Transfer Learning Analysis for Subvisible Particle Flow Imaging of Pharmaceutical Formulations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. FlowCam Imaging
2.3. Data Analyses
2.3.1. Image Preprocessing
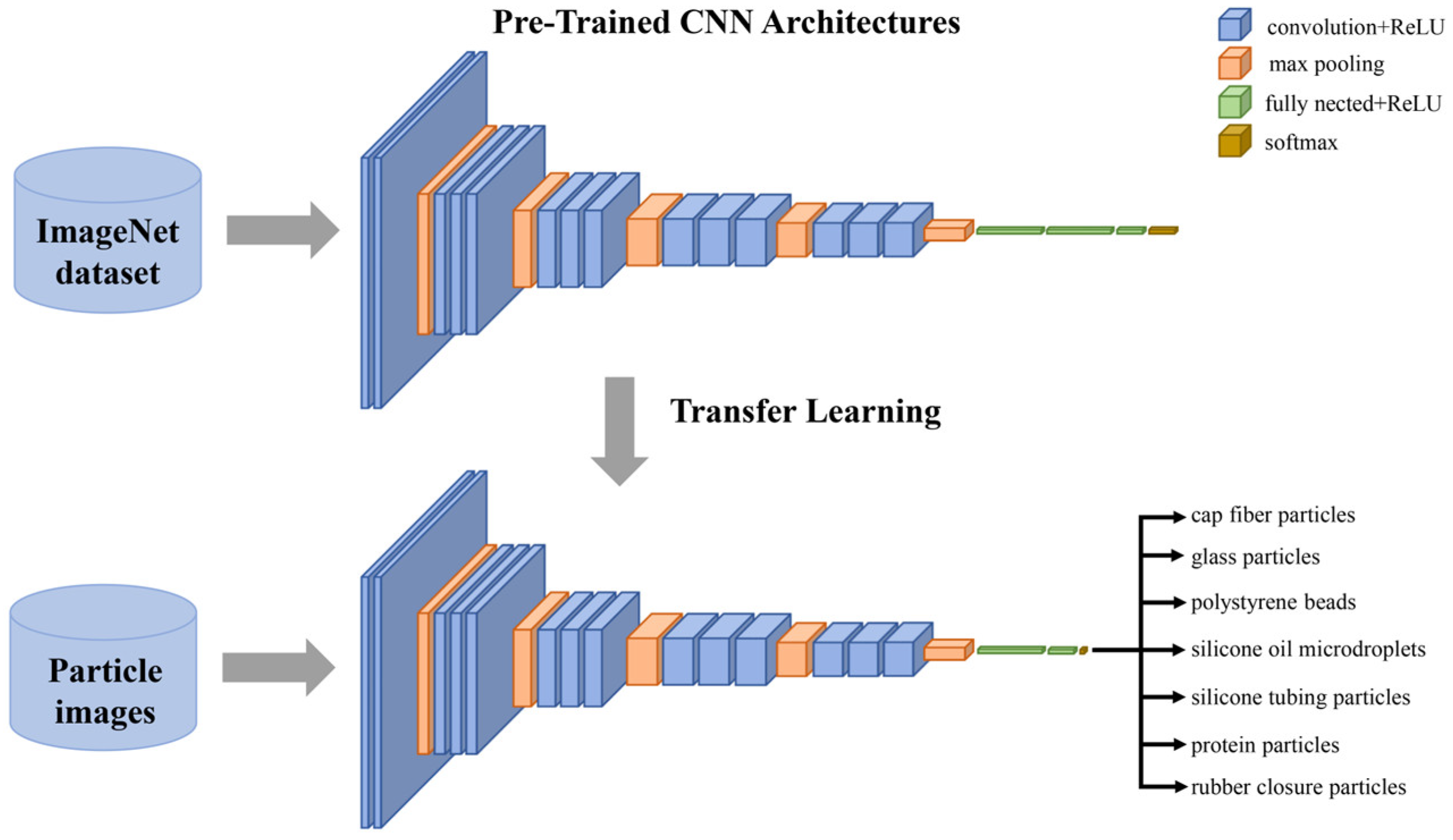
