1. Introduction
Psoriasis is a skin disease which is a chronic inflammatory skin condition [
1,
2]. The appearance of psoriasis on skin causes anxiety and social obstacles for patients. It results in a low quality of life for patients. To assess psoriasis severity, the Psoriasis Area Severity Index (PASI) composed of erythema (redness), area (the percent of area of skin involved), desquamation (scaling), and induration (thickness) is accepted as a golden standard [
1,
3].
Figure 1 illustrates some psoriasis images taken in outpatient rooms. As shown in
Figure 1, the symptoms of patients with psoriasis are erythema, desquamation, and induration. In addition, the psoriasis regions shown in
Figure 1 have different sizes and arbitrary shapes. There is no doubt that observing and evaluating the patients’ skin to obtain PASI scores is difficult for physicians. On the other hand, the area factor is more important among the PASI parameters to evaluate the psoriasis severity according to physicians’ evaluations. For instance, the ratio of psoriasis area to total skin surface less than 10% could be considered as the mild-level severity. Therefore, efficiently measuring the ratio of psoriasis areas to total skin surface is very helpful to assess the psoriasis severity for physicians.
In dermatology, the symptoms of psoriasis diseases appearing on the skin surfaces can be recorded using digital devices with cameras. It is expected that an image-based computer-aided diagnosis (CAD) developed to analyze psoriasis images can reduce the working load of physicians and obtain a consistent and efficient assessment. To develop an image-based CAD method for clinical psoriasis images, image segmentation is a key process for further analyses [
2,
3,
4,
5,
6]. As we know, there are many traditional image segmentation algorithms such as thresholding, region growing, watersheds, active contour model, graph cuts, etc. [
7]. However, these traditional image segmentation algorithms may not deal with noisy or blurred images well. In addition to traditional image segmentation algorithms, some machine-learning-based image segmentation algorithms have been used to develop CAD methods for dermatology. To date, some CAD methods for psoriasis images have been developed [
3,
4,
5,
8,
9]. For example, Taur et al. proposed a psoriasis segmentation method [
4]. For a psoriasis image, the texture and color features were extracted and combined with a multiresolution-based signature subspace classifier for psoriasis segmentation. Juang et al. presented an image processing algorithm with K-means clustering approach followed by morphological operations for psoriasis segmentation [
5]. The K-means clustering algorithm was used to obtain the coarse segmentation, and then morphological operations were exploited to refine the coarse results. The authors of [
3] developed a machine-learning-based method where a scaling contrast map and the texture features were measured and combined with a support vector machine (SVM), and the Markov random field was developed to identify scaling boundaries in psoriasis skin images. Shrivastava et al. proposed a CAD system for psoriasis image classification [
8] where texture and color features were often extracted, and the authors combined a SVM classifier for psoriasis image classification using many existing methods. In the existing CAD system [
8], the high-order spectra-based (HOS) features, texture features, and color features are extracted. To reduce the feature dimensions, principal component analysis (PCA) is used to find the dominant ones from these extracted features. The dominant features are used with a SVM classifier to achieve psoriasis image classification. Unfortunately, the existing method [
8] seems not to be suitable for psoriasis segmentation.
