Featured Application
The workflow can be used to train other models with other datasets. The trained model can be used to create as many synthetic MRI scans of the rat brain as required for different purposes without the need for further scanning, thus reducing animal suffering and complying with the ethical 3R rule.
Abstract
Translational brain research using Magnetic Resonance Imaging (MRI) is becoming increasingly popular as animal models are an essential part of scientific studies and more ultra-high-field scanners are becoming available. Some disadvantages of MRI are the availability of MRI scanners and the time required for a full scanning session. Privacy laws and the 3Rs ethics rule also make it difficult to create large datasets for training deep learning models. To overcome these challenges, an adaptation of the alpha Generative Adversarial Networks (GANs) architecture was used to test its ability to generate realistic 3D MRI scans of the rat brain in silico. As far as the authors are aware, this was the first time a GAN-based approach was used to generate synthetic MRI data of the rat brain. The generated scans were evaluated using various quantitative metrics, a Turing test, and a segmentation test. The last two tests proved the realism and applicability of the generated scans to real problems. Therefore, by using the proposed new normalisation layer and loss functions, it was possible to improve the realism of the generated rat MRI scans, and it was shown that using the generated data improved the segmentation model more than using the conventional data augmentation.
1. Introduction
Translational Magnetic Resonance Imaging (MRI) of the brain is becoming increasingly popular, as evidenced by the growing number of “MRI”, “Translational” and “Brain” publications in various research databases such as Scopus and PubMed. Rodents are often used as starting points for various complex studies, e.g., in the field of neuroscience. The ability to obtain multimodal information in the same animal, e.g., anatomy, function, and metabolism, makes MRI a powerful imaging modality for neuroscience research in humans, as it allows non-invasive in vivo studies from disease diagnosis to treatment monitoring, the study of anatomy, functions and metabolism, and has no adverse effects [1,2].
The use of MRI allows a simplified transfer of results from animal models to humans, as the same acquisition scheme can be used to generate datasets in both humans and animals. Although it is known that there are differences between species, and the parameters measured are not identical due to structural and functional differences, some homologies may be significant to allow translation. Therefore, the use of rodents remains essential for studying the brain in physiological or pathological states to develop new brain models or new therapeutic strategies. The development of specific rodent models makes it possible to work under more controlled conditions and goes beyond what is possible in human studies [1,2,3].
A variety of preclinical studies have already shown the importance of using rodents, e.g., Magalhães (2018); Magalhães et al. (2018, 2019) [4,5,6] studied the structural and functional modifications of the rat brain induced by the exposure to stress, Boucher et al. (2017) [7] investigated in glioblastoma-bearing mice the possibility of using genetically tailored magnetosomes as MRI probes for molecular imaging of brain tumours, and Vanhoutte et al. (2005) [8] used MRI and mice to detect amyloid plaques for the detection of Alzheimer’s disease in vivo.
Although the use of rodents has many advantages, MRI acquisitions on small animals require special scanners with a high magnetic field and special high-frequency coils to ensure an optimal signal-to-noise ratio and thus high spatial and temporal resolution. However, these types of scanner are becoming more common nowadays [1,2]. The relative lack of appropriate methods and pipelines for data processing, especially considering the unique anatomy, is another limitation that has resulted in a slow increase in the use of MRI to study the rodent brain. The lack of available data impedes the potential innovation of DL algorithms for segmentation and classification of the rodent brain, which could subsequently enable better preclinical studies [4]. The availability of well-trained and tested models for tasks such as segmentation could save several hours of manual segmentation in various works, e.g., [7,9,10].
One of the disadvantages of MRI is the time needed to perform a scan with good resolution and satisfactory data quality. A typical MRI session takes more than 30 min and up to an hour. The availability of scanners is also a problem, as they are not always operational for use or their use is restricted by government regulations. Data protection laws can also lead to a lack of data [11]. For preclinical experiments, the number of scanning sessions must be defined in strict accordance with the 3R ethical rule [12]—Replace, Reduce, Refine—which limits the number of animals scanned and reduces the number of scans available. In addition, the MRI scanner is very expensive and so is each scan. Some efforts have been made to overcome these problems, but the cost per available data ratio remains too high.
Machine learning is on the rise in the field of medical imaging. These techniques can be used to train various machine learning models such as Deep Learning (DL) models to assist specialists in image processing, e.g., segmentation, disease detection, decision support and other tasks. The main limitation is that they usually require a huge amount of data to train properly and perform well.
To mitigate this problem, conventional data augmentation [13] and generative models have been used. These techniques have improved the ability of DL models to achieve the goals for which they were designed, but only slightly, as they do not generate new and realistic data and may even be harmful by creating samples that are anatomically incorrect [14,15,16]. Conventional data augmentation does not fill all existing gaps in the dataset, and some generative models, for example, AutoEncoder and Variational AutoEncoder (VAE), cannot produce sufficiently realistic scans, and they are characterised by blur and low detail in each structure [17]. To deal with this situation, Goodfellow et al. (2014) [18] introduced a new approach to improve generative models, which is called Generative Adversarial Networks (GANs).
GANs are a type of deep neural architecture with two simultaneously trained networks, one generating the image and the other specialised in discrimination. The first aims to generate fake images from an input vector and to fool the discriminator, which is a classifier that evaluates whether the images generated by the generator are realistic. The discriminator gives a probability of truth each time it receives an image, with higher values corresponding to images that are closer to reality. A probability close to 0.5 is the optimal solution, as this means that the discriminator is not able to distinguish the real images from the fake ones [19,20]. The whole process is illustrated in Figure 1.
