Metagenomic Research of Infectious Diseases in Archaeological Contexts: Evidence from the Hospital Real de Todos-os-Santos (Portugal)
Abstract
:1. Introduction
The Royal Hospital of All Saints Bioanthropological Collection
2. Materials and Methods
3. Results and Discussion
3.1. Overall Profile of the Microbiomes
3.2. Human Pathogenic Microorganisms
3.3. The Absence of Treponema Pallidum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buikstra, J.E. (Ed.) Ortner’s Identification of Pathological Conditions in Human Skeletal Remains; Academic Press: London, UK, 2019. [Google Scholar]
- Alves-Cardoso, F. Palaeopathology. In The SAS Encyclopaedia of Archaeological Sciences; Sandra, L., López Varela, Eds.; John Wiley & Sons, Inc.: London, UK, 2019. [Google Scholar]
- Buikstra, J.E.; Cook, D.C.; Bolhofner, K.L. Introduction: Scientific rigor in paleopathology. Int. J. Paleopathol. 2017, 19, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Snoddy, A.M.E.; Beaumont, J.; Buckley, H.R.; Colombo, A.; Halcrow, S.E.; Kinaston, R.L.; Vlok, M. Sensationalism and speaking to the public: Scientific rigour and interdisciplinary collaborations in palaeopathology. Int. J. Paleopathol. 2020, 28, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Blevins, K.E.; Crane, A.E.; Lum, C.; Furuta, K.; Fox, K.; Stone, A.C. Evolutionary history of Mycobacterium leprae in the Pacific Islands. Philos. Trans. R. Soc. B 2020, 375, 20190582. [Google Scholar] [CrossRef] [PubMed]
- Bos, K.I.; Harkins, K.M.; Herbig, A.; Coscolla, M.; We, N.; Comas, I.; Forrest, S.A.; Bryant, J.M.; Harris, S.R.; Schuenemann, V.J.; et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 2014, 514, 494–497. [Google Scholar] [CrossRef]
- Bos, K.I.; Herbig, A.; Sahl, J.; Waglechner, N.; Fourment, M.; Forrest, S.A.; Klunk, J.; Schuenemann, V.J.; Poinar, D.; Kuch, M.; et al. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. Elife 2016, 5, e12994. [Google Scholar] [CrossRef]
- Schuenemann, V.J.; Kumar Lankapalli, A.; Barquera, R.; Nelson, E.A.; Hernández, D.I.; Alonzo, V.A.; Boss, K.I.; Morfín, L.M.; Herbig, A.; Krause, J. Historic Treponema pallidum genomes from Colonial Mexico retrieved from archaeological remains. PLoS Negl. Trop. Dis. 2018, 12, e0006447. [Google Scholar] [CrossRef] [Green Version]
- Schuenemann, V.J.; Singh, P.; Mendum, T.A.; Krause-Kyora, B.; Jäger, G.; Bos, K.I.; Herbig, A.; Economou, C.; Benjak, A.; Busso, P.; et al. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science 2013, 341, 179–183. [Google Scholar] [CrossRef]
- Ortner, D. Identification of Pathological Conditions in Human Skeletal Remains; Academic Press: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Hackett, C.J. Diagnosis Criteria of Syphilis Yaws and Treponarid (Treponematosis) and Some other Diseases in Dry Bones; Springer: Heidelberg, Germany, 1976. [Google Scholar]