Since visual features in an image play an important role in many applications, researchers have paid more attention to visual feature extraction to solve the image classification, object detection, and image segmentation problems. Convolution Neural Networks (CNNs) [
10,
11], one of deep learning models, have been demonstrated to be useful to learn the multilevel visual features from images. The main advantage of CNN is that strong features invariant to distortion and position at the same time can be extracted for image classification. Since 2012, the ability of the deep learning method AlexNet has been demonstrated for image classification compared with traditional machine-learning methods. In addition to AlexNet, there are some popular CNNs such as VGGNet, ResNet, GoogleNet, Inception, and DarkNet. For example, VGGNet stacks more than ten 3 × 3 convolution layers to generate a deeper network. To reduce the effect of the vanishing gradient problem, the core idea of ResNet is the identity shortcut connection for designing a deeper network. It is expected that CNNs can be exploited to extract useful features to develop CAD methods for medical image analysis. For instance, an existing method [
12] developed based on a sliding-window approach was proposed for psoriasis images. The authors of [
12] used a CNN to extract useful features from a local rectangle region. Then, these features were exploited to determine whether one pixel was a psoriasis pixel. After evaluating each pixel based on these features, the psoriasis regions can be separated from the others in a psoriasis image. Though the existing method [
12] can provide a better performance, its computational complexity is a little high. Actually, some state-of-the-art deep learning-based medical image segmentation methods such as the Fully Convolution Neural Network (FCN) and U-Net have been proposed [
11,
13,
14,
15,
16,
17,
18,
19,
20]. To achieve pixel-wise classification, the fully connected layer is replaced by a fully convolutional layer in FCNs [
19,
20]. Another popular approach is the U-Net, which was developed based on the encoder-decoder structure [
13,
19,
20]. In U-Net, the encoder compresses the input into a feature-space (also called latent-space) representation and the decoder predicts the network’s output from the feature-space representation. Some existing methods based on the U-Net have been proposed to analyze mammogram, computed tomography, and Magnetic Resonance Imaging (MRI) images.
From the view of segmentation output, image segmentation can be categorized into semantic segmentation and instance segmentation [
11,
17]. Semantic segmentation achieves pixel-level classification with a set of object categories such as buildings, vehicles, and humans for all image pixels. Compared with semantic segmentation, instance segmentation can not only assign the pixel-level label information but also identify each individual region on the basis of specific categories in an image. Actually, the skin image shown in
Figure 1 may have several psoriasis regions, and physicians often prefer to observe and evaluate the status of psoriasis regions for further treatment research in real applications [
3]. This means that detecting and identifying each psoriasis regions in a skin image is helpful in conducting further treatment research for physicians. Although the U-net can achieve semantic segmentation, it cannot distinguish different regions of the same category. This means that an instance segmentation scheme is suitable for psoriasis image segmentation compared with semantic segmentation.
So far, most existing methods have been designed to deal with psoriasis skin images captured under a simple background or without a background, i.e., constrained psoriasis images [
1,
3,
21]. Furthermore, images captured by smartphones or digital cameras may have a poor visual quality to factors such as motion blurring, image resolution, noise, etc. Unfortunately, the psoriasis images captured by smartphones or digital cameras in outpatient rooms may also have a complex background. This means that the captured clinical psoriasis images are often unconstrained in real applications. However, the existing image segmentation algorithms may not be robust to deal with unconstrained psoriasis images well. Therefore, these motivate us to develop a robust instance segmentation scheme to deal with the clinical unconstrained psoriasis skin images in dermatology.
The rest of this paper is organized as follows.
Section 2 describes the proposed instance segmentation scheme systematically. In
Section 3, we elaborate each part of the proposed instance segmentation scheme based on transfer learning.
Section 4 demonstrates the experimental results. Finally,
Section 5 concludes this paper.
2. System Description
As shown in
Figure 1, a clinical psoriasis skin image often has normal skin regions, psoriasis areas, and a simple background. Consider an input image of size
, where
and
represent the width and height of the input image, respectively. The goal of the proposed scheme is to separate some objects or instances (normal skin regions and psoriasis areas) from the background in an unconstrained image. Then, these instances are classified into three categories: Normal
, psoriasis
, and background
. This means that each pixel of an unconstrained image will be assigned with one of the class information (
,
, and
) after image segmentation.
Generally, an instance segmentation method can be divided into two parts: object detection and pixel classification. Currently, the instance segmentation methods are usually developed based on object detection algorithms such as single-shot detector (SSD) [
22] and Faster Region-Based Convolutional Neural Networks (Faster R-CNN) [
23]. According to the different types of object detection architecture, instance segmentation methods can be divided into two categories: Single-stage [
24,
25] and two-stage [
26,
27,
28].