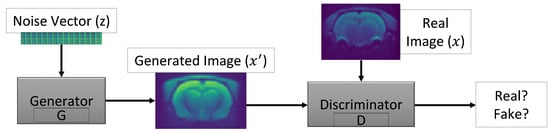
Figure 1.
Representation of the basic Generative Adversarial Networks (GANs) architecture with noise vector (z), generator (G), synthetic data (), real data (x) and discriminator (D).
Given the results that GANs have achieved in various tasks [21,22], it will be possible to disseminate a larger dataset than the original one without protection restrictions once GANs have the ability to anonymise [23]. Additionally, data augmentation with GANs is more robust than traditional data augmentation alone. This reduces the number of real magnetic resonance scans needed to train good medical DL models or even improve some existing models. The fact that the entire 3D MRI volume is used exponentially increases the training difficulty, but by using the entire scan, it is possible to save time and resources by reducing the number of samples required.
The use of MRI for this study is advantageous from a practical and ethical point of view, as high-resolution images can be obtained in a non-invasive way, reducing animal suffering and allowing them to be observed over a longer period. As far as the authors are aware, the use of GANs in preclinical research is a little addressed topic, but it is necessary, as it is fully in line with the ethical 3R rule by reducing the number of scanning sessions. The use of synthetic data generated with GANs in this work improved a DL model for the segmentation of Grey Matter (GM), White Matter (WM), and CerebroSpinal Fluid (CSF), as can be seen in the Section 3.
2. Materials and Methods
2.1. Sigma Dataset of Rat Brains
An MRI dataset of the Wistar rat brain from the Sigma project was used to test the ability of GANs to produce synthetic scans. A total of 210 scanning sessions were performed using a Bruker (Bruker Corporation, Ettlingen, Germany) preclinical ultra-high field scanner 11.7 Tesla and a 4 × 4 surface coil dedicated to the rat head. A T2-weighted Echo Planar Imaging sequence was implemented to acquire resting-state data, with a spatial resolution of 0.375 mm × 0.375 mm × 0.5 mm over a matrix of 64 × 64 × 40, a TR of 2000 ms, a TE of 17.5 ms, and nine averages. Figure 2 shows slices from three different planes—coronal, sagittal and axial—of a rat brain. The dataset consists of 210 scans with a resolution of 64 × 64 × 40, which have been preprocessed to avoid complex values. For more information on the rat brain sigma dataset, see Barrière et al. (2019) and Magalhães et al. (2018) [3,5].
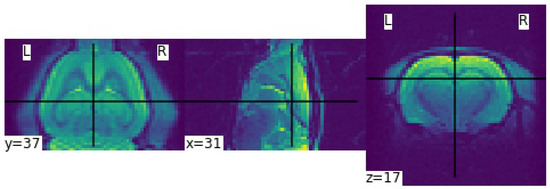
Figure 2.
Example of one Magnetic Resonance Imaging (MRI) scan with resolution of 64 × 64 × 40 from the Sigma rat brain dataset displayed in all three planes—coronal, sagittal and axial planes, respectively.
In this work, a different number of scans and sources—real or synthetic—were used in some steps of the experiment. Therefore, each dataset is formally defined as DsN = (xi, yi)i=1N, where s indicates whether the scans are synthetic (s) or real (r), x indicates the scans, y indicates the respective labels—WM, GM and CSF masks—and N indicates the number of scans. The original sigma dataset of rat brains is formally defined as Dr210 = (xi, yi)i=1210.
2.2. Overall Process Workflow
This work was developed in Python (version 3.7.7, Python Software Foundation), drawing on two important DL frameworks, PyTorch [24] (version 1.7.1+cu101, Facebook AI Research team) and MONAI [25] (version 0.5.0, Project MONAI). The specifications of the workstations are given in Table 1. The code is available at https://github.com/ShadowTwin41/alpha-WGAN-SigmaRat (accessed on 4 January 2022).

Table 1.
Workstation Specifications.
The entire training and evaluation process flow is described in Figure 3. Figure 3A, the Image Resources block, represents the acquisition of the MRI scans and the creation of the dataset. Figure 3B, the Development Environment block, describes the development environment with all frameworks, libraries, and other dependencies for training the models. Figure 3C, the Preprocessing block, corresponds to the preprocessing of the dataset—always working with the whole 3D scan—such as resizing to 64 × 64 × 64 with constant padding of zero values, intensity normalisation between −1 and 1, and some conventional data augmentation. Figure 3D, the Deep Learning application block, is the final step where the training and evaluation of the models are done and where the best model is selected.
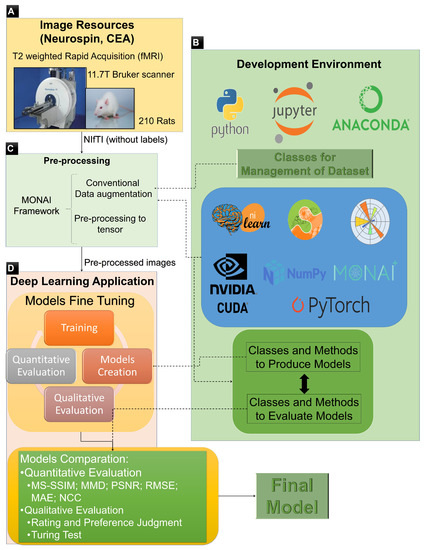
Figure 3.