2.3.2. Convolutional Neural Networks
2.3.3. Machine Learning
3. Results and Discussion
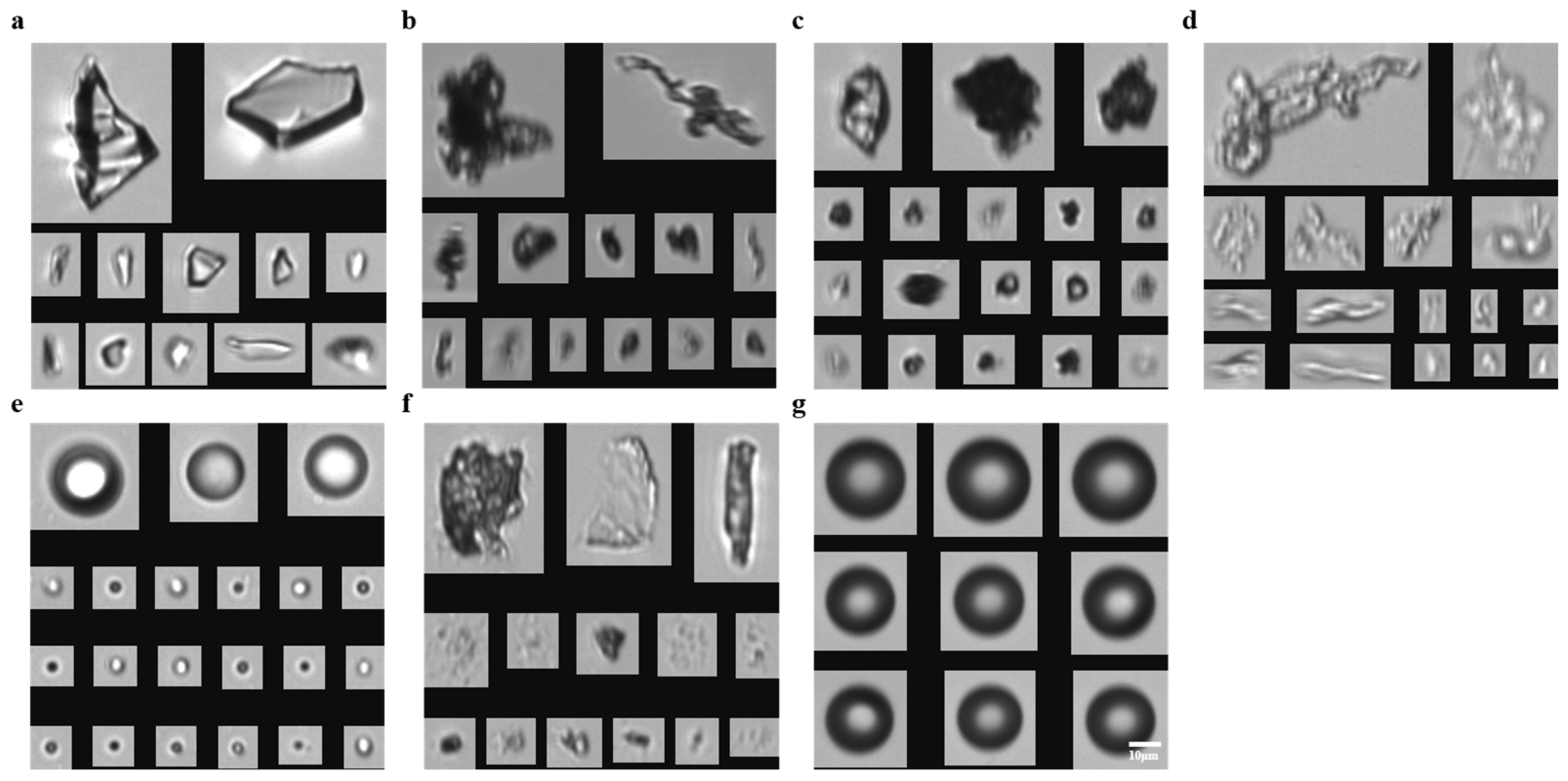
3.1. Particle FIM Imaging
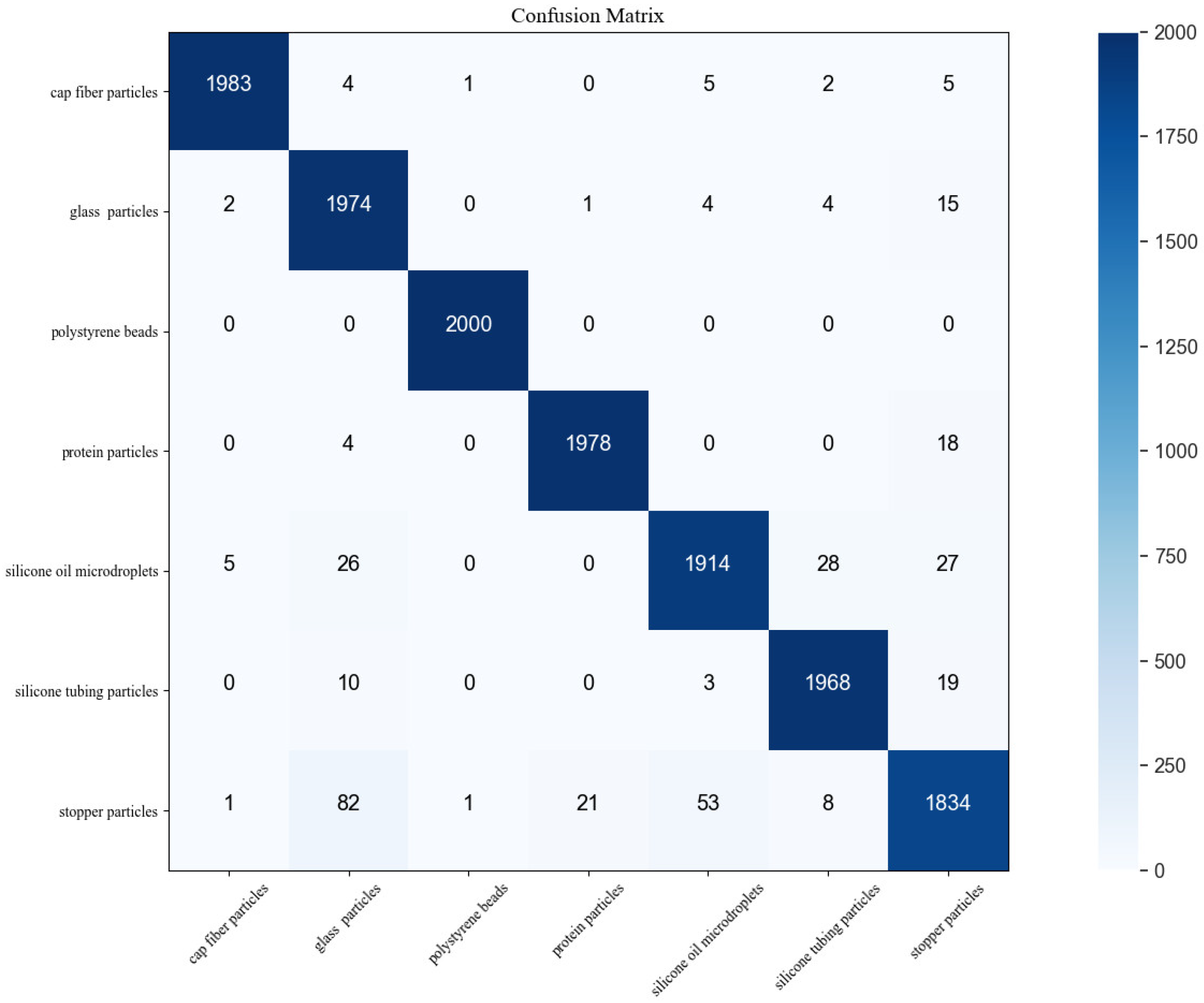
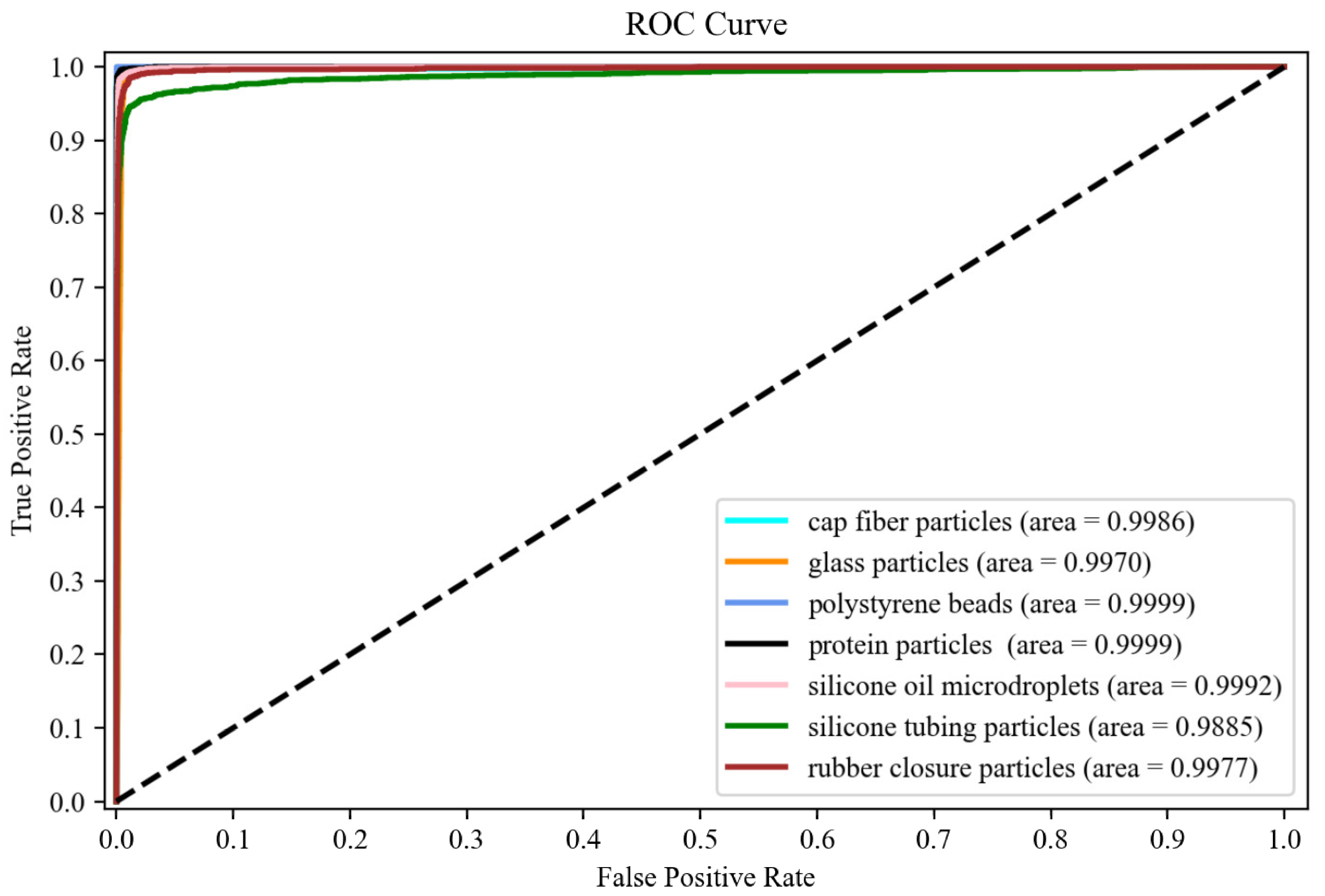
3.2. CNN Transfer Learning for Subvisible Particle Classification
