Comprehensive Review on Development of Early Diagnostics on Oral Cancer with a Special Focus on Biomarkers
Abstract
1. Introduction
2. The Pathogenesis
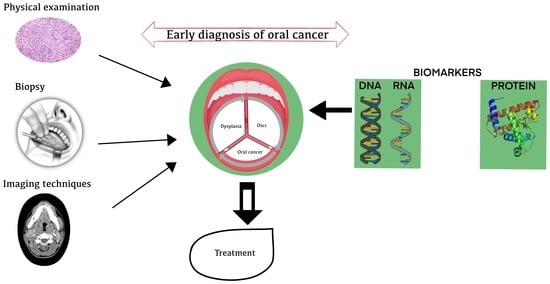
3. Process Flow in the Molecular Diagnosis of OC
4. Need for Early Diagnosis
5. Current Diagnostic Methods for Detection of OC
5.1. Visual Oral Examination
5.2. Physical Examination
5.3. Histopathological Examination
5.4. Vital Staining Techniques
5.5. Biopsy
5.6. Imaging Techniques
5.6.1. Magnetic Resonance Imaging
5.6.2. Computed Tomography
5.6.3. Positron Emission Tomography
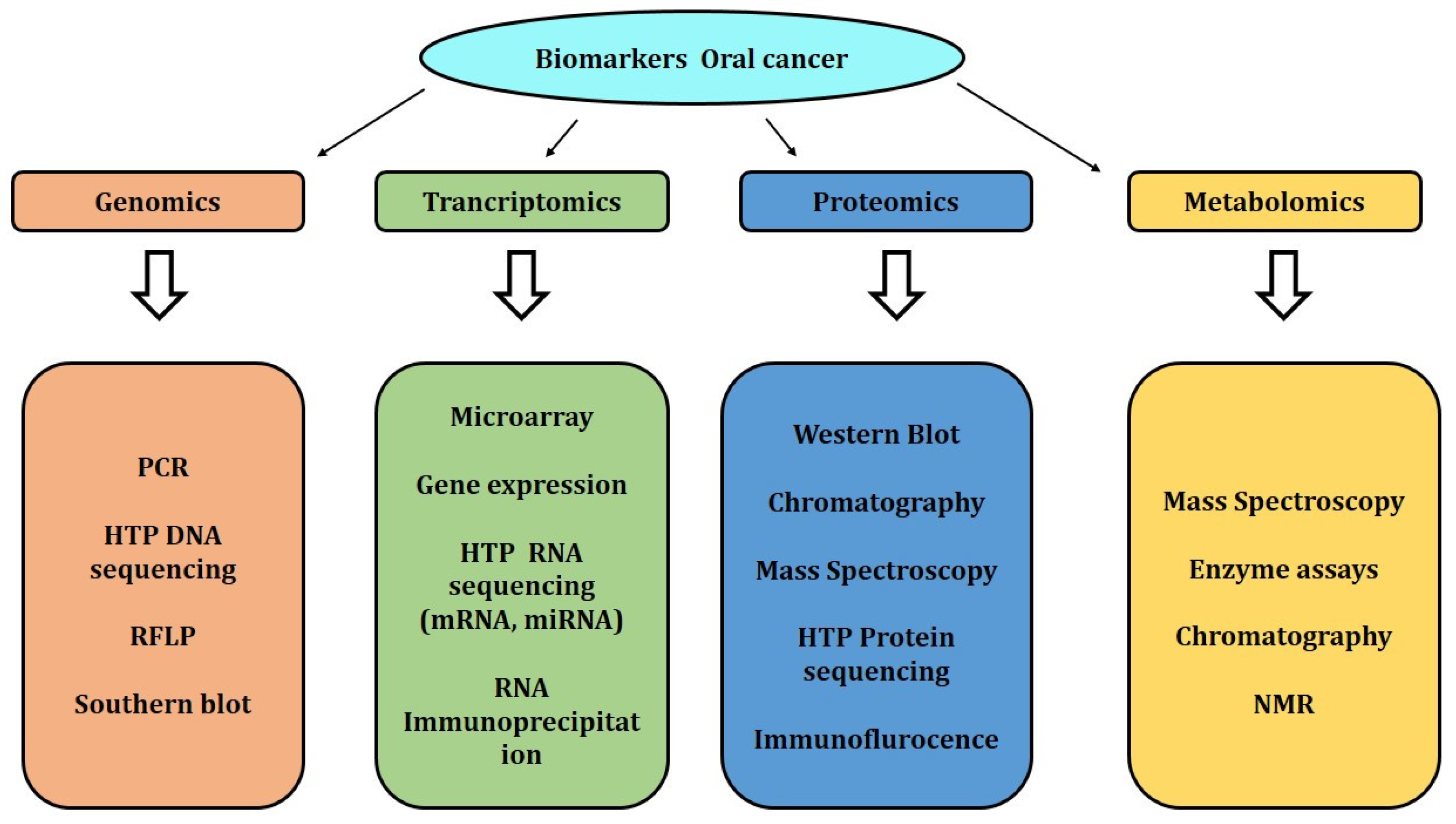
6. Biomarker Detection
6.1. Next-Generation-Sequencing-Based Biomarkers
6.2. Transcriptomics-Based Biomarkers
6.3. Proteomic-Based Biomarkers
6.4. Advantage of Use of Biomarker over the Traditional Technique
7. Future Implications
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gupta, B.; Bray, F.; Kumar, N.; Johnson, N.W. Associations between oral hygiene habits, diet, tobacco and alcohol and risk of oral cancer: A case–control study from India. Cancer Epidemiol. 2017, 51, 7–14. [Google Scholar] [CrossRef]
- Sankaranarayanan, R.; Ramadas, K.; Thomas, G.; Muwonge, R.; Thara, S.; Mathew, B.; Rajan, B. Effect of screening on oral cancer mortality in Kerala, India: A cluster randomised controlled trial. Lancet 2005, 365, 1927–1933. [Google Scholar] [CrossRef]
- Ajay, P.; Ashwinirani, S.; Nayak, A.; Suragimath, G.; Kamala, K.; Sande, A.; Naik, R. Oral cancer prevalence in Western population of Maharashtra, India, for a period of 5 years. J. Oral Res. Rev. 2018, 10, 11. [Google Scholar]
- Gupta, S.; Gupta, R.; Sinha, D.N.; Mehrotra, R. Relationship between type of smokeless tobacco & risk of cancer: A systematic review. Indian J. Med. Res. Suppl. 2018, 148, 56–76. [Google Scholar]
- Kadashetti, V.; Shivakumar, K.; Chaudhary, M.; Patil, S.; Gawande, M.; Hande, A. Influence of risk factors on patients suffering from potentially malignant disorders and oral cancer: A case-control study. J. Oral Maxillofac. Pathol. 2017, 21, 455–456. [Google Scholar] [CrossRef]
- Malik, A.; Mishra, A.; Garg, A.; Shetty, R.; Mair, M.; Chakrabarti, S.; Nair, D.; Balasubramaniam, G.; Chaturvedi, P. Trends of oral cancer with regard to age, gender, and subsite over 16 years at a tertiary cancer center in India. Indian J. Med. Paediatr. Oncol. 2018, 39, 297. [Google Scholar] [CrossRef]
- Murugesan, A.; Sabarinath, S. Awareness of oral cancer among medical students in Chennai. J. Med. Radiol. Pathol. Surg. 2016, 2, 18–22. [Google Scholar] [CrossRef]
- Singh, V.; Singh, A.K.; Dutta, K.D.; Kumar, N.; Kumari, A. Evaluation of quality of life and the nutritional status of oral cancer treated patients as compared with the control group in Varanasi district: A cross sectional study. Int. J. Community Med. Public Health 2019, 6, 4804. [Google Scholar] [CrossRef]
- Thavarool, S.B.; Muttath, G.; Nayanar, S.; Duraisamy, K.; Bhat, P.; Shringarpure, K.; Nayak, P.; Tripathy, J.P.; Thaddeus, A.; Philip, S.; et al. Improved survival among oral cancer patients: Findings from a retrospective study at a tertiary care cancer centre in rural Kerala, India. World J. Surg. Oncol. 2019, 17, 1–7. [Google Scholar] [CrossRef]
- Singh, M.; Prasad, C.P.; Singh, T.D.; Kumar, L. Cancer research in India: Challenges & opportunities. Indian J. Med. Res. 2018, 148, 362–365. [Google Scholar]