Overall training and evaluation process workflow, where (A) describes the image resources, (B) the development environment, (C) the pre-processing steps, and (D) the deep learning application.
Some conventional data augmentations such as zooming, rotating, adding Gaussian noise, flipping, shifting, and scaling the intensity were done, as this can fill some gaps in the dataset. The size of the input random vector is an important consideration, as it should be large enough to represent the dataset but not too large to avoid overfitting. Therefore, the sizes 500/1000 were tested.
The creation of the final files is the last step, which is shown in Figure 4. To generate a new scan, a random vector, e.g., a noise vector from a Gaussian distribution, and the final model from the training process are needed. Afterwards, the generated scan is subjected to an automatic post-processing to reverse the preprocessing steps—flipping, cropping to 64 × 64 × 40, and normalising—resulting in an image with the same properties as the original file. Cropping is necessary because the input data were padded as mentioned above. Flipping was done because the input dataset was inverted. Finally, it is possible to generate a 3D NIfTI file from the dataset using the generated scan and metadata—headers and affines—from an original file—from the Dr210.
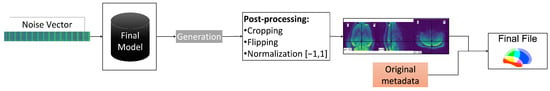
Figure 4.
Overall generation workflow.
Eleven models were created, each trained for 200,000 iterations, i.e., 952 epochs, with a batch size of four, but only the two best and the baseline model for comparison are presented in this article. The first model was the baseline based on the Kwon et al. (2019) [26] model, where the generator, discriminator, encoder, and code discriminator networks are identical, and the latent space is of size 1000. The use of this architecture is more computationally intensive than some traditional GANs architectures because it involves four networks instead of just two. However, this choice was justified because the -GAN architecture avoids mode collapse and fuzziness by adding a VAE and a code discriminator to the GAN network [26]. The loss functions used in this training process were (1) and (2) for generator/encoder, (3) for code discriminator, and (4) for discriminator with —based on the experiments of Kwon et al. (2019) [26]:
where denotes the feedback from the Discriminator, D denotes the Discriminator, G denotes the Generator, C denotes the Code Discriminator, denotes the latent vector of the encoder, denotes the input random vector, denotes a real scan, denotes the gradient penalty of Discriminator, denotes the gradient penalty of Code Discriminator, denotes the L1 loss function, and E denotes the total distribution. Vertical flipping was the only conventional data augmentation used to train this first model. The Adam optimiser [27] was used with a learning rate of 0.0002, betas of 0.9, 0.999 and eps of 10−8. This architecture was called -WGAN_ADNI.
The new loss functions and normalisation layer used in this work are described below. The first proposed architecture, based on Sun et al. (2020) [28] is shown in Figure 5 in which Spectral Normalisation (SN) [29] was added after each convolution to stabilise training, especially discriminator training, batch normalisation layers were replaced by instance normalisation layers to avoid some artefacts, and the activation function LeakyReLU was used instead of ReLU to speed up the training and improve the results [30].
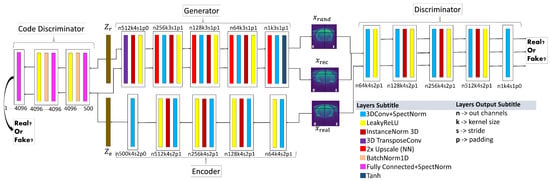
Figure 5.
-WGANSigmaRat1 architecture. Near each layer, there is a code that explains the result of each convolution, e.g., n256k3s1p1: n256, size of the output channels (in this case 256); k3, kernel size (cubic dimension 3 × 3 × 3); s1, stride (1 × 1 × 1); p1, padding (1 × 1 × 1). Please use the virtual version to get a more detailed visualisation.
This architecture was called -WGANSigmaRat1. The loss functions used to train this model were the same as those used for the -WGAN_ADNI model—except for the generator—for which a new loss function was introduced (5) with :
For this training process, all previously mentioned conventional data augmentations—zoom, rotation, Gaussian noise, flip, translation and scaling intensity—as well as the Adam optimiser and a random vector size of 500 were used.
The last proposed model architecture is shown in Figure 6. The main changes compared to the -WGANSigmaRat1 architecture were the removal of SN after each convolution only in the generator and encoder, since in the original work by Miyato et al. (2018) [29] the SN was created to stabilise discriminator training. The instance normalisation layers were also removed in the discriminator and in the code discriminator to avoid artefacts and reduce computational costs. In the new loss function (6), the L1 loss function was replaced by the Gradient Difference Loss (GDL) [31], which is described in the latest works on super-resolution [31,32,33]:
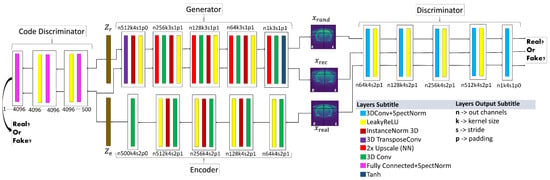
Figure 6.
-WGANSigmaRat2 architecture. Near each layer is displayed a code that explains the result of each convolution, e.g., n256k3s1p1: n256, size of the output channels (in this case 256); k3, kernel size (cubic dimension 3 × 3 × 3); s1, stride (1 × 1 × 1); p1, padding (1 × 1 × 1). Please use the virtual version to get a more detailed visualisation.