- Baker, B.; Armelagos, G.; Becker, M.; Brothwell, D.; Drusini, A.; Geise, M.; Kelley, M.; Moritoto, I.; Morris, A.; Nurse, G.; et al. The origin and antiquity of syphilis. Curr. Anthopol. 1988, 29, 703–737. [Google Scholar] [CrossRef]
- Rothschild, B. History of syphilis. Clin. Infec. Dis. 2005, 40, 1454–1463. [Google Scholar] [CrossRef]
- Rothschild, B.; Calderon, F.; Coppa, A.; Rothschild, C. First European exposure to syphilis: The Dominican Republic at the time of Columbian contact. Clin. Infect. Dis. 2000, 31, 936–941. [Google Scholar] [CrossRef] [Green Version]
- Saunders, L.; Rothschild, B.; Rothschild, C. Old world origins of syphilis in New York. Chungara 2000, 32, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Harper, K.N.; Ocampo, P.S.; Steiner, B.M.; George, R.W.; Silverman, M.S.; Bolotin, S.; Pillay, A.; Saunders, N.J.; Armelagos, G.J. On the origin of the treponematoses: A phylogenetic approach. PLoS Negl. Trop. Dis. 2008, 2, e148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harper, K.N.; Zuckerman, M.K.; Armelagos, G.J. Syphilis: Then and now. Scientist Magazine, 1 February 2014; 1–11. [Google Scholar]
- Sousa, J. Impacto social da sífilis: Alguns aspectos historicos. Med. Interna 1996, 3, 184–192. [Google Scholar]
- Casimiro, S.; Alves-Cardoso, F. Death in the Royal Hospital: An 18th Century Testimony. In Hospital Real de Todos-os-Santos/All Saints Royal Hospital: Lisbon and Public Health; Alberto, E.M., Silva, R.B., Teixeira, A., Eds.; Câmara Municipal de Lisboa/Santa Casa da Misericórdia: Lisboa, Portugal, 2021; pp. 515–517. [Google Scholar]
- Alberto, E.M.; Silva, R.B.; Teixeira, A. (Eds.) All Saints Royal Hospital: Lisbon and Public Health; Câmara Municipal de Lisboa/Santa Casa da Misericórdia: Lisboa, Portugal, 2021. [Google Scholar]
- Assis, S.; Casimiro, S.; Alves-Cardoso, F. A possible case of acquired syphilis at the former Royal Hospital of All-Saints (RHAS) in Lisbon, Portugal (18th century): A comparative methodological approach to differential diagnosis. Anthropol. Anz. 2015, 72, 427–449. [Google Scholar] [CrossRef] [Green Version]
- de Melo, F.L.; de Mello, J.C.M.; Fraga, A.M.; Nunes, K.; Eggers, S. Syphilis at the crossroad of phylogenetics and paleopathology. PLoS Negl. Trop. Dis. 2010, 4, e575. [Google Scholar] [CrossRef]
- Gaul, J.S.; Winter, E.; Grossschmidt, K. Ancient pathogens in museal dry bone specimens: Analysis of paleocytology and aDNA. Wien. Med. Wochenschr. 2015, 165, 133–139. [Google Scholar] [CrossRef]