- Laprise, H.P.; Shahul, S.A.; Madathil, A.S.; Thekkepurakkal, G.; Castonguay, I.; Varghese, S.; Shiraz, P.; Allison, N.F.; Schlecht, M.C.; Rousseau, E.L.; et al. Periodontal diseases and risk of oral cancer in Southern India: Results from the HeNCe Life study. Int. J. Cancer 2016, 139, 1512–1519. [Google Scholar] [CrossRef]
- Asthana, S.; Patil, R.S.; Labani, S. Tobacco-related cancers in India: A review of incidence reported from population-based cancer registries. Indian J. Med. Paediatr. Oncol. 2016, 37, 152–157. [Google Scholar] [CrossRef]
- Lukesova, E.; Boucek, J.; Rotnaglova, E.; Salakova, M.; Koslabova, E.; Grega, M.; Eckschlager, T.; Rihova, B.; Prochazka, B.; Klozar, J.; et al. High level of tregs is a positive prognostic marker in patients with HPV-Positive oral and oropharyngeal squamous cell carcinomas. Biomed. Res. Int. 2014, 2014, 303929. [Google Scholar] [CrossRef]
- Rivera, C.; Venegas, B. Histological and molecular aspects of oral squamous cell carcinoma (Review). Oncol. Lett. 2014, 8, 7–11. [Google Scholar] [CrossRef]
- Koontongkaew, S. The tumor microenvironment contribution to development, growth, invasion and metastasis of head and necksquamous cell carcinomas. J. Cancer 2013, 4, 66–83. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Watanabe, M.; Wato, M.; Tanaka, A.; Kakudo, K.; Nozaki, M. Cyclin D1 expression is correlated with cell differentiation and cell proliferation in oral squamous cell carcinomas. Oncol. Lett. 2014, 7, 1123–1127. [Google Scholar] [CrossRef]
- Titford, M. A short history of histopathology technique. J. Histotechnol. 2006, 29, 99–110. [Google Scholar] [CrossRef]
- Varshitha, A. Prevalence of oral cancer in India. J. Pharmaceut. Sci. Res. 2015, 7, 845–848. [Google Scholar]
- Farooqi, A.; Shu, C.-W.; Huang, H.-W.; Wang, H.-R.; Chang, Y.-T.; Fayyaz, S.; Yuan, S.-S.F.; Tang, J.-Y.; Chang, H.-W. TRAIL, Wnt, sonic hedgehog, TGFβ, and miRNA signalings are potential targets for oral cancer therapy. Int. J. Mol. Sci. 2017, 18, 1523. [Google Scholar] [CrossRef]
- Downer, M.C.; Moles, D.R.; Palmer, S.; Speight, P.M. A systematic review of test performance in screening for oral cancer and precancer. Oral Oncol. 2004, 40, 264–273. [Google Scholar] [CrossRef]
- Edwards, P.C. Oral cancer screening for asymptomatic adults: Do the United States preventive services task force draft guidelines miss the proverbial forest for the trees? Oral Sur. Oral Med. Oral Path. Oral Radiol. 2013, 116, 131–134. [Google Scholar] [CrossRef]
- Liu, S.Y.; Lu, C.L.; Chiou, C.T.; Yen, C.-Y.; Liaw, G.-A.; Chen, Y.-C.; Liu, Y.-C.; Chiang, W.-F. Surgical outcomes and prognostic factors of oral cancer associated with betel quid chewing and tobacco smoking in Taiwan. Oral Oncol. 2010, 46, 276–282. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Brinkman, B.M.; Wong, D.T. Disease mechanism and biomarkers of oral squamous cell carcinoma. Curr. Opin. Oncol. 2006, 18, 228–233. [Google Scholar] [CrossRef]
- Mehrotra, R.; Gupta, D.K. Exciting new advances in oral cancer diagnosis: Avenues to early detection. Head Neck Oncol. 2011, 3, 33. [Google Scholar] [CrossRef]
- Speight, P.M.; Epstein, J.; Kujan, O.; Lingen, M.W.; Nagao, T.; Ranganathan, K.; Vargas, P. Screening for oral cancer—A perspective from the Global Oral Cancer Forum. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017, 123, 680–687. [Google Scholar] [CrossRef]
- Mascolo, M.; Siano, M.; Ilardi, G.; Russo, D.; Merolla, F.; De Rosa, G.; Staibano, S. Epigenetic disregulation in oral cancer. Int. J. Mol. Sci. 2012, 13, 2331–2353. [Google Scholar] [CrossRef]