The loss functions used to train this model were (3) for the code discriminator, (4) for the discriminator and (6) for the generator/encoder with and . The chosen values for , and proved to be more stable than other values after some experimental tests.
All conventional data augmentations were performed, the size of the input random vector was 500, and a new optimiser was used, AdamW [34] with a learning rate of 0.0002, betas of 0.9, 0.999, eps of 10−8 and a weight decay of 0.01. The AdamW is known to have a more stable weight decay than the Adam. This architecture was named -WGANSigmaRat2.
The difference between the synthesis of rat and human MRI brains is the resolution of the input scans. Here, scans with a resolution of 64 × 64 × 40 were used, but nowadays—with the arrival of better GPUs—it is possible to process scans with higher resolutions, e.g., 256 × 256 × 256. Since an alpha-GAN architecture with new loss functions and a special normalisation (SN) is used, the training can be performed with human MRI scans without much difference, except for the longer runtime.
3. Results
As explained in the Section 2, eleven different models were trained, but only the baseline model for comparison and the two best models are examined and evaluated here, as the other models are not realistic enough.
To select the best metrics to evaluate the different models, various MRI-related articles dealing with 3D scans and GANs were reviewed. The evaluation metrics were then divided into two categories: quantitative—Multi-Scale Structural Similarity Index Measure (MS-SSIM), Mean Absolute Error (MAE), Normalized Cross-Correlation (NCC), Maximum–Mean Discrepancy (MMD) and Dice score—and qualitative—Visual Turing Test performed by experts. The most commonly used metrics for evaluating GANs, e.g., Frechet Inception Distance, Inception Score, and Kernel Inception Distance, were not used due to the complexity of their adaptation to 3D volumes. The mean and standard deviation were calculated for each metric. For MMD, MAE, and NCC metrics, 21,000—the number of scans in dataset *100—comparisons were made between a random real scan and a generated scan; for MS-SSIM, only 2100 comparisons were made for both real and generated data, since the computational power required was much higher than for the other metrics. The MS-SSIM value was calculated under the same conditions as in Kwon et al. (2019) [26], i.e., with a batch size of 8. The MS-SSIM value should be similar to the MS-SSIM value of the real dataset to prove that the distribution is similar. There is no consensus on which metrics should be used to evaluate the best GAN models; however, the use of the quantitative metrics helped to identify the worst models in the first step [35,36,37].
From Table 2, it can be seen that the new models outperformed the baseline model in all quantitative metrics. The -WGANSigmaRat1 had the best overall quantitative results, but after a quick analysis of several scans by some specialists and the authors, the -WGANSigmaRat2 model seems to produce more realistic scans with less blur and fewer structural anomalies, as shown in Figure 7. Therefore, these two models were used for the Turing test. The -WGAN_ADNI model was discarded for the Turing test because it was too easy to distinguish between real and generated scans. This architecture, adapted from [26], was unable to properly learn the distribution of the dataset, and the generated scans contain artefacts, as seen in Figure 7.

Table 2.
Quantitative results. For each cell, the first value is the mean and the second is the standard deviation. The best results are in bold for each metric. An up arrow ↑ next to the metric name means that larger values are better, and a down arrow ↓ means the opposite.
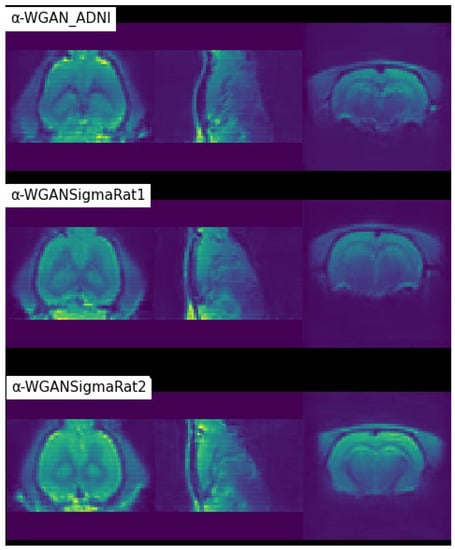
Figure 7.
Coloured visualisation of scans generated by different models in the coronal, sagittal and axial planes, respectively.
The Turing test was performed by four MRI experts on 50 scans, of which 20 were real and 15 + 15 were generated by each model, -WGANSigmaRat1 and -WGANSigmaRat2. Only 50 scans were selected because each evaluation is very time consuming. Therefore, it was agreed with the evaluators that 50 scans were a reasonable number. In addition, the experts had to follow some rules, such as: do not open more than one scan at a time; do not change any answer; consider only the original slices—axial plane; do not look at the dataset Dr210 while performing the test. Table 3 presents the results of the Turing test and the respective expertise of each expert. The level of expertise was categorised as a function of the amount of time each expert had spent working with MRI scans of rat brains. For each rater, the first three values are the number of failed answers, e.g., -WGANSigmaRat1 = 13 means that 13 scans generated by the -WGANSigmaRat1 model were misclassified, and the last value is the number of correct answers.

Table 3.
Turing test results. The first two rows are the classification results of the scans generated by the -WGANSigmaRat1 and -WGANSigmaRat2 models, respectively, the third row is the classification results of the real scans, and in the last row are the number of right classifications. Syn are the Synthetic scans. The correct answers are in bold.