- Majander, K.; Pfrengle, S.; Kocher, A.; Neukamm, J.; Plessis, L.; Pla-Díaz, M.; Arora, N.; Akgül, G.; Salo, K.; Schats, R. Ancient bacterial genomes reveal a high diversity of Treponema pallidum strains in early modern Europe. Curr. Biol. 2020, 30, 3788–3803. [Google Scholar] [CrossRef]
- Guedes, L.; Dias, O.; Neto, J.; Ribeiro da Silva, L.D.P.; Mendonça de Souza, S.M.F.; Iñiguez, A.M. First Paleogenetic Evidence of Probable Syphilis and Treponematoses Cases in the Brazilian Colonial Period. BioMed Res. Int. 2018, 2018, 8304129. [Google Scholar] [CrossRef] [Green Version]
- Ortner, D. Differential diagnosis and issues in disease classification. In A Companion to Paleopathology; Grauer, A., Ed.; Blackwell Publishing Ltd.: Chichester, UK, 2012; pp. 250–267. [Google Scholar]
- Bruzek, J. A method for visual determination of sex using the human hip bone. Am. J. Phys. Anthropol. 2002, 117, 157–168. [Google Scholar] [CrossRef]
- Buikstra, J.E.; Ubelaker, D.H. Standards for Data Collection from Human Skeletal Remains; Arkansas Archaeological Survey: Fayetteville, AR, USA, 1994. [Google Scholar]
- White, T.; Folkens, P. The Human Bone Manual; Academic Press: London, UK, 2005. [Google Scholar]
- Gomes, C.; Palomo-Díez, S.; Roig, J.; López-Parra, A.M.; Baeza-Richer, C.; Esparza-Arroyo, A.; Gibaja, J.; Arroyo-Pardo, E. Nondestructive extraction DNA method from bones or teeth, true or false? Forensic Sci. Int. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e279–e282. [Google Scholar] [CrossRef] [Green Version]
- Rohland, N.; Hofreiter, M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007, 2, 1756–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohland, N.; Hofreiter, M. Comparison and optimization of ancient DNA extraction. Biotechniques 2007, 42, 343–352. [Google Scholar] [CrossRef] [Green Version]
- Rohland, N.; Siedel, H.; Hofreiter, M. A rapid column-based ancient DNA extraction method for increased sample throughput. Mol. Ecol. Resour. 2010, 10, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Pääbo, S.; Poinar, H.; Serre, D.; Jaenicke-Despres, V.; Hebler, J.; Rohland, N.; Kuch, M.; Krause, J.; Vigilant, L.; Hofreiter, M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palomo-Díez, S. Caracterización Genética de las Poblaciones de las Edades del Cobre y del Bronce en la Submeseta Norte de la Península Ibérica. Ph.D. Thesis, Universidad Complutense de Madrid, Madrid, Spain, 2015. [Google Scholar]
- Becares, A.A.; Fernandez, A.F. Microbiome Based Identification, Monitoring and Enhancement of Fermentation Processes and Products. U.S. Patent Application 15/779,531, 20 December 2018. [Google Scholar]
