15):7562. https://doi.org/10.3390/app12157562
Chicago/Turabian StyleArísticas Ribalta, Roberto Carlos, Lisandra Martínez Valdés, Meinardo Lafargue Gámez, Sonia Rodríguez Davydenko, Eric Dubreucq, Veronique Perrier, Benoît Moreau, and Reinaldo Fraga Vidal. 2022. "Constitutive High Expression Level of a Synthetic Deleted Encoding Gene of Talaromyces minioluteus Endodextranase Variant (r–TmDEX49A–ΔSP–ΔN30) in Komagataella phaffii (Pichia pastoris)" Applied Sciences 12, no. 15: 7562. https://doi.org/10.3390/app12157562
APA StyleArísticas Ribalta, R. C., Valdés, L. M., Lafargue Gámez, M., Rodríguez Davydenko, S., Dubreucq, E., Perrier, V., Moreau, B., & Vidal, R. F. (2022). Constitutive High Expression Level of a Synthetic Deleted Encoding Gene of Talaromyces minioluteus Endodextranase Variant (r–TmDEX49A–ΔSP–ΔN30) in Komagataella phaffii (Pichia pastoris). Applied Sciences, 12(15), 7562. https://doi.org/10.3390/app12157562





