Network Analysis to Identify the Risk of Epidemic Spreading
Abstract
1. Introduction
2. Materials and Methods