As for the two-stage category, these instance segmentation methods are developed based on existing two-stage object detection algorithms. For example, Mask R-CNN [
24] is an extension of Faster R-CNN. To achieve instance segmentation, Mask R-CNN is composed of backbone network, Regional Proposal Network (RPN), feature pyramid network (FPN), ROIAlign, and FCN. The backbone network composed of several convolutional layers is utilized to extract multilevel feature maps from an image, and FPN is designed to generate multiscale feature maps for effectively classifying and localizing objects with multiple sizes. RPN is used to find regions of interest (ROIs) from these multiscale feature maps. ROIAlign is exploited to reduce the misalignment of an object position between the feature map and the spatial coordinate. The last key component, the FCN network, is used to find the precise boundary of each ROI. Another existing method [
25] predicts a set of position-sensitive output score maps which simultaneously address object classes, boxes, and masks. The above state-of-the-art two-stage methods can achieve satisfying performance, but they are time-consuming.
As for single-stage instance segmentation, the existing methods are usually proposed based one-stage object detection methods. Polarmask [
27] formulates the instance segmentation problem as instance center classification and dense distance regression in a polar coordinate. SPRNet [
28] has an encoder-decoder structure where classification, regression, and mask branches are processed in parallel. SPRNet generates each instance mask from a single pixel, and then resizes the mask to fit the corresponding box to gain the final instance level prediction. In the decoding part, each pixel is exploited as an instance carrier to create the instance mask on which consecutive deconvolutions are applied to gain the final predictions. The authors of [
26] proposed an instance segmentation network, You Only Look At CoefficienTs (YOLACT), for real-time applications. Compared with the existing image segmentation methods, YOLACT is more efficient because one-stage object detector is adopted as the base.
As mentioned in
Section 1, the goal of the proposed instance segmentation scheme is to effectively detect and identify psoriasis regions and normal skin areas in clinical psoriasis images. Currently, most deep learning frameworks are developed to deal with natural images, and only a few deep-learning-based frameworks are designed for psoriasis images [
12,
21]. Though these existing learning-based segmentation methods are developed for dealing with natural images, they can be modified to develop a CAD scheme for medical images. Unfortunately, it may be difficult to collect many medical images and corresponding labeling information for training deeper networks. On the other hand, transfer learning [
20,
29] is to transfer the knowledge from a related domain (source domain) to increase the performance in a specific domain (target domain). It is expected that transfer learning is a time-saving approach for building a machine-learning-based method when only a small training dataset is available in real applications. Therefore, an instance segmentation scheme based on single-stage instance segmentation can be developed via transfer learning for psoriasis images.