- Galeano, M.; Altavilla, D.; Cucinotta, D.; Russo, G.T.; Calò, M.; Bitto, A.; Marini, H.; Marini, R.; Adamo, E.B.; Seminara, P.; et al. Recombinant human erythropoietin stimulates angiogenesis and wound healing in the genetically diabetic mouse. Diabetes 2004, 53, 2509–2517. [Google Scholar] [CrossRef]
- Siriwardena, S.B.S.M.; Tsunematsu, T.; Qi, G.; Ishimaru, N.; Kudo, Y. Invasion-related factors as potential diagnostic and therapeutic targets in oral squamous cell carcinoma—A review. Int. J. Mol. Sci. 2018, 19, 1462. [Google Scholar] [CrossRef]
- Sahibzada, H.A.; Khurshid, Z.; Khan, R.S.; Naseem, M.; Siddique, K.M.; Mali, M.; Zafar, M.S. Salivary IL-8, IL-6 and TNF-α as potential diagnostic biomarkers for oral cancer. Diagnostics 2017, 7, 21. [Google Scholar] [CrossRef]
- Chuang, J.-Y.; Huang, Y.-L.; Yen, W.-L.; Chiang, I.-P.; Tsai, M.-H.; Tang, C.-H. Syk/JNK/AP-1 signaling pathway mediates interleukin-6-promoted cell migration in oral squamous cell carcinoma. Int. J. Mol. Sci. 2014, 15, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.T.; Kang, H.S.; Huh, J.S.; Kim, Y.M.; Lim, Y.; Cho, M. Effects of the novel compound DK223 ([1E,2E-1,2-bis(6-methoxy-2H-chromen-3-yl)methylene]hydrazine) on migration and proliferation of human keratinocytes and primary dermal fibroblasts. Int. J. Mol. Sci. 2014, 15, 13091–13110. [Google Scholar] [CrossRef] [PubMed]
- Hameedaldeen, A.; Liu, J.; Batres, A.; Graves, G.S.; Graves, D.T. FOXO1, TGF-β regulation and wound healing. Int. J. Mol. Sci. 2014, 15, 16257–16269. [Google Scholar] [CrossRef] [PubMed]
- Cervino, G.; Fiorillo, L.; Laino, L.; Herford, A.S.; Lauritano, F.; Giudice, G.L.; Famà, F.; Santoro, R.; Troiano, G.; Iannello, G.; et al. Oral health impact profile in celiac patients: Analysis of recent findings in a literature review. Gastroenterol. Res. Pract. 2018, 2018, 7848735. [Google Scholar] [CrossRef] [PubMed]
- Majtan, J.; Jesenak, M. β-Glucans: Multi-functional modulator of wound healing. Molecules 2018, 23, 806. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.Y.; Hyun, C.; Koh, D.; Park, S.; Lim, Y.; Kim, Y.M.; Cho, M. A novel synthetic material, BMM, accelerates wound repair by stimulating re-epithelialization and fibroblast activation. Int. J. Mol. Sci. 2018, 19, 1164. [Google Scholar] [CrossRef]
- Ligi, L.; Croce, G.; Mosti, J.; Raffetto, D.; Mannello, F. Chronic venous insufficiency: Transforming growth factor-β isoforms and soluble endoglin concentration in different states of wound healing. Int. J. Mol. Sci. 2017, 18, 2206. [Google Scholar] [CrossRef]
- Horng, H.-C.; Chang, W.-H.; Yeh, C.-C.; Huang, B.-S.; Chang, C.-P.; Chen, Y.-J.; Tsui, K.-H.; Wang, P.-H. Estrogen effects on wound healing. Int. J. Mol. Sci. 2017, 18, 2325. [Google Scholar] [CrossRef]
- Cicciù, M.; Herford, A.S.; Cervino, G.; Troiano, G.; Lauritano, F.; Laino, L. Tissue fluorescence imaging (VELscope) for quick non-invasive diagnosis in oral pathology. J. Craniofacial Surg. 2017, 28, e112–e115. [Google Scholar] [CrossRef]
- Weinberg, M.A.; Estefan, D.J. Assessing oral malignancies. Am. Fam. Physician 2002, 65, 1–5. [Google Scholar]
- Mangalath, U.; Mikacha, M.K.; Abdul Khadar, A.H.; Aslam, S.; Francis, P.; Kalathingal, J. Recent trends in prevention of oral cancer. J. Int. Soc. Prev. Community Dent. 2014, 4, 131. [Google Scholar] [CrossRef] [PubMed]
- Dascalu, I.T. Histopathological aspects in oral squamous cell carcinoma. J. Dent. Sci. 2018, 3, 000173. [Google Scholar] [CrossRef]