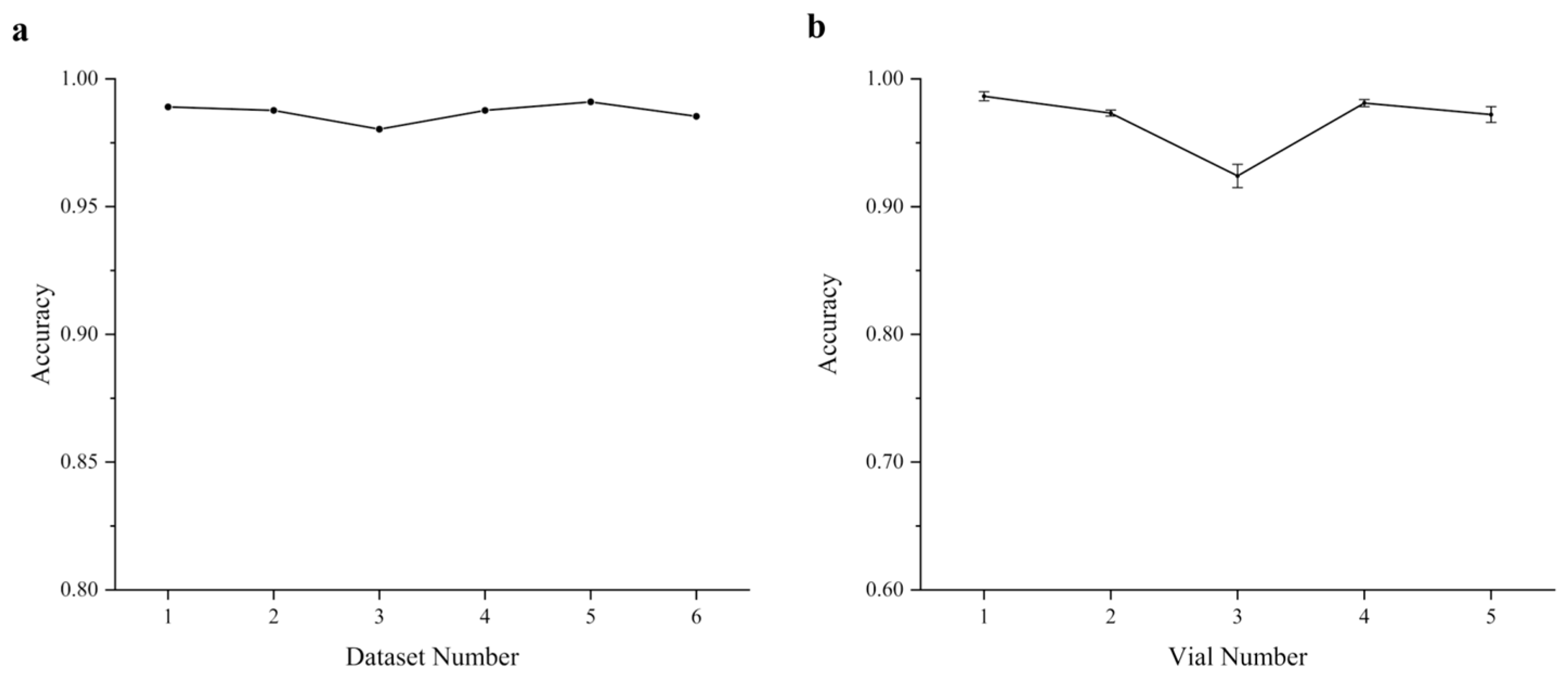
3.3. Inter-Vial and Intra-Vial Variability
3.4. Machine Learning Classifiers for Particle Classification
3.5. Pre-Trained and Scratch-Trained CNN
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, S.K.; Afonina, N.; Awwad, M.; Bechtold-Peters, K.; Blue, J.T.; Chou, D.; Cromwell, M.; Krause, H.-J.; Mahler, H.-C.; Meyer, B.K.; et al. An industry perspective on the monitoring of subvisible particles as a quality attribute for protein therapeutics. J. Pharm. Sci. 2010, 99, 3302–3321. [Google Scholar] [CrossRef]
- Visual Inspection of Injections. The United States Pharmacopeial Convention. 2017. Available online: https://www.pharmout.net/wp-content/uploads/2018/02/NGVF-2016-Visual-Inspections-of-Injection.pdf (accessed on 27 March 2022).