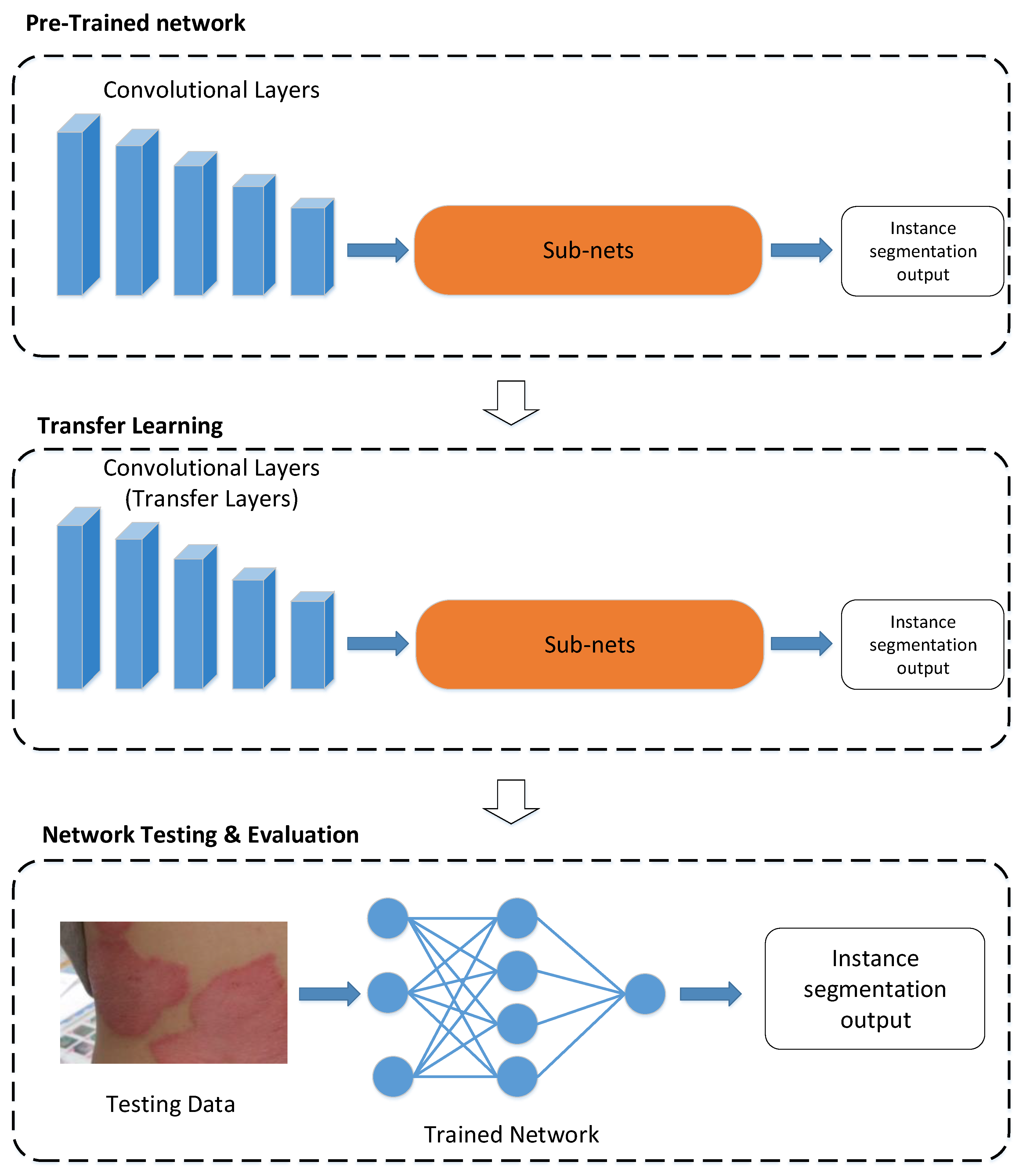
Figure 2 shows the methodological illustration of the proposed scheme via transfer learning. As shown in
Figure 2, a pretrained deep neural network is selected from the source domain and can be retrained on the target domain to generate a CAD system via transfer learning. In real applications, the source domain
with the corresponding task
is often collected as follows:
where
and
denote the
i-th instance and the corresponding label information in
, respectively, and
means the number of instances in
. For example, there are some popular datasets such as ImageNet [
10] and MSCOCO [
30], and these datasets are used for computer vision tasks such as image classification, object detection, image segmentation, and so on. For instance, in segmentation, the label space
contains the class, the bounding box, and the mask information of the
i-th instance in
. Similarly, the data in the target domain
with the corresponding task
are mentioned below:
where
and
represent the
i-th instance and the corresponding label information in
, respectively, and
is the number of instances in
. Here, the target task
is instance segmentation for psoriasis images. Then the following problem is how to efficiently learn a network model with high accuracy for
according to
or trained models in
.