The generated scans were also evaluated in a segmentation task to determine if the synthetic data could improve the results and if it performed better than traditional data augmentation. This test is more important than tricking experts, since synthetic data should be created with a finality, and here, it is the segmentation task. Using a DL model developed by Rodrigues (2018) [38] for GM, WM and CSF segmentation, an experiment was conducted using the following datasets: Dr174 = (xi, yi)i=1174; Dr87 = (xi, yi)i=187; Ds174 = (xi, yi)i=1174; Ds87 = (xi, yi)i=187; Ds261 = (xi, yi)i=1261; Ds348 = (xi, yi)i=1348. The number of scans in each dataset is a multiple of 174, since the maximum of labelled real scans—with existing segmentation masks of GM, WM and CSF—is the dataset Dr174. Only the -WGANSigmaRat2 model was used to generate synthetic scans because the raters involved in the Turing test, especially the first one, indicated that the scans generated with this model were more realistic, had less blur and more detail, and could perform better on the segmentation task than the -WGANSigmaRat1 model. The same 25 real scans were used to test all models, with 80% of the remaining scans used for training and 20% used for validation. The Statistical Parametric Mapping software—SPM12, Welcome Centre for Human Neuroimaging [39]—was used to generate the GM, WM and CSF labels for the generated scans. SPM12 was unable to synthesise the labels of some synthetic scans, so these were discarded, and new scans were generated, i.e., the SPM12 software was used in a final step to assess the quality of the generated scans before segmentation. Table 4 presents the results of this segmentation task performed on different combinations of the previously mentioned datasets. The Dice score results of the first test are shown, i.e., segmentation using only real scans, such as the one presented by Rodrigues (2018) [38]. The use of the conventional data augmentation with the dataset Dr174 was also tested, and the results are presented in Table 5, where tests 10 and 11, random mirroring, random rotation by 3 degrees in all three axes, random zooming between 1.0 and 1.1 and a translation range of 4 voxels were used. For test 12, only random mirroring and a translation range of 4 voxels were used. These conventional data augmentation techniques were recommended by the experts who created the original dataset due to the fact that it is very difficult to place all rat heads in the same position between scanning sessions, so some rotation, some brains larger than others and some translation are normal. The mirroring was also recommended by the experts because the rat brain is almost symmetrical.

Table 4.
Segmentation of Grey Matter (GM), White Matter (WM), and CerebroSpinal Fluid (CSF) dice score, where r refers to real and s refers to synthetic scans. If two datasets are included in the same test, this means that the test was performed with the union of both datasets. The best results are in bold.

Table 5.
Results for comparison between data augmentations techniques. The best results are in bold.
4. Discussion
The initial evaluation of the models was based on the quantitative results of Table 2 to discard the worse models more quickly, e.g., the MS-SSIM value should not be very close to one, as it is important that the model can generate a large variety of scans. Table 2 shows that the -WGANSigmaRat1 model has the second highest MS-SSIM value and the value closest to the real distribution that is higher than the real distribution, which means that the generated scans are more repetitive than the real ones. This is not necessarily a problem, as the value is far from one, but it does not correspond to the real distribution of the sigma dataset of rat brains. This value is even higher for the -WGANSigmaRat2 model, but the generated scans appear to be more realistic after a qualitative visual inspection by the experts. The remaining metrics were too close to each other, so it was necessary to use other tools to further compare these two models.
In the loss function plots of the -WGANSigmaRat2 model (Figure 8), it can be seen that the training of the discriminator was very stable and after 60,000 iterations, the training of the generator also stabilised.
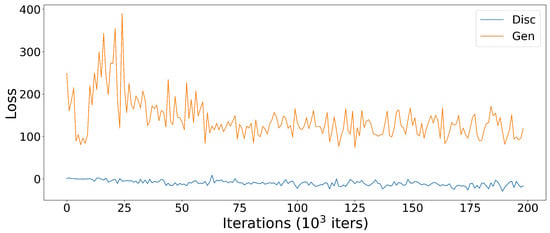
Figure 8.
Discriminator (blue) and Generator (orange) loss function plots of the -WGANSigmaRat2 model training process.
It is not good practice to compare different generative models using loss function diagrams, as they do not correspond to human perception. These diagrams are only a good tool to check for the presence of mode collapse, which is characterised by the divergence between the discriminator and the generator loss function diagrams, one tending to and the other to +∞. Normally, the training runs until this divergence occurs, but since an -GAN architecture with VAE was used, this would never happen or it would require many more iterations, so it was decided to run 200,000 iterations. Comparison of the scans generated after 100,000 and 200,000 training iterations confirmed that the learning process is not directly related to the representation of the loss functions. This can be seen in Figure 9 by the lack of detail in the generated scans after 100,000 training iterations—first row—and a significant improvement after 200,000 training iterations—second row.
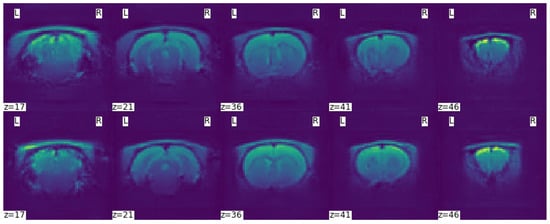
Figure 9.
Axial slices of a generated scan with the -WGANSigmaRat2 model after 100,000 iterations (first row) and after 200,000 iterations (second row) of training. L and R refer to the left and right sides of the brain respectively.