- Wang, W.; Nema, S.; Teagarden, D. Protein aggregation—Pathways and influencing factors. Int. J. Pharm. 2010, 390, 89–99. [Google Scholar] [CrossRef]
- Simler, B.R.; Hui, G.; Dahl, J.E.; Perez-Ramirez, B. Mechanistic complexity of subvisible particle formation: Links to protein aggregation are highly specific. J. Pharm. Sci. 2012, 101, 4140–4154. [Google Scholar] [CrossRef]
- Kumru, O.; Liu, J.; Ji, J.A.; Cheng, W.; Wang, Y.J.; Wang, T.; Joshi, S.B.; Middaugh, C.R.; Volkin, D.B. Compatibility, physical stability, and characterization of an IgG4 monoclonal antibody after dilution into different intravenous administration bags. J. Pharm. Sci. 2012, 101, 3636–3650. [Google Scholar] [CrossRef]
- Pardeshi, N.N.; Zhou, C.; Randolph, T.W.; Carpenter, J.F. Protein nanoparticles promote microparticle formation in intravenous immunoglobulin solutions during freeze-thawing and agitation stresses. J. Pharm. Sci. 2018, 107, 852–857. [Google Scholar] [CrossRef]
- Joubert, M.K.; Hokom, M.; Eakin, C.; Zhou, L.; Deshpande, M.; Baker, M.P.; Goletz, T.J.; Kerwin, B.A.; Chirmule, N.; Narhi, L.O.; et al. Highly Aggregated Antibody Therapeutics can enhance the in vitro innate and late-stage T-cell immune responses. J. Biol. Chem. 2012, 287, 25266–25279. [Google Scholar] [CrossRef] [Green Version]
- Chisholm, C.F.; Soucie, K.R.; Song, J.S.; Strauch, P.; Torres, R.M.; Carpenter, J.F.; Ragheb, J.A.; Randolph, T.W. Immunogenicity of structurally perturbed hen egg lysozyme adsorbed to silicone oil microdroplets in wild-type and transgenic mouse models. J. Pharm. Sci. 2017, 106, 1519–1527. [Google Scholar] [CrossRef]
- Fathallah, A.M.; Chiang, M.; Mishra, A.; Kumar, S.; Xue, L.; Middaugh, C.R.; Balu-Iyer, S.V. The effect of small oligomeric protein aggregates on the immunogenicity of intravenous and subcutaneous administered antibodies. J. Pharm. Sci. 2015, 104, 3691–3702. [Google Scholar] [CrossRef]
- Kotarek, J.; Stuart, C.; De Paoli, S.H.; Simak, J.; Lin, T.-L.; Gao, Y.; Ovanesov, M.; Liang, Y.; Scott, D.; Brown, J.; et al. Subvisible particle content, formulation, and dose of an erythropoietin peptide mimetic product are associated with severe adverse postmarketing events. J. Pharm. Sci. 2016, 105, 1023–1027. [Google Scholar] [CrossRef] [Green Version]
- Particulate Matter in Injections, the United States Pharmacopeial Convention. 2006. Available online: https://www.uspnf.com/sites/default/files/usp_pdf/EN/USPNF/revisionGeneralChapter788.pdf (accessed on 27 March 2022).
- Zölls, S.; Weinbuch, D.; Wiggenhorn, M.; Winter, G.; Friess, W.; Jiskoot, W.; Hawe, A. Flow imaging microscopy for protein particle analysis—A comparative evaluation of four Different Analytical Instruments. AAPS J. 2013, 15, 1200–1211. [Google Scholar] [CrossRef] [Green Version]
- Demeule, B.; Messick, S.; Shire, S.J.; Liu, J. Characterization of particles in Protein Solutions: Reaching the limits of current technologies. AAPS J. 2010, 12, 708–715. [Google Scholar] [CrossRef] [Green Version]
- Methods for Detection of Particulate Matter in Injections and Ophthalmic Solutions. The United States Pharmacopeial Convention. 2018. Available online: https://www.usp.org/ (accessed on 27 March 2022).