- Feld, L.; Nielsen, T.K.; Hansen, L.H.; Aamand, J.; Albers, C.N. Establishment of bacterial herbicide degraders in a rapid sand filter for bioremediation of phenoxypropionate-polluted groundwater. Appl. Environ. Microbiol. 2016, 82, 878–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albers, C.N.; Ellegaard-Jensen, L.; Hansen, L.H.; Sørensen, S.R. Bioaugmentation of rapid sand filters by microbiome priming with a nitrifying consortium will optimize production of drinking water from groundwater. Water Res. 2018, 129, 1–10. [Google Scholar] [CrossRef]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From Vineyard Soil to Wine Fermentation: Microbiome Approximations to Explain the “terroir”. Concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, D.; Bhagwat, M. BLAST quickstart. In Comparative Genomics; Bergman, N.H., Ed.; Human Press: Totowa, NJ, USA, 2007; Volume 1 and 2, pp. 149–175. [Google Scholar]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 386, 9. [Google Scholar] [CrossRef] [Green Version]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a metagenomics service for analysis of microbial community structure and function. In Microbial Environmental Genomics (MEG); Martin, F., Uroz, S., Eds.; Humana Press: New York, NY, USA, 2016; pp. 207–233. [Google Scholar]
- Kircher, M.; Kelso, J. High-throughput DNA sequencing-concepts and limitations. Bioessays 2010, 326, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.M. Encyclopaedia of Microbiology; Academic Press: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Schaechter, M. Encyclopaedia of Microbiology; Academic Press: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Madigan, M.T.; Bender, K.S.; Buckley, D.H.; Sattley, W.M.; Stahl, D.A. Brock Biology of Microorganisms, 16th ed.; Pearson Education: Hoboken, NJ, USA, 2021. [Google Scholar]
- Hardham, J.M.; King, K.W.; Dreier, K.; Wong, J.; Strietzel, C.; Eversole, R.R.; Sfintescu, C.; Evans, R.T. Transfer of Bacteroides splanchnicus to Odoribacter gen. nov. as Odoribacter splanchnicus comb. nov., and description of Odoribacter denticanis sp. nov., isolated from the crevicular spaces of canine periodontitis patients. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 1, 103–109. [Google Scholar] [CrossRef] [Green Version]
- Morgan, X.C.; Tickle, T.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Werner, H.; Rintelen, G.; Kunstek-Santos, H. A new butyric acid-producing Bacteroides species: B. splanchnicus n. sp. (author’s transl). Zentralbl. Bakteriol. Orig. A 1975, 231, 133–144. [Google Scholar] [PubMed]
- Heitor, S.; Miranda, J.M.; Miranda, P.; Mendes-Victor, L. The 1755 Lisbon tsunami; evaluation of the tsunami parameters. J. Geodyn. 1998, 25, 143–157. [Google Scholar]
- Mendes-Victor, L. (Ed.) The 1755 Lisbon Earthquake: Revisited; Springer Science & Business Media: Lisbon, Portugal, 2008. [Google Scholar]
- Oliveira, E.F. Elementos Para a História do Município de Lisboa; Typographia Universal: Lisboa, Portugal, 1882. [Google Scholar]