- Hasan, S.; Elongovan, S. Conventional and advanced diagnostic aids in oral cancer screening—The journey so far. Int. J. Pharm. Pharmaceut. Sci. 2015, 7, 29–33. [Google Scholar]
- Sreeshyla, H.; Sudheendra, U.; Shashidara, R. Vital tissue staining in the diagnosis of oral precancer and cancer: Stains, technique, utility, and reliability. Clin. Cancer Investig. J. 2014, 3, 141. [Google Scholar] [CrossRef]
- Nagaraju, K.; Prasad, S.; Ashok, L. Diagnostic efficiency of toluidine blue with Lugol’s iodine in oral premalignant and malignant lesions. Indian J. Dent. Res. 2010, 21, 223. [Google Scholar] [CrossRef]
- Chaudhari, A.; Hegde-Shetiya, S.; Shirahatti, R.; Agrawal, D. Comparison of different screening methods in estimating the prevalence of precancer and cancer amongst male inmates of a jail in Maharashtra, India. Asian Pac. J. Cancer Prev. 2013, 14, 859–864. [Google Scholar] [CrossRef]
- Mehrotra, R.; Mishra, S.; Singh, M.; Singh, M. The efficacy of oral brush biopsy with computer-assisted analysis in identifying precancerous and cancerous lesions. Head Neck Oncol. 2011, 3, 39. [Google Scholar] [CrossRef]
- Jaitley, S.; Agarwal, P.; Upadhyay, R. Role of oral exfoliative cytology in predicting premalignant potential of oral submucous fibrosis: A short study. J. Cancer Res. Ther. 2015, 11, 471–474. [Google Scholar] [CrossRef]
- Shaila, M.; Shetty, P.; Pai, P. A new approach to exfoliative cytology: A comparative cytomorphometric study. Indian J. Cancer 2016, 53, 193. [Google Scholar] [CrossRef]
- Sylvie, A.L.; Hagen, K.B.E. Oral soft tissue biopsy:an overview. J. Can. Dent. Assoc. 2012, 78, c75. [Google Scholar]
- Zargaran, M. A review of biopsy in dentistry: Principles, techniques, and considerations. J. Dent. Mater. Tech. 2014, 3, 47–54. [Google Scholar]
- Pałasz, P.; Adamski, Ł.; Gorska-Chrząstek, M.; Starzynska, A.; Studniarek, M. Contemporary diagnostic imaging of oral squamous cell carcinoma – a review of literature. Pol. J. Radiol. 2017, 82, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Thukral, C.L.; Gupta, K.; Sood, A.S.; Singla, H.; Singh, K. Role of MRI in evaluation of malignant lesions of tongue and oral cavity. Pol. J. Radiol. 2017, 82, 92–99. [Google Scholar] [CrossRef]
- Schoder, H.; Carlson, D.L.; Kraus, D.H.; Stambuk, H.E.; Gönen, M.; Erdi, Y.E.; Yeung, H.W.D.; Huvos, A.G.; Shah, J.P.; Larson, S.M.; et al. 18F-FDG PET/CT for detecting nodal metastases in patients with oral cancer staged N0 by clinical examination and CT/MRI. J. Nucl. Med. 2006, 47, 755–762. [Google Scholar]
- Masthan, K.M.K.; Aravindha Babu, N.; Dash, K.C.; Elumalai, M. Advanced diagnostic aids in oral cancer. Asian Pac. J. Cancer Prev. 2012, 13, 3573–3576. [Google Scholar] [CrossRef] [PubMed]
- Padmavathi, G.; Bordoloi, D.; Banik, K.; Kunnumakkara, A.B. Cancer biomarkers: Important tools for cancer diagnosis and prognosis. In Next Generation Point-of-Care Biomedical Sensors Technologies for Cancer Diagnosis; Springer: Singapore, 2017; pp. 1–29. [Google Scholar]
- Cristaldi, M.; Mauceri, R.; Di Fede, O.; Giuliana, G.; Campisi, G.; Panzarella, V. Salivary biomarkers for oral squamous cell carcinoma diagnosis and follow-up: Current status and perspectives. Front. Physiol. 2019, 10, 1476. [Google Scholar] [CrossRef]