The results presented in Table 3 show that some experts had several difficulties distinguishing between real and fake scans, although some of them had no problems due to their experience with MRI acquisition and the visualisation of rat brains. Rater 1, who was very familiar with the MRIs of rat brains, said that the generated scans looked very grainy, without sufficient anatomical detail, and that the signal distribution outside the skull did not look right. He was the expert best able to distinguish between synthetic and real scans, so his input and comments were very important. Raters 3 and 4 reported some distortions, some strange blurs and low signal homogeneity. Rater 2 is used to analyse MRI scans from humans but not from rats. This could be the reason why he rejected many responses because they were too homogeneous and symmetrical.
After generating the scans for the final test, a selection was made to ensure that all scans had correct anatomy, i.e., whether the SPM12 software could—or could not—generate the GM, WM, and CSF labels. About half of the randomly generated scans had structural defects, and the SPM software was not able to create the GM, WM and CSF labels correctly. This is the main weakness of this work and should be improved in a future work; however, it can easily be overcome by a quick visual inspection to discard the imperfect scans. It was also tested whether SPM could create the labels of the synthetic data generated by the remaining models. It was found that almost all the scans generated by -WGAN_ADNI had problems in creating the labels, so the proposed methods, i.e., the new loss functions and the normalisation layer were able to improve the quality of the generated scans, as can be seen in Figure 10.
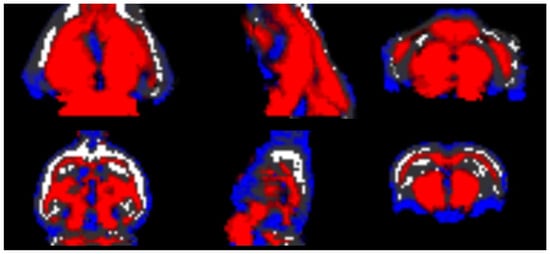
Figure 10.
Semantic masks built with the SPM12 software. The semantic mask in the first row was built using a scan generated by the -WGAN_ADNI model, and the semantic mask in the second row was built using a scan generated by the -WGANSigmaRat2 model. The blue colour is the CSF, the red colour is the WM, and the grey scale is the GM.
The segmentation results using synthetic data are shown in Table 4. The best results for global and white matter segmentation were obtained when a dataset combining Dr174 and Ds348 was used, with an improvement in the Dice score of 0.0172 and 0.0129, respectively. For grey matter and CSF segmentation, the use of the Dr174 and the Ds87 resulted in an improvement of 0.0038 and 0.0764, respectively. The introduction of only 87 synthetic scans—Ds87—into the Dr174 dataset had a big impact, particularly on CSF segmentation, evidencing that the use of GANs can significantly improve results without the need for more scanning sessions.
The use of the conventional data augmentation was also tested, and the results are shown in Table 5. These tests were conducted to assess the impact of this type of data augmentation in this situation. It was found that in this case, the use of conventional data augmentation is detrimental to the training process. This situation occurred because the data augmentation used affects the quality of the data, e.g., interpolations are used when rotating and zooming, which affected the quality of the scans. In Test 10, where 826 conventional augmented samples were used, it can be seen that results were worse due to the aggressive data augmentation used. In Tests 11 and 12, the results improved when fewer augmented samples with conventional techniques were used. However, the results were worse compared to baseline, so data augmentation with synthetic samples was better in this case. It should be noted that the exclusive use of synthetic data was harmful, so real scans are mandatory.
The results obtained with rats may indicate that this technique can also be used to create synthetic 3D MRI scans of healthy and/or unhealthy humans. The next step is therefore to adapt this work to MRI scans of humans.
5. Conclusions
In this work, improved alpha-GAN architectures were proposed to generate synthetic MRI scans of the rat brain. The new proposed loss functions used to train the generator and the new proposed normalisation layer applied to the discriminator enabled more realistic results on rat MRI data. The use of traditional data augmentation also helped to generate more diverse and realistic scans. Different techniques were used to evaluate the different architectures and strategies. The -WGANSigmaRat1 model performed better than the others in the quantitative metrics. However, with the help of some MRI experts, it was determined that the best qualitative model was -WGANSigmaRat2. The Turing test proved that it is possible to trick experts with moderate or low expertise, but it is very difficult to trick experts who are familiar with the dataset. Unfortunately, it was not possible to use this test to check which model generated the best scans. Brain structure segmentation also improved when synthetic scans were added to the original dataset, with the global Dice score improving from 0.8969 to 0.9141. The improvements in CSF segmentation were even more significant, from 0.7468 to 0.8232. It was also found that using synthetic data generated by the new data augmentation model produced better than traditional data augmentation. The segmentation task proved that this method is capable of improving segmentation tasks and therefore other DL tasks.
This work has shown that it is possible to adapt tools developed mainly for processing human MR images to preclinical research. From a methodological point of view, it was possible to extend methods that work with 2D images to 3D images, and it was shown that it is possible to obtain improved segmentation results by adding synthetic scans to the original dataset. The use of data augmentation techniques in the context of preclinical research is interesting, particularly the ability to create larger datasets without scanning more animals, which contributes to the ethical 3R rule, i.e., the reducing part in particular. It is important to note that the use of GANs for data augmentation is not a substitute for traditional data augmentation but a complement. Both together contribute to increasing the amount of data.