- Bargão, A. Vivências do Quotidiano do Hospital Real de Todos-os-Santos (Lisboa): Os contextos do poço SE do claustro NE.; Tese de Mestrado em Arqueologia, Universidade Nova de Lisboa Faculdade de Ciências Sociais e Humanas: Lisboa, Portugal, 2015. [Google Scholar]
- Cardoso, J.L. Água, iluminação e esgotos em Lisboa nos finais do século XVIII. Análise Soc. 2000, 35, 495–509. [Google Scholar]
- Diekema, D.J.; Pfaller, M.A.; Jones, R.N.; Doern, G.V.; Winokur, P.L.; Gales, A.C.; Sader, H.S.; Kugler, K.; Beach, M. Survey of bloodstream infections due to gram-negative bacilli: Frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY Antimicrobial Surveillance Program, 1997. Clin. Infect. Dis. 1999, 29, 595–607. [Google Scholar]
- Kidd, J.M.; Kuti, J.L.; Nicolau, D.P. Novel pharmacotherapy for the treatment of hospital-acquired and ventilator-associated pneumonia caused by resistant gram-negative bacteria. Expert Opin. Pharmacother. 2018, 19, 397–408. [Google Scholar] [CrossRef]
- Tordecilla Echenique, Y.; Salamanca Bautista, M.P.; Arias Jiménez, J.L.; Guisado Espartero, E.; Ortega Calvo, M.; Pérez Cano, R. Hematogenous sternal osteomyelitis and community acquired pneumonia in a methicillin-susceptible Staphilococcus aureus sepsis. An. De Med. Interna 2005, 22, 91–193. [Google Scholar]
- Akinkugbe, O.; Stewart, C.; Mckenna, C. Presentation and investigation of pediatric bone and joint infections in the pediatric emergency department. Pediatr. Emerg. Care 2019, 35, 700–704. [Google Scholar] [CrossRef]
- Suligoy, C.M.; Lattar, S.M.; Llana, M.N.; González, C.D.; Alvarez, L.P.; Ashley Robinson, D.; Gómez, M.I.; Buzzola, F.R.; Sordelli, D.O. Mutation of Agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front. Cell Infect. Microbiol. 2018, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Romani Vestman, N.; Chen, T.; Lif Holgerson, P.; Öhman, C.; Johansson, I. Oral microbiota shift after 12-week supplementation with Lactobacillus reuteri DSM 17938 and PTA 5289; a randomized control trial. PLoS ONE 2015, 10, e0125812. [Google Scholar] [CrossRef] [PubMed]
- Scapoli, L.; Girardi, A.; Palmieri, A.; Martinelli, M.; Cura, F.; Lauritano, D.; Carinci, F. Quantitative analysis of periodontal pathogens in periodontitis and gingivitis. J. Biol. Regul. Homeost. Agents 2015, 29 (Suppl. 1), 101–110. [Google Scholar] [PubMed]
- Wasiela, M.; Brzezińska-Błaszczyk, E. The influence of anaerobic bacteria and mycoplasma isolated from the genital tract on mast cell activation. Med. Dosw. Mikrobiol. 2000, 52, 389–396. [Google Scholar] [PubMed]