To date, some approaches such as model finetuning, multi-task learning, and domain-adversarial learning have been developed in transfer learning [
20,
29]. As for model finetuning, layer transfer is a common approach for network-based methods. Since the proposed psoriasis segmentation scheme is a network-based method, as shown in
Figure 2, layer transfer is then a suitable approach to train the network model in the proposed scheme. In addition, it is worth mentioning that overfitting may happen if a small dataset is used to train a deep neural network. Unfortunately,
may be less than
in some applications, e.g., medical image analysis and computer-aided diagnosis. As we know, data augmentation is a very powerful method to prevent the model overfitting issue [
31]. Due to the limited size of psoriasis images, data augmentation was adopted to enlarge the size of the training image dataset here.
4. Experimental Results
To evaluate the proposed instance segmentation scheme, YOLACT++ [
33] was implemented in a PC with CPU i7-9700, 32 GB RAM, and NVIDIA Tesla T4. The backbone was ResNet-101, which had 100 convolutional layers and 1 fully connected layer. The output feature maps of the backbone were combined to yield multilevel visual feature maps in YOLACT++. The parameters of YOLACT++ are listed in
Table 1. Since the input size of the proposed scheme was 550 × 550 pixels, each input image was resized. Moreover, in line with our experiences, the weights in Equation (5) were predefined as follows:
= 1,
= 1.5, and
= 6.125. Here, the optimizer for network model learning was the stochastic gradient descent algorithm where the values of the learning rate and momentum were 0.0001 and 0.9, respectively. The parameter, IOU_THRESHOLD, was a threshold exploited in NMS.
4.1. Data Augmentation
The psoriasis images were captured using smartphones or digital cameras in outpatient rooms. For performance evaluation, there were 52 unconstrained psoriasis images with spatial resolutions of 3000 × 4000 pixels. These high-resolution images were partitioned to obtain 400 psoriasis images with 640 × 480 pixels. Some unconstrained psoriasis images are shown in
Figure 5. As shown in
Figure 5, we can observe some factors. These psoriasis images have different backgrounds, the sizes of psoriasis regions are different, and the psoriasis regions have different severity levels and arbitrary boundaries. For example, the left image in the top row of
Figure 5 has a background including clothes and a chair. Furthermore, these psoriasis regions have different sizes and severity levels, as shown in
Figure 5. After image partition, these 400 images were divided into training, validation, and testing sets for model training and performance evaluation.
As we know, the more data a machine learning-based scheme can access, the more powerful and robust the scheme can be. Here, data augmentation [
31] was adopted to increase the variability of the input images for model training, enabling the proposed scheme to process unseen images well. The generic ways for augmenting images are to perform geometric transformation [
7] and photometric change [
7]. Here, some geometric transformations such as flipping and translation, two photometric changes, and noise insertion were adopted. A combination of these operations was also used for data augmentation. Thus, we obtained 38,400 augmented psoriasis images for model training.
Figure 6 shows some examples of augmented psoriasis images. In
Figure 6, the first column (
Figure 6(a1,a2)) shows the original images and the second column (
Figure 6(b1,b2)) shows the results of brightness adjustment.
Figure 6(c1,c2) are the resulting images after combining three operations: Flipping, brightness increasing, and translation.
Figure 6(d1,d2) are the resulting images after combining three operations: Flipping, brightness decreasing, and translation.
4.2. Performance Indices
For objective evaluation, several performance indices are used to assess the performance of image segmentation. The recall and precision rates are widely used in many applications such as object detection, shot change detection, etc. We utilized them to evaluate the performance of the proposed scheme. The recall rate is the ratio of correct detections to the total number of pixels in the
i-th class (
,
, and
), the precision rate is the ratio of correct detections to total number of detected pixels in the
i-th class, and F1_score is the weighted mean of recall and precision for the
i-th class [
14,
34,
35,
36]. The definitions of the performance indices are described as follows:
where
TP,
FP,
TN, and
FN represent the true positive, false positive, true negative, and false negative for classifying pixels into the
i-th class. Theoretically, if an image segmentation scheme achieves high recall and precision rates, its performance is considered effectively. In addition, we also utilize the accuracy rate to evaluate the proposed scheme [
14,
37]. The accuracy rate can be calculated as follows:
As we can see in Equation (9), the higher the value of the accuracy rate, the better the scheme’s performance.