Author Contributions
Conceptualization, A.F., R.M., S.M. and V.A.; methodology, A.F. and V.A.; software, A.F.; validation, A.F., R.M., S.M. and V.A.; formal analysis, A.F. and R.M.; investigation, A.F.; resources, R.M., S.M. and V.A.; data curation, A.F., R.M., S.M. and V.A.; writing—original draft preparation, A.F.; writing—review and editing, A.F., R.M., S.M. and V.A.; visualization, A.F.; supervision, R.M., S.M. and V.A.; project administration, V.A. All authors have read and agreed to the published version of the manuscript.
Funding
The MRI dataset of rat brains was acquired in the context of the Sigma project with the reference FCT-ANR/NEU-OSD/0258/2012. This project was co-financed by the French public funding agency ANR (Agence Nationale pour la Recherche, APP Blanc International II 2012), the Portuguese FCT (Fundação para a Ciência e Tecnologia) and the Portuguese North Regional Operational Program (ON.2-O Novo Norte) under the National Strategic Reference Framework (QREN), through the European Regional Development Fund (FEDER), as well as the Projecto Estratégico cofunded by FCT (PEst-C/SAU/LA0026/2013) and the European Regional Development Fund COMPETE (FCOMP-01-0124-FEDER-037298). France Life Imaging is acknowledged for its support in funding the NeuroSpin platform of preclinical MRI scanners. This work of André Ferreira and Victor Alves has been supported by FCT-Fundação para a Ciência e a Tecnologia within the R&D Units Project Scope: UIDB/00319/2020.
Institutional Review Board Statement
All experiments were performed in accordance with the recommendations of the European Union Directive (2010/63/EU) and the French legislation (Decree no. 87/848) for the use and care of laboratory animals. The protocols have been approved by the Comité d’Éthique en Expérimentation Animale du Comissariat à l’Énergie Atomique et aux Énergie Alternatives–Direction des Sciences du Vivant Île de France (CETEA/CEA/ DSV IdF) under protocol ID 12-058.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CSF | Cerebrospinal Fluid |
| DL | Deep Learning |
| GAN | Generative Adversarial Networks |
| GDL | Gradient Difference Loss |
| GM | Grey Matter |
| MAE | Mean Absolute Error |
| MMD | Maximum–Mean Discrepancy |
| MRI | Magnetic Resonance Imaging |
| MSE | Mean Squared Error |
| MS-SSIM | Multi-Scale Structural Similarity Index Measure |
| NCC | Normalised Cross Correlation |
| SN | Spectral Normalisation |
| VAE | Variational AutoEncoder |
| WM | White Matter |
References
- Denic, A.; Macura, S.I.; Mishra, P.; Gamez, J.D.; Rodriguez, M.; Pirko, I. MRI in rodent models of brain disorders. Neurotherapeutics 2011, 8, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Brockmann, M.A.; Kemmling, A.; Groden, C. Current issues and perspectives in small rodent magnetic resonance imaging using clinical MRI scanners. Methods 2007, 43, 79–87. [Google Scholar] [CrossRef]
- Barrière, D.A.; Magalhães, R.; Novais, A.; Marques, P.; Selingue, E.; Geffroy, F.; Marques, F.; Cerqueira, J.; Sousa, J.C.; Boumezbeur, F.; et al. The SIGMA rat brain templates and atlases for multimodal MRI data analysis and visualization. Nat. Commun. 2019, 10, 5699. [Google Scholar] [CrossRef] [Green Version]
- Magalhães, R.J.d.S. An Imaging Characterization of the Adaptive and Maladaptive Response to Chronic Stress. Ph.D. Thesis, University of Minho, Braga, Portugal, 2018. [Google Scholar]
- Magalhães, R.; Barrière, D.A.; Novais, A.; Marques, F.; Marques, P.; Cerqueira, J.; Sousa, J.C.; Cachia, A.; Boumezbeur, F.; Bottlaender, M.; et al. The dynamics of stress: A longitudinal MRI study of rat brain structure and connectome. Mol. Psychiatry 2018, 23, 1998–2006. [Google Scholar] [CrossRef]
- Magalhães, R.; Ganz, E.; Rodrigues, M.; Barrière, D.A.; Mériaux, S.; Jay, T.M.; Sousa, N. Biomarkers of resilience and susceptibility in rodent models of stress. In Stress Resilience: Molecular and Behavioral Aspects; Academic Press: Cambridge, MA, USA, 2019; pp. 311–321. [Google Scholar] [CrossRef]
- Boucher, M.; Geffroy, F.; Prévéral, S.; Bellanger, L.; Selingue, E.; Adryanczyk-Perrier, G.; Péan, M.; Lefèvre, C.T.; Pignol, D.; Ginet, N.; et al. Genetically tailored magnetosomes used as MRI probe for molecular imaging of brain tumor. Biomaterials 2017, 121, 167–178. [Google Scholar] [CrossRef]
- Vanhoutte, G.; Dewachter, I.; Borghgraef, P.; Van Leuven, F.; Van Der Linden, A. Noninvasive in vivo MRI detection of neuritic plaques associated with iron in APP[V717I] transgenic mice, a model for Alzheimer’s disease. Magn. Reson. Med. 2005, 53, 607–613. [Google Scholar] [CrossRef]
- Jamgotchian, L.; Vaillant, S.; Selingue, E.; Doerflinger, A.; Belime, A.; Vandamme, M.; Pinna, G.; Ling, W.L.; Gravel, E.; Meriaux, S.; et al. Tumor-targeted superfluorinated micellar probe for sensitive in vivo 19 F-MRI. Nanoscale 2021, 13, 2373–2377. [Google Scholar] [CrossRef]