- Ulaganathan, G.; Mohamed Niazi, K.; Srinivasan, S.; Balaji, V.; Manikandan, D.; Shahul Hameed, K.; Banumathi, A. A clinicopathological study of various oral cancer diagnostic techniques. J. Pharm. Bioallied Sci. 2017, 9, S4–S10. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Soon, W.W.; Hariharan, M.; Snyder, M.P. High-throughput sequencing for biology and medicine. Mol. Syst. Biol. 2013, 9, 640. [Google Scholar] [CrossRef]
- Li, R.; Faden, D.L.; Fakhry, C.; Langelier, C.; Jiao, Y.; Wang, Y.; Wilkerson, M.D.; Pedamallu, C.S.; Old, M.; Lang, J.; et al. Clinical, genomic and metagenomic characterization of oral tongue squamous cell carcinoma in patients who do not smoke. Head Neck 2014, 37, 1642–1649. [Google Scholar] [CrossRef]
- Pickering, C.R.; Zhang, J.; Yoo, S.Y.; Bengtsson, L.; Moorthy, S.; Neskey, D.M.; Zhao, M.; Alves, M.V.O.; Chang, K.; Drummond, J.; et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013, 3, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Mane, S.P.; Ji, X.; Li, Y.; Evans, C.; Crasta, O.R.; Morse, D.; Meagher, R.; Singh, A.; Saxena, D. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol. Med. Microbiol. 2011, 61, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef] [PubMed]
- Shamaa, A.A.; Zyada, M.M.; Wagner, M.; Awad, S.S.; Osman, M.M.; Abdel Azeem, A.A. The significance of Epstein Barr virus (EBV) & DNA topoisomerase II alpha (DNA-Topo II alpha) immunoreactivity in normal oral mucosa, oral epithelial dysplasia (OED) and oral squamous cell carcinoma (OSCC). Diagn. Pathol. 2008, 3, 45. [Google Scholar]
- Kim, K.Y.; Cha, I.H. Possibility of the use of public microarray database for identifying significant genes associated with oral squamous cell carcinoma. Genom. Inform. 2012, 10, 23–32. [Google Scholar] [CrossRef]
- Neville, B.W.; Day, T.A. Oral cancer and precancerous lesions. CA Cancer J. Clin. 2002, 52, 195–215. [Google Scholar] [CrossRef]
- Rentoft, M.; Coates, P.J.; Loljung, L.; Wilms, T.; Laurell, G.; Nylander, K. Expression of CXCL10 is associated with response to radiotherapy and overall survival in squamous cell carcinoma of the tongue. Tumor Biol. 2014, 35, 4191–4198. [Google Scholar] [CrossRef]
- Lajer, C.B.; Nielsen, F.C.; Friis-Hansen, L.; Norrild, B.; Borup, R.; Garnaes, E.; Rossing, M.A.; Specht, L.; Therkildsen, M.H.; Nauntofte, B.; et al. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: A prospective translational study. Br. J. Cancer 2011, 104, 830–840. [Google Scholar] [CrossRef]
- Wiklund, E.D.; Gao, S.; Hulf, T.; Sibbritt, T.; Nair, S.; Costea, D.E.; Villadsen, S.B.; Bakholdt, V.; Bramsen, J.B.; Sørensen, J.A.; et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS ONE 2011, 6, e27840. [Google Scholar] [CrossRef]
- Gibb, E.A.; Enfield, K.S.; Stewart, G.L.; Lonergan, K.M.; Chari, R.; Ng, R.T.; Zhang, L.; MacAulay, C.E.; Rosin, M.P.; Lam, W.L. Long non-coding RNAs are expressed in oral mucosa and altered in oral premalignant lesions. Oral Oncol. 2011, 47, 1055–1061. [Google Scholar] [CrossRef]
- Herrmann, P.C.; Liotta, L.A.; Petricoin, E.F. Cancer proteomics: The state of the art. Dis. Markers 2001, 17, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Turhani, D.; Krapfenbauer, K.; Thurnher, D.; Langen, H.; Fountoulakis, M. Identification of differentially expressed, tumor-associated proteins in oral squamous cell carcinoma by proteomic analysis. Electrophoresis 2006, 27, 1417–1423. [Google Scholar] [CrossRef]