- Narhi, L.O.; Corvari, V.; Ripple, D.C.; Afonina, N.; Cecchini, I.; Defelippis, M.R.; Garidel, P.; Herre, A.; Koulov, A.V.; Lubiniecki, T.; et al. Subvisible (2–100 Μm) particle analysis during biotherapeutic drug product development: Part 1, Considerations and Strategy. J. Pharm. Sci. 2015, 104, 1899–1908. [Google Scholar] [CrossRef]
- Strehl, R.; Rombach-Riegraf, V.; Diez, M.; Egodage, K.; Bluemel, M.; Jeschke, M.; Koulov, A.V. Discrimination between silicone oil droplets and protein aggregates in biopharmaceuticals: A novel multiparametric image filter for sub-visible particles in microflow imaging analysis. Pharm. Res. 2012, 29, 594–602. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Deep Learning, H.G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Rawat, W.; Wang, Z. Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Kerr, T.; Clark, J.R.; Fileman, E.S.; Widdicombe, C.E.; Pugeault, N. Collaborative deep learning models to handle class imbalance in flowcam plankton imagery. IEEE Access 2020, 8, 170013–170032. [Google Scholar] [CrossRef]
- Calderon, C.P.; Daniels, A.L.; Randolph, T.W. Deep convolutional neural network analysis of flow imaging microscopy data to classify subvisible particles in protein formulations. J. Pharm. Sci. 2018, 107, 999–1008. [Google Scholar] [CrossRef] [Green Version]
- Daniels, A.L.; Calderon, C.P.; Randolph, T.W. Machine learning and statistical analyses for extracting and characterizing “fingerprints” of antibody aggregation at container interfaces from flow microscopy images. Biotechnol. Bioeng. 2020, 117, 3322–3335. [Google Scholar] [CrossRef]
- Gambe-Gilbuena, A.; Shibano, Y.; Krayukhina, E.; Torisu, T.; Uchiyama, S. Automatic identification of the stress sources of protein aggregates using flow imaging microscopy images. J. Pharm. Sci. 2020, 109, 614–623. [Google Scholar] [CrossRef] [Green Version]
- Umar, M.; Krause, N.; Hawe, A.; Simmel, F.; Menzen, T. Towards quantification and differentiation of protein aggregates and silicone oil droplets in the low micrometer and submicrometer size range by using oil-immersion flow imaging microscopy and convolutional neural networks. Eur. J. Pharm. Biopharm. 2021, 169, 97–102. [Google Scholar] [CrossRef]
- Huh, M.; Agrawal, P.; Efros, A.A. What makes ImageNet good for transfer learning? arXiv 2016, arXiv:1608.08614. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2015, arXiv:1409.1556. Available online: https://arxiv.org/abs/1409.1556 (accessed on 27 March 2022).
| Lay No. | Layer Type | No. of Features | Feature Size | Activation | Input Shape | Output Shape |
|---|---|---|---|---|---|---|
| 1 | Convolutional | 64 | 3 × 3 | ReLU | 150 × 150 × 3 | 150 × 150 × 64 |
| 2 | Convolutional | 64 | 3 × 3 | ReLU | 150 × 150 × 64 | 150 × 150 × 64 |
| 3 | Max pooling (2 × 2) | – | – | – | 150 × 150 × 64 | 75 × 75 × 64 |
| 4 | Convolutional | 128 | 3 × 3 | ReLU | 75 × 75 × 64 | 75 × 75 × 128 |
| 5 | Convolutional | 128 | 3 × 3 | ReLU | 75 × 75 × 128 | 75 × 75 × 128 |