- Okamoto-Shibayama, K.; Sekino, J.; Yoshikawa, K.; Saito, A.; Ishihara, K. Antimicrobial susceptibility profiles of oral Treponema species. Anaerobe 2017, 48, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.A.; Unakal, C.G. Staphylococcus Aureus; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Chandranesan, A. Actinomycosis in Cancer patients: Dilemma in Differentiating Colonization Versus True Disease. Infect. Dis. Clin. Pract. 2018, 26, 303. [Google Scholar] [CrossRef]
- Farazmand, C.; Klinkova, O.; Denham, J.; Nanjappa, S.; Greene, J. Actinomycosis in cancer patients: A single-center chart review study. Infect. Dis. Clin. Pract. 2018, 26, 339–343. [Google Scholar] [CrossRef]
- Lamrabet, O.; Drancourt, M. Mycobacterium gilvum illustrates size-correlated relationships between mycobacteria and Acanthamoeba polyphaga. Appl. Environ. Microbiol. 2013, 79, 1606–1611. [Google Scholar] [CrossRef] [Green Version]
- Matos, V. O diagnóstico retrospectivo da lepra: Complementaridade clínica e paleopatológica no arquivo médico do Hospital-colónia Rovisco Pais (Século XX, Tocha, Portugal) e na colecção de Esqueletos da Leprosaria Medieval De St. Jørgen’s (Odense, Dinamarca). Ph.D. Thesis, University of Coimbra, Coimbra, Portugal, 2009. [Google Scholar]
- Monde, N.; Munyeme, M.; Muwonge, A.; Muma, J.B.; Malama, S. Characterization of non-tuberculous mycobacterium from humans and water in an Agropastoral area in Zambia. BMC Infect. Dis. 2018, 18, 20. [Google Scholar] [CrossRef] [Green Version]
- Lavania, M.; Turankar, R.; Singh, I.; Nigam, A.; Sengupta, U. Detection of Mycobacterium gilvum first time from the bathing water of leprosy patient from Purulia, West Bengal. Int. J. Mycobacteriol. 2014, 3, 286–289. [Google Scholar] [CrossRef] [Green Version]
- Feleke, D.G.; Yemanebrhane, N. Trichomonas vaginalis infection in Ethiopia: A systematic review and meta-analysis. Int. J. STD AIDS 2022, 33, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.F.; Saragadam, S.D.; DiMaria, C.N.; Delgado-Daza, A. Infection of a prosthetic knee joint with Clostridium bifermentans. Oxf. Med. Case Rep. 2020, 8, omaa057. [Google Scholar] [CrossRef] [PubMed]
- Edagiz, S.; Lagace-Wiens, P.; Embil, J.; Karlowsky, J.; Walkty, A. Empyema caused by Clostridium bifermentans: A case report. Can. J. Infect. Dis. Med. Microbiol. 2015, 26, 105–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlan, D.R.; Smith, M.A.; Isenberg, H.D.; Engrassia, S.; Hilton, E. Clostridium bifermentans bacteremia with metastatic osteomyelitis. J. Clin. Microbiol. 1994, 32, 2867–2868. [Google Scholar] [CrossRef] [Green Version]
- Henneberg, M.; Holloway-Kew, K.; Lucas, T. Human major infections: Tuberculosis, treponematoses, leprosy—A paleopathological perspective of their evolution. PLoS ONE 2021, 16, e0243687. [Google Scholar] [CrossRef]