- Wu, J.Y.; Yi, C.; Chung, H.R.; Wang, D.J.; Chang, W.C.; Lee, S.-Y.; Lin, C.-T.; Yang, Y.-C.; Yang, W.-C.V. Potential biomarkers in saliva for oral squamous cell carcinoma. Oral Oncol. 2010, 46, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Mlynarek, A.M.; Balys, R.L.; Su, J.; Hier, M.P.; Black, M.J.; Alaoui-Jamali, M.A. A cell proteomic approach for the detection of secretable biomarkers of invasiveness in oral squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2007, 133, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.Y.; Tsai, M.H.; Tsai, Y.; Hua, C.H.; Tsai, F.J.; Huang, S.Y.; Tsai, C.-H.; Lai, C.-C. Identification of over-expressed proteins in oral squamous cell carcinoma (OSCC) patients by clinical proteomic analysis. Clin. Chim. Acta 2007, 376, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.Y.; Lai, C.C.; Hua, C.H.; Tsai, M.H.; Huang, S.Y.; Tsai, C.H.; Tsai, F.-J. S100A8 is identified as a biomarker of HPV18-infected oral squamous cell carcinomas by suppression subtraction hybridization, clinical proteomics analysis, and immunohistochemistry staining. J. Proteome Res. 2007, 6, 2143–2151. [Google Scholar] [CrossRef]
- Lai, C.H.; Chang, N.W.; Lin, C.F.; Lin, C.D.; Lin, Y.J.; Wan, L.; Lin, C.W. Proteomics-based identification of haptoglobin as a novel plasma biomarker in oral squamous cell carcinoma. Clin. Chim. Acta 2010, 411, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Shintani, S.; Hamakawa, H.; Ueyama, Y.; Hatori, M.; Toyoshima, T. Identification of a truncated cystatin SA-I as a saliva biomarker for oral squamous cell carcinoma using the SELDI ProteinChip platform. Int. J. Oral Maxillofac. Surg. 2010, 39, 68–74. [Google Scholar] [CrossRef]
- Negishi, A.; Masuda, M.; Ono, M.; Honda, K.; Shitashige, M.; Satow, R.; Sakuma, T.; Kuwabara, H.; Nakanishi, Y.; Kanai, Y.; et al. Quantitative proteomics using formalin-fixed paraffin-embedded tissues of oral squamous cell carcinoma. Cancer Sci. 2009, 100, 1605–1611. [Google Scholar] [CrossRef]
- Jancsik, V.A.; Gelencser, G.; Maasz, G.; Schmidt, J.; Molnar, G.A.; Wittmann, I.; Olasz, L.; Mark, L. Salivary proteomic analysis of diabetic patients for possible oral squamous cell carcinoma biomarkers. Pathol. Oncol. Res. 2014, 20, 591–595. [Google Scholar] [CrossRef]
- Sivadasan, P.; Palit, P.; Pinto, S.; Sekhar, N.; Harsha, H.C.; Pandey, A.; Suresh, A.; Hicks, W., Jr.; Gupta, V.; Sirdeshmukh, R.; et al. Salivary proteome for diagnosis and prognosis of oral cancer. Oral Oncol. 2013, 49, S27. [Google Scholar] [CrossRef]
- Sharma, S.; Zuniga, F.; Rice, G.E.; Perrin, L.C.; Hooper, J.D.; Salomon, C. Tumor-derived exosomes in ovarian cancer - liquid biopsies for early detection and realtime monitoring of cancer progression. Oncotarget 2017, 8, 104687–104703. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Du, D.; Wang, X.; Liu, D.; Xu, W.; Luo, Y.; Lin, Y. Recent advances in biosensors for detecting cancer-derived exosomes. Trends Biotechnol. 2019, 37, 1236–1254. [Google Scholar] [CrossRef] [PubMed]
| Method | Sample Type | Highlight |
|---|---|---|
| Physical examination (visual) | Oral cavity (in situ) | Low cost and decreased death and morbidity |
| Vital staining (visual) | Oral tissue | Low cost and non-invasive |
| Histopathology | Oral tissue | Large sample can be analyzed |
| Biopsy | ||
| Brush biopsy | Oral cells | Non-invasive, painless, and low cost |
| Liquid biopsy | Biofluids | Fast, less invasive, and complete sample profile |
| Incisional biopsy | Oral tissue | Less sample; specific and accurate |
| Exfoliative biopsy | Epithelial cells | Low cost and minimum skills |
| Imaging Techniques | ||
| CT scan | Tissue (in situ) | Rapid, good visualization |