- Richard, S.; Boucher, M.; Lalatonne, Y.; Mériaux, S.; Motte, L. Iron oxide nanoparticle surface decorated with cRGD peptides for magnetic resonance imaging of brain tumors. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 1515–1520. [Google Scholar] [CrossRef]
- Foroozandeh, M.; Eklund, A. Synthesizing brain tumor images and annotations by combining progressive growing GAN and SPADE. arXiv 2020, arXiv:2009.05946. [Google Scholar]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen: London, UK, 1959. [Google Scholar]
- Nalepa, J.; Marcinkiewicz, M.; Kawulok, M. Data Augmentation for Brain-Tumor Segmentation: A Review. Front. Comput. Neurosci. 2019, 13, 83. [Google Scholar] [CrossRef] [Green Version]
- Sandfort, V.; Yan, K.; Pickhardt, P.J.; Summers, R.M. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019, 9, 16884. [Google Scholar] [CrossRef]
- Motamed, S.; Rogalla, P.; Khalvati, F. Data Augmentation Using Generative Adversarial Networks (GANs) for GAN-Based Detection of Pneumonia and COVID-19 in Chest X-Ray Images. Inform. Med. Unlocked 2020, 27, 100779. [Google Scholar] [CrossRef]
- Mok, T.C.; Chung, A.C. Learning data augmentation for brain tumor segmentation with coarse-to-fine generative adversarial networks. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2018; pp. 70–80. [Google Scholar] [CrossRef] [Green Version]
- El-Kaddoury, M.; Mahmoudi, A.; Himmi, M.M. Deep generative models for image generation: A practical comparison between variational autoencoders and generative adversarial networks. In International Conference on Mobile, Secure, and Programmable Networking; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–8. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 3, 2672–2680. [Google Scholar]
- Alqahtani, H.; Kavakli-Thorne, M.; Kumar, G. Applications of Generative Adversarial Networks (GANs): An Updated Review. Arch. Comput. Methods Eng. 2019, 28, 525–552. [Google Scholar] [CrossRef]
- Gui, J.; Sun, Z.; Wen, Y.; Tao, D.; Ye, J. A Review on Generative Adversarial Networks: Algorithms, Theory, and Applications. IEEE Trans. Knowl. Data Eng. 2020, 14, 1–28. [Google Scholar] [CrossRef]
- Brock, A.; Donahue, J.; Simonyan, K. Large scale GaN training for high fidelity natural image synthesis. arXiv 2018, arXiv:1809.11096. [Google Scholar]
- Karras, T.; Laine, S.; Aittala, M.; Hellsten, J.; Lehtinen, J.; Aila, T. Analyzing and improving the image quality of stylegan. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 8107–8116. [Google Scholar]
- Shin, H.C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–11. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- Consortium, T.M. Project MONAI. 2020. Available online: https://zenodo.org/record/4323059#.YXaMajgzaUk (accessed on 25 May 2020).
- Kwon, G.; Han, C.; shik Kim, D. Generation of 3D Brain MRI Using Auto-Encoding Generative Adversarial Networks. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2019; pp. 118–126. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Sun, Y.; Yuan, P.; Sun, Y. MM-GAN: 3D MRI data augmentation for medical image segmentation via generative adversarial networks. In Proceedings of the 11th IEEE International Conference on Knowledge Graph, ICKG 2020, Nanjing, China, 9–11 August 2020; pp. 227–234. [Google Scholar] [CrossRef]
- Miyato, T.; Kataoka, T.; Koyama, M.; Yoshida, Y. Spectral normalization for generative adversarial networks. arXiv 2018, arXiv:1802.05957. [Google Scholar]
- Agrawal, N.; Katna, R. Applications of Computing, Automation and Wireless Systems in Electrical Engineering; Springer: Singapore, 2019; Volume 553, pp. 859–863. [Google Scholar] [CrossRef]
- Mathieu, M.; Couprie, C.; LeCun, Y. Deep multi-scale video prediction beyond mean square error. arXiv 2015, arXiv:1511.05440. [Google Scholar]
- Chen, Y.; Shi, F.; Christodoulou, A.G.; Xie, Y.; Zhou, Z.; Li, D. Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 91–99. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, I.; Vilaplana, V. Brain MRI super-resolution using 3D generative adversarial networks. arXiv 2018, arXiv:1812.11440. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Borji, A. Pros and Cons of GAN Evaluation Measures: New Developments. Comput. Vis. Image Underst. 2021, 215, 103329. [Google Scholar] [CrossRef]
- Borji, A. Pros and cons of GAN evaluation measures. Comput. Vis. Image Underst. 2019, 179, 41–65. [Google Scholar] [CrossRef] [Green Version]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training gans. Adv. Neural Inf. Process. Syst. 2016, 29, 2234–2242. [Google Scholar]
- Rodrigues, M.F. Brain Semantic Segmentation: A DL Approach in Human and Rat MRI Studies. Ph.D. Thesis, Universidade do Minho, Braga, Portugal, 2018. [Google Scholar]
- Penny, W.D.; Friston, K.J.; Ashburner, J.T.; Kiebel, S.J.; Nichols, T.E. Statistical Parametric Mapping: The Analysis of Functional Brain Images; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).