- Tió-Coma, M.; Wijnands, T.; Pierneef, L.; Schilling, A.K.; Alam, K.; Roy, J.C.; Faber, W.R.; Menke, H.; Pieters, T.; Stevenson, K.; et al. Detection of Mycobacterium leprae DNA in soil: Multiple needles in the haystack. Sci. Rep. 2019, 9, 3165. [Google Scholar] [CrossRef] [Green Version]
- Hagge, D.; Parajuli, P.; Kunwar, C.; Rana, D.; Thapa, R.; Neupane, K.; Nicholls, P.; Adams, L.; Geluk, A.; Shah, M.; et al. Opening a Can of Worms: Leprosy Reactions and Complicit Soil-Transmitted Helminths. EBioMedicine 2017, 23, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Bratschi, M.W.; Steinmann, P.; Wickenden, A.; Gillis, T.P. Current knowledge on Mycobacterium leprae transmission: A systematic literature review. Lepr. Rev. 2015, 86, 142–155. [Google Scholar] [CrossRef]
- da Silva, M.B.; Portela, J.M.; Li, W.; Jackson, M.; Gonzalez-Juarrero, M.; Hidalgo, A.S.; Belisle, J.T.; Bouth, R.C.; Gobbo, A.R.; Barreto, J.G.; et al. Evidence of zoonotic leprosy in Pará, Brazilian Amazon, and risks associated with human contact or consumption of armadillos. PLoS Negl. Trop. Dis. 2018, 12, e0006532. [Google Scholar] [CrossRef]
- Sakamoto, M.; Takeuchi, Y.; Umeda, M.; Ishikawa, I.; Benno, Y.; Nakase, T. Detection of Treponema socranskii associated with human periodontitis by PCR. Microbiol. Immunol. 1999, 43, 485–490. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Umeda, M.; Sakamoto, M.; Benno, Y.; Huang, Y.; Ishikawa, I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J. Periodontol. 2001, 72, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.K.; Ohk, S.H.; Lee, H.J.; Kang, J.H.; Jeong, G.J.; Yoo, Y.J. Effects of whole cell sonicates of Treponema lecithinolyticum on osteoclast differentiation. J. Periodontol. 2001, 72, 1172–1177. [Google Scholar] [CrossRef] [PubMed]
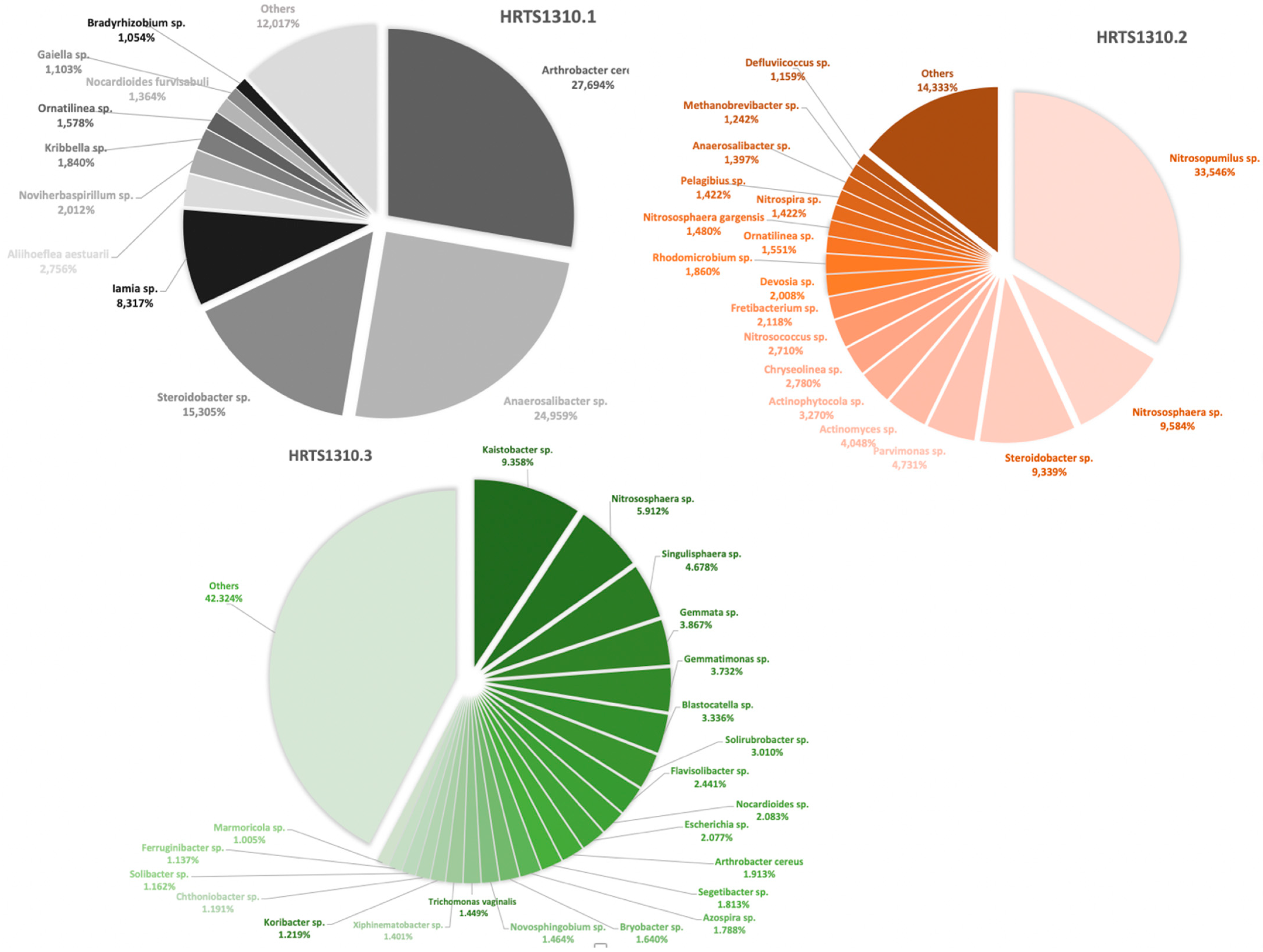
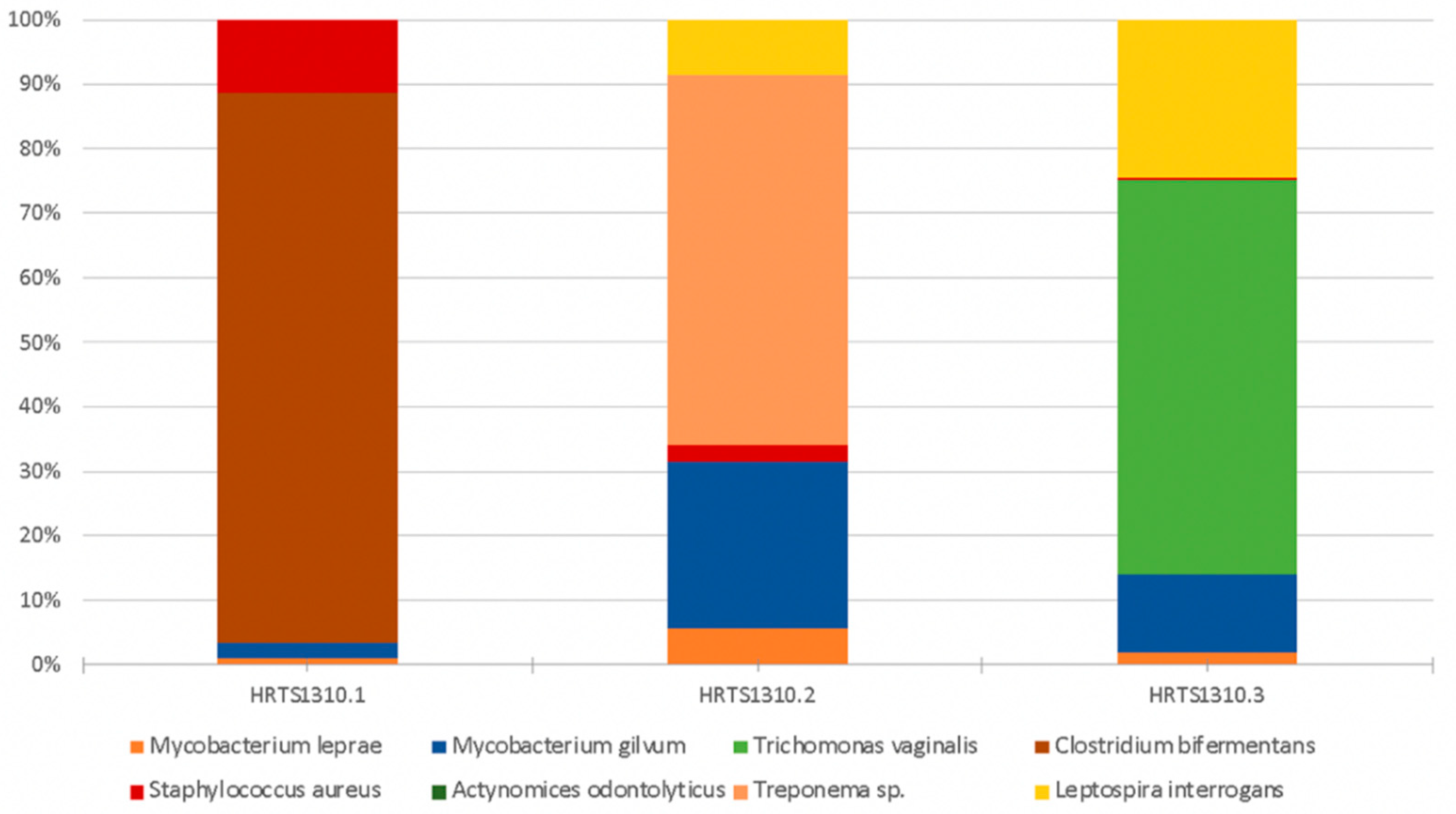

| Microorganism | Observations | HRTS1310.1 a | HRTS1310.2 b | HRTS1310.3 c |
|---|---|---|---|---|
| Mycobacterium leprae | Mycobacterium leprae causes leprosy (Hansen’s disease). This disease can leave its signature in bones if untreated. It primarily leads to the destruction of peripheral nerves and muscles and their atrophy, and the motor function is impaired. The loss of sensation in the extremities causes accidental injuries with subsequent infections. In addition, loss of bone calcium contributes to a slow shrinking of the digits and their transition to claw/pencil-like forms in late-stage leprosy [10,48,70]. | 0.005 | 0.033 | 0.043 |
| Mycobacterium gilvum | This is an environmental mycobacterium isolated from river sediments. M. gilvum has rarely been isolated as an opportunistic pathogen [69] and is viewed as a nontuberculous mycobacterium and as nonpathogenic and found in water and humans [71]. Its presence has been detected in accumulated water associated with a bathing place for leprosy patients [72]. | 0.011 | 0.148 | 0.290 |