| MRI scan | Tissue (in situ) | Non-invasive and high-resolution image |
| Optical coherence topography | Oral tissue | Non-invasive and high-resolution image |
| Molecular method | ||
| PCR | Biofluids, cells | High sensitivity and reproducibility |
| Mass spectroscopy | Tissue (proteins/lipids) | Accurate and high specificity |
| Sl. No. | Biomarker | Sample Type | Technique Used |
|---|---|---|---|
| Protein biomarkers | |||
| 1 | Collagen | Oral tissue | Diffuse reflectance spectroscopy |
| 2 | Elastin | ||
| 3 | Keratin | ||
| 4 | FAD | ||
| 5 | NADH | ||
| 6 | Cytokeratin-19 | Oral tissue | Incisional biopsy |
| 7 | Secretory leukocyte protease inhibitor | Oral cells | Brush biopsy |
| 8 | Epidermal growth factor receptor (EGFR) | Oral epithelial cell linings | Exfoliative biopsy |
| 9 | Serum proteins | Serum | HPLC |
| 10 | Proteases | Saliva | ELISA |
| 11 | ANXA2 | ||
| 12 | CA2 | ||
| 13 | CD44 | ||
| 14 | CRNN | ||
| 15 | CST3 | ||
| 16 | CSTA | ||
| 17 | DSG3 | ||
| 18 | FLNA | ||
| 19 | FSCN1 | ||
| 20 | GANAB | ||
| 21 | GSTP1 | ||
| 22 | HMGCS1 | ||
| 23 | HSPA5 | ||
| 24 | IGFBP3 | ||
| 25 | ISG15 | ||
| 26 | KNG1 | ||
| 27 | LDHA | ||
| 28 | LGALS3BP | ||
| 29 | MMP1 | ||
| 30 | MMP3 | ||
| 31 | MMP9 | ||
| 32 | PRDX2 | ||
| 33 | S100A9 | ||
| 34 | SPARC | ||
| 35 | STAT1 | ||
| 36 | TIMP1 | ||
| 37 | TYMP | ||
| 38 | YWHAB | ||
| 39 | Interleukin-6 (IL-6) | ||
| 40 | Interleukin-8 (IL-8) | ||
| 41 | Interleukin 1a (IL-1a) | ||
| 42 | Interleukin 1b (IL-1b) | ||
| 43 | TNF-a | ||
| 44 | Tissue polypeptide antigen (TPA) | ||
| 45 | Cyfra 21-1 | ||
| 46 | Cancer antigen 125 (CA 125) | ||
| 47 | Telomerase | ||
| 48 | Mac-2 binding protein (M2BP) | Saliva | ELISA, shotgun proteomics |
| 49 | CD44 | Saliva | Immunoblot analysis |
| 50 | CD59 | ||
| 51 | Profilin | ||
| 52 | MRP14 | ||
| 53 | Glutathione | Saliva | HPLC |
| 54 | Squamous cell carcinoma antigen 2 | Saliva | ELISA, shotgun proteomics |
| 55 | Involucrin | ||
| 56 | Calcyclin | ||
| 57 | Cathepsin-G | ||
| 58 | Azurocidin | ||
| 59 | Transaldolase | ||
| 60 | Carbonic anhydrase I | ||
| 61 | Calgizzarin | ||
| 62 | Myeloblastin | ||
| 63 | Vitamin D–binding protein | ||
| 64 | Immunoglobulin heavy-chain constant region gamma (IgG) | Saliva | LC/MS |
| 65 | S100 calcium-binding protein | ||
| 66 | Cofilin-1 | ||
| 67 | Transferrin | ||
| 68 | Fibrin | ||
| 69 | Alpha-1-antitrypsin (AAT) | Saliva | 2DE |
| 70 | Secretory leukocyte peptidase inhibitor (SLPI) | Saliva | MS-based approaches |
| 71 | Cystatin A | ||
| 72 | Keratin 36 | ||
| 73 | Thioredoxin | ||
| 74 | Haptoglobin (HAP) | ||
| 75 | Salivary zinc finger | ||
| 76 | Protein 510 peptide | ||
| 77 | A-amylase | ||
| 78 | Albumin | ||
| 79 | OPN (osteopontin) | Human biopsy | Western blot |
| 80 | EZM2 | Tissue-bank sample | |
| 81 | DEPDC1B | Human biopsy | Immunoprecipitation, Northern blot, Western blot |
| 82 | P63 | Human biopsy | Microarray gene expression |
| 83 | DeltaNp63 | Human biopsy | Cell membrane immunoreactivity, microscope |
| 84 | EIC | ||
| 85 | Podoplanin | ||
| 86 | EGFR | Human biopsy | FISH |
| DNA-based biomarkers | |||
| 87 | DNA (promoter hypermethylation) | Saliva | PCR, qPCR, microarrays |
| 88 | Histone family 3 (HA3) | ||
| 89 | S100 calcium-binding protein P (S100P) | ||
| 90 | Spermidine/spermine N1- acetyltransferase EST (SAT) | ||
| 91 | Ornithine decarboxylase antizyme 1 (OAZ) | ||
| 92 | P53 gene codon 63 | ||
| 93 | CDH1 | Oral tissue | DNA Microarray |