As for object detection, Mean Average Precision (mAP) [
22,
23] was utilized to assess whether a scheme can simultaneously detect several kinds of objects successfully. In addition, intersection over union (IoU) [
22,
23] was also adopted as a performance index to evaluate whether the proposed scheme can locate objects precisely. We expected that the higher the mAP and IoU, the better the performance.
4.3. Performance Analysis
Here, we conducted the qualitative and quantitative evaluation for performance analysis.
4.3.1. Qualitative Evaluation
Figure 7 illustrates the instance segmentation results.
Figure 7a,b are the input images and the ground truth, respectively. As shown in
Figure 7(a1–a4), the input images contained some psoriasis areas with different sizes, arbitrary boundaries, and different severity levels. For example, the psoriasis region in
Figure 7(a4) is larger than that in
Figure 7(a1,a4) and has more blur effect compared with
Figure 7(a1). In addition, the input images also have some background regions with different sizes. For example, the images in
Figure 7(a1,a2) have a bigger background, and those in
Figure 7(a4) only include a small part of background. The color information of some background areas is a little similar than the skin region in
Figure 7(a3), and a little background region (the seal) is similar to the psoriasis area in
Figure 7(a1).
The instance segmentation results of the proposed scheme are shown in the third column of
Figure 7. In
Figure 7c,d, the background remains unchanged, different colors are used to represent each detected individual psoriasis region and the skin area, and each bounding box represents an instance. Compared with
Figure 7b,c, the psoriasis regions and normal skin areas were localized and classified well using the proposed scheme. The results show that the proposed scheme located the psoriasis regions well despite their different sizes, severity levels, and arbitrary boundaries. Furthermore, although the input images contained different background regions, the proposed scheme not only located psoriasis regions but also distinguished the psoriasis regions from the normal skin and the background well. These experimental results show that the instance segmentation scheme can deal with unconstrained psoriasis images well.
To evaluate the robustness of the proposed scheme, two common operators (blurring and noise insertion) were selected as postprocessing for testing.
Figure 8 illustrates the instance segmentation results of the proposed scheme for the blurred and noisy images. The first and second columns of
Figure 8 are the input images and the ground truth, respectively. In the third column of
Figure 8, the first two images are blurred versions, and the others are noisy versions. As we can observe in
Figure 8(c2), a part of the background was similar to the skin and the blur effect was obvious. The last column of
Figure 8 shows the instance segmentation results. As we can see in
Figure 8d, the proposed scheme detected these psoriasis regions and normal skin areas in blurred and noisy images. The results show that the proposed scheme was robust to blurring and noise insertion. Furthermore, according to
Figure 7 and
Figure 8, these experimental results show that the proposed scheme processed unconstrained psoriasis images effectively even when blurring and noise insertion occurred.
4.3.2. Quantitative Evaluation
Cross validation (CV) is a common method to evaluate the performance of machine-learning-models on limited data samples [
38]. Then, the holdout CV is adopted to evaluate the performance of the proposed scheme. For each holdout CV, these 400 psoriasis images were partitioned into training, validation, and testing sets. The ratio among the training, validation, and testing sets was 80:10:10, respectively. Here, we performed the holdout CV three times.
Figure 9 illustrates the image segmentation results of the proposed scheme after CV. As shown in
Figure 9, the average precision, recall, accuracy, and F1-score values of the proposed scheme were at least 94.74%, 93.17%, 96.12%, and 93.77%, respectively. These performance indexes were higher than 93% after CV. Since the F1-score values of the proposed scheme were at least 93.77%, the result shows that most pixels in the unconstrained psoriasis images were correctly classified using the proposed scheme. These experimental results demonstrate that the proposed scheme can not only detect the psoriasis and normal skin regions but also classify these pixels into three categories effectively for unconstrained psoriasis images.