| Trichomonas vaginalis | May cause vaginitis or sexually transmitted infections [48,73]. | 0 | 0 | 1.449 |
| Clostridium bifermentans | It is a commensal in the gut, oral cavity, and female genital tract and can be found in soil, sewage, feces, and marine sedimentations. It was once considered to be nonpathogenic; however, it has been recently associated with septic arthritis, osteomyelitis, soft tissue infection, brain abscess, bacteremia, and endocarditis cases [74,75]. | 0.405 | 0 | 0 |
| Staphylococcus aureus | Staphylococcus aureus is a highly virulent, life-threatening pathogen that can cause diseases such as deep abscesses, osteomyelitis, meningitis, sepsis, endocarditis, or pneumonia (to name but a few examples) [62,66]. | 0.055 | 0.032 | 0.007 |
| Treponema sp. | The genus Treponema includes a wide diversity of commensal and pathogen species, most of which exist in complex bacterial communities [47,48]. | 0 | 0.328 | 0 |
| Leptospira interrogans | This pathogen evolves from environmental organisms and a human parasite [47,48]. The most common leptospiral syndrome in humans is leptospirosis, a disorder that localizes in the kidneys and can cause renal failure and ultimately death [48]. | 0 | 0.049 | 0.580 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves-Cardoso, F.; Palomo-Díez, S.; Conde, A.A.; Gomes, C.; Casimiro, S.; da Silva, R.B.; Arroyo-Pardo, E. Metagenomic Research of Infectious Diseases in Archaeological Contexts: Evidence from the Hospital Real de Todos-os-Santos (Portugal). Appl. Sci. 2022, 12, 6096. https://doi.org/10.3390/app12126096
Alves-Cardoso F, Palomo-Díez S, Conde AA, Gomes C, Casimiro S, da Silva RB, Arroyo-Pardo E. Metagenomic Research of Infectious Diseases in Archaeological Contexts: Evidence from the Hospital Real de Todos-os-Santos (Portugal). Applied Sciences. 2022; 12(12):6096. https://doi.org/10.3390/app12126096
Chicago/Turabian StyleAlves-Cardoso, Francisca, Sara Palomo-Díez, Alejandro Alonso Conde, Cláudia Gomes, Silvia Casimiro, Rodrigo Banha da Silva, and Eduardo Arroyo-Pardo. 2022. "Metagenomic Research of Infectious Diseases in Archaeological Contexts: Evidence from the Hospital Real de Todos-os-Santos (Portugal)" Applied Sciences 12, no. 12: 6096. https://doi.org/10.3390/app12126096
APA StyleAlves-Cardoso, F., Palomo-Díez, S., Conde, A. A., Gomes, C., Casimiro, S., da Silva, R. B., & Arroyo-Pardo, E. (2022). Metagenomic Research of Infectious Diseases in Archaeological Contexts: Evidence from the Hospital Real de Todos-os-Santos (Portugal). Applied Sciences, 12(12), 6096. https://doi.org/10.3390/app12126096