| 6 | Max pooling (2 × 2) | – | – | – | 75 × 75 × 128 | 37 × 37 × 128 |
| 7 | Convolutional | 256 | 3 × 3 | ReLU | 37 × 37 × 128 | 37 × 37 × 256 |
| 8 | Convolutional | 256 | 3 × 3 | ReLU | 37 × 37 × 256 | 37 × 37 × 256 |
| 9 | Convolutional | 256 | 3 × 3 | ReLU | 37 × 37 × 256 | 37 × 37 × 256 |
| 10 | Max pooling (2 × 2) | – | – | – | 37 × 37 × 256 | 18 × 18 × 256 |
| 11 | Convolutional | 512 | 3 × 3 | ReLU | 18 × 18 × 256 | 18 × 18 × 512 |
| 12 | Convolutional | 512 | 3 × 3 | ReLU | 18 × 18 × 512 | 18 × 18 × 512 |
| 13 | Convolutional | 512 | 3 × 3 | ReLU | 18 × 18 × 512 | 18 × 18 × 512 |
| 14 | Max pooling (2 × 2) | – | – | – | 18 × 18 × 512 | 9 × 9 × 512 |
| 15 | Convolutional | 512 | 3 × 3 | ReLU | 9 × 9 × 512 | 9 × 9 × 512 |
| 16 | Convolutional | 512 | 3 × 3 | ReLU | 9 × 9 × 512 | 9 × 9 × 512 |
| 17 | Convolutional | 512 | 3 × 3 | ReLU | 9 × 9 × 512 | 9 × 9 × 512 |
| 18 | Max pooling (2 × 2) | – | – | – | 9 × 9 × 512 | 4 × 4 × 512 |
| 19 | Flatten | – | – | – | 4 × 4 × 512 | 8192 |
| 20 | Dense | 1024 | n/a | ReLU | 8192 | 1024 |
| 21 | Dropout (50% rate) | – | – | – | 1024 | 1024 |
| 22 | Dense | 512 | n/a | ReLU | 1024 | 512 |
| 21 | Dropout (30% rate) | – | – | – | 512 | 512 |
| 22 | Dense | 7 | n/a | Softmax | 512 | 7 |
| Particle Type | Precision | Recall | F-Score |
|---|---|---|---|
| Cap fiber particles | 99.60% | 99.15% | 99.37% |
| Glass particles | 94.00% | 98.70% | 96.29% |
| Polystyrene beads | 99.90% | 100.00% | 99.95% |
| Protein particles | 98.90% | 98.90% | 98.90% |
| Silicone oil microdroplets | 97.91% | 98.40% | 98.15% |
| Silicone tubing particles | 95.62% | 91.70% | 93.62% |
| Rubber closure particles | 96.72% | 95.70% | 96.21% |
| Accuracy Rank | Model | Train (Validation) Accuracy |
|---|---|---|
| 1 | SVM—Fine Gaussian SVM | 87.70% |
| 2 | KNN—Weighted KNN | 86.60% |
| 3 | SVM—Medium Gaussian SVM | 86.30% |
| 4 | KNN—Three-time KNN | 85.40% |
| 5 | KNN—Cosine KNN | 84.40% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, X.; Ma, C.; Sheng, H.; Chen, L.; Fei, Y.; Mi, L.; Han, D.; Ma, J. Transfer Learning Analysis for Subvisible Particle Flow Imaging of Pharmaceutical Formulations. Appl. Sci. 2022, 12, 5843. https://doi.org/10.3390/app12125843
Long X, Ma C, Sheng H, Chen L, Fei Y, Mi L, Han D, Ma J. Transfer Learning Analysis for Subvisible Particle Flow Imaging of Pharmaceutical Formulations. Applied Sciences. 2022; 12(12):5843. https://doi.org/10.3390/app12125843
Chicago/Turabian StyleLong, Xiangan, Chongjun Ma, Han Sheng, Liwen Chen, Yiyan Fei, Lan Mi, Dongmei Han, and Jiong Ma. 2022. "Transfer Learning Analysis for Subvisible Particle Flow Imaging of Pharmaceutical Formulations" Applied Sciences 12, no. 12: 5843. https://doi.org/10.3390/app12125843
APA StyleLong, X., Ma, C., Sheng, H., Chen, L., Fei, Y., Mi, L., Han, D., & Ma, J. (2022). Transfer Learning Analysis for Subvisible Particle Flow Imaging of Pharmaceutical Formulations. Applied Sciences, 12(12), 5843. https://doi.org/10.3390/app12125843