| 94 | MMP3 | ||
| 95 | SPARC | ||
| 96 | POSTN | ||
| 97 | TNC | ||
| 98 | TGM3 | ||
| 99 | HMGA1 | ||
| 100 | PABPC1 | ||
| 101 | NT5E | ||
| 102 | FOS | ||
| 103 | FASN | ||
| 104 | P53 | ||
| 105 | Cytochrome co-oxidase I | Saliva | PCR, qPCR, microarrays |
| 106 | Cytochrome co-oxidase II | ||
| 107 | DAPK | ||
| 108 | DCC | ||
| 109 | TIMP-31 | ||
| 110 | TIMP-3 | ||
| 111 | MGMT | ||
| 112 | CCNA1 | ||
| 113 | MINT-31 | ||
| 114 | DNMT3B | Human biopsy | Microarray gene expression |
| RNA-based biomarkers | |||
| 115 | miR-21 | Tissue-bank sample | mirVana™, microarray gene expression, qPCR |
| 116 | Has-miR-101 | Human biopsy | Microarray gene expression |
| 117 | miR-155-5p | Tissue | Gene expression, micro-RNA expression |
| 118 | miR-216a | ||
| 119 | miR-21-3p | ||
| 120 | miR-96-5p | ||
| 121 | miR-141-3p | ||
| 122 | miR-130b-3p | ||
| 123 | miR-21-5p | ||
| 124 | miR-483-5p | Serum | Gene expression, micro-RNA expression |
| 125 | miR-31-5p | Saliva | Gene expression, micro-RNA expression |
| 126 | miR-31 | Human tissue | qRT-PCR |
| 127 | miR-21 | ||
| 128 | miR-92b | ||
| 129 | miR-34a | Human tissue | Human biopsy, TaqMan miRNA assays |
| 130 | miR-139-5p | Saliva | Agilent miRNA microarray, qPCR |
| 131 | miR-203 | Saliva | qPCR |
| 132 | IL-1b, IL-8 | Saliva | ELISA |
| 133 | Dual-specificity phosphatase 1 (DUSP1) | Saliva | qPCR and microarrays |
| 134 | H3 histone family 3A(H3F3A) | ||
| 135 | Long noncoding HOTAIR | ||
| 136 | miR-125a, miR-200a, miR-31 | ||
| Metabolomics-based biomarkers | |||
| 137 | Cadaverine | Saliva | Capillary electrophoresis–time-of-flight mass spectrometry (CE–TOF-MS) and HPLC with quadrupole/TOF-MS. |
| 138 | Alanine | ||
| 139 | Serine | ||
| 140 | Glutamine | ||
| 141 | Piperidine | ||
| 142 | Taurine piperidine | ||
| 143 | Choline | ||
| 144 | Pyrroline hydroxycarboxylic acid | ||
| 145 | Beta-alanine | ||
| 146 | Alpha-aminobutyric acid betaine | ||
| 147 | Tyrosine | ||
| 148 | Leucine þ isoleucine | ||
| 149 | Histidine | ||
| 150 | Tryptophan | ||
| 151 | Glutamic acid | ||
| 152 | Threonine | ||
| 153 | Carnitine | ||
| 154 | Pipercolic acid | ||
| 155 | Lactic acid | ||
| 156 | Phenylalanine | ||
| 157 | Valine | ||
| 158 | Hypoxanthine | Saliva | Capillary electrophoresis–time-of-flight mass spectrometry |
| 159 | Guanine | ||
| 160 | Guanosine | ||
| 161 | Trimethylamine N-oxide | ||
| 162 | Spermidine | ||
| 163 | Pipecolate | ||
| 164 | Methionine | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umapathy, V.R.; Natarajan, P.M.; Swamikannu, B. Comprehensive Review on Development of Early Diagnostics on Oral Cancer with a Special Focus on Biomarkers. Appl. Sci. 2022, 12, 4926. https://doi.org/10.3390/app12104926
Umapathy VR, Natarajan PM, Swamikannu B. Comprehensive Review on Development of Early Diagnostics on Oral Cancer with a Special Focus on Biomarkers. Applied Sciences. 2022; 12(10):4926. https://doi.org/10.3390/app12104926
Chicago/Turabian StyleUmapathy, Vidhya Rekha, Prabhu Manickam Natarajan, and Bhuminathan Swamikannu. 2022. "Comprehensive Review on Development of Early Diagnostics on Oral Cancer with a Special Focus on Biomarkers" Applied Sciences 12, no. 10: 4926. https://doi.org/10.3390/app12104926
APA StyleUmapathy, V. R., Natarajan, P. M., & Swamikannu, B. (2022). Comprehensive Review on Development of Early Diagnostics on Oral Cancer with a Special Focus on Biomarkers. Applied Sciences, 12(10), 4926. https://doi.org/10.3390/app12104926