As for object localization, IoU was determined as 0.5 for computing the mAP rate. The mAP rates of the proposed scheme were 92.53%, 96.16%, and 85.9% after CV. The results show that the proposed scheme can locate and recognize the psoriasis regions and the normal skin areas successfully for unconstrained psoriasis skin images.
During CV2, these images in the testing dataset were also used to perform robustness evaluation.
Table 2 shows the experimental results of the proposed scheme for robustness evaluation. As shown in
Table 2, the average precision, recall, accuracy, and F1-score values of the proposed scheme were at least 93.05%, 81.62%, 97.34%, and 84.34%, respectively, for psoriasis regions after noise insertion or blurring. Since these performance indices were at least more than 81.6%, the results show that the proposed scheme still located psoriasis regions effectively even when blurring or noise insertion occurred. For normal skin areas, the average precision, recall, accuracy, and F1-score values of the proposed scheme were at least 96.31%, 97.14%, 96.72%, and 96.57%, respectively, after noise insertion or blurring. These performance indices were at least more than 96.3%, so the results show that the proposed scheme located the normal skin areas correctly for blurred or noisy psoriasis images. For psoriasis regions and normal skin areas, the average precision, recall, accuracy, and F1-score values of the proposed scheme were 95.86%, 95.62%, 97.03%, and 95.73%, respectively. The F1_score reached up to 95.73% for psoriasis regions and normal skin areas even when blurring or noise insertion occurred. These experimental results demonstrate that the proposed scheme can distinguish pixels in psoriasis and normal skin regions from the background pixels regardless of whether blurring or noise insertion exists.
4.4. Comparison with Mask R-CNN-Based Method
As mentioned in
Section 2, Mask R-CNN is one of the state-of-the-art instance segmentation methods [
11,
24]. To make a comparison, we retrained Mask R-CNN for instance psoriasis segmentation. A pr-trained Mask R-CNN model based on the MSCOCO dataset [
30] was retrained for psoriasis image segmentation via transfer learning.
Figure 7d shows the results of Mask R-CNN. As shown in
Figure 7(d2,d3), the boundaries of skin and background may not be located well using Mask R-CNN. Compared with
Figure 7c,d, the normal skin regions can be localized and identified well using the proposed scheme. Furthermore, although the input images contain background regions, the proposed scheme can not only detect psoriasis regions but also distinguish the psoriasis regions and the normal skin areas from the background well.
Figure 10 illustrates the performance indices of the proposed scheme and the Mask R-CNN-based method. As shown in
Figure 10, the average precision, recall, accuracy, and F1-score values of the proposed scheme were 90.88%, 96.14%, 97.30%, and 96.08%, respectively. The results show that the proposed scheme performed instance segmentation well for unconstrained psoriasis images. In addition, the average precision, recall, accuracy, and F1-score values of the Mask R-CNN-based method were 92.83%, 90.88%, 94.08%, and 91.82%, respectively. Compared with the Mask R-CNN-based method, the recall rate was better and the increment of F1_score was 4.2% for psoriasis regions and normal skin areas using the proposed scheme. Therefore, according to the above experimental results, the proposed scheme can provide a better instance segmentation performance compared with the Mask R-CNN-based method for unconstrained psoriasis images.
To evaluate the proposed scheme in psoriasis images with different sizes, the test images with 3000 × 4000 pixels were tested.
Figure 11 illustrates the instance segmentation results of Mask R-CNN and the proposed scheme.
Figure 11a,b are the input images and the ground truth, respectively. As shown in
Figure 11a,b, the test images had psoriasis regions with different severity levels, different sizes, and arbitrary boundaries.
Figure 11c,d are the segmentation results of the proposed scheme and Mask R-CNN, respectively. As we can observe in
Figure 11c, the proposed scheme located psoriasis regions with different severity levels and then provided good object segmentation boundaries. There is a false classified region in the bottom of
Figure 11(c3). As we can see in
Figure 11d, the boundaries between normal skin and background may not be separated well using the Mask-R-CNN-based method. There are false classified regions in the bottom of
Figure 11(d2,d3). Compared with Mask R-CNN, the proposed scheme can find the boundaries between normal skin and background well. The main reason is that DCN was adopted in YOLACT++ to enhance the capability of handling objects with arbitrary boundaries and then reduce the effect of geometric variations. Furthermore,
Figure 12 illustrates the instance segmentation results for local regions. The first and second columns of
Figure 12 are the original regions and the label information, respectively. The third and fourth columns of
Figure 12 are the results of the proposed scheme and Mask RCNN-based method, respectively. As shown in
Figure 12, both methods detected psoriasis regions effectively.
To evaluate the execution speed, 300 psoriasis images were used for testing. The performance index, frame (i.e., image) per second (FPS), is adopted here. The FPSs were 0.44 and 15.1 for the Mask-RCNN-based method and the proposed scheme, respectively. The result shows that the proposed scheme is much faster than the Mask-RCNN-based method. This means that the proposed scheme is suitable for real applications.
According to the abovementioned experimental results, the proposed scheme not only had a higher F1_Score but also a larger FPS compared with the Mask-RCNN-based method for psoriasis image segmentation. Therefore, these results demonstrate that the proposed instance segmentation scheme based on YOLACT++ is superior to the Mask-RCNN-based method in terms of the F1_score and the FPS for dealing with unconstrained psoriasis images.
5. Conclusions
To assess psoriasis severity, the Psoriasis Area Severity Index (PASI), composed of erythema, area, desquamation, and induration, is accepted as a golden standard. Actually, the area factor is more important among the PASI parameters to evaluate the psoriasis severity according to physicians’ evaluations. Furthermore, these clinical skin images captured in outpatient rooms are often unconstrained. To efficiently measure the area factor, an efficient instance segmentation scheme based on deep convolutional neural networks was proposed to deal with unconstrained psoriasis images for computer-aided diagnosis. To achieve instance segmentation, the YOLACT network composed of backbone, feature pyramid network (FPN), Protonet, and prediction head was utilized to deal with psoriasis images. The backbone network was used to extract feature maps from an image, and FPN was designed to generate multiscale feature maps for effectively classifying and localizing objects with multiple sizes. The prediction head was exploited to predict the classification information and bounding box information of objects and mask coefficients. Some prototypes generated by Protonet were combined with mask coefficients to estimate the pixel-level shapes for objects. To achieve instance segmentation for unconstrained psoriasis images, YOLACT++ with a pretrained model was retrained via transfer learning.
To evaluate the performance of the proposed scheme, unconstrained psoriasis images with different severity levels were collected for testing. For objective evaluation, some performance indices, including recall, precision, accuracy, F1-score, and mAP values, are adopted. As for subjective testing, the psoriasis region and normal skin areas could be located and classified well. For testing images, the four performance indices of the proposed scheme were higher than 93% after cross validation. About object localization, the mAP rates were at least 85.9% after cross validation. As for efficiency, the FPS rate of the proposed scheme reached up to 15. In addition, the F1_score and the execution speed of the proposed scheme were higher than those of the Mask R-CNN-based method. These experimental results demonstrate that the proposed scheme based on YOLACT++ and transfer learning can not only locate the psoriasis regions but also distinguish psoriasis pixels from background and normal skin pixels well. Furthermore, the proposed instance segmentation scheme is superior to the Mask R-CNN-based method in terms of the F1_score and the execution speed for dealing with unconstrained psoriasis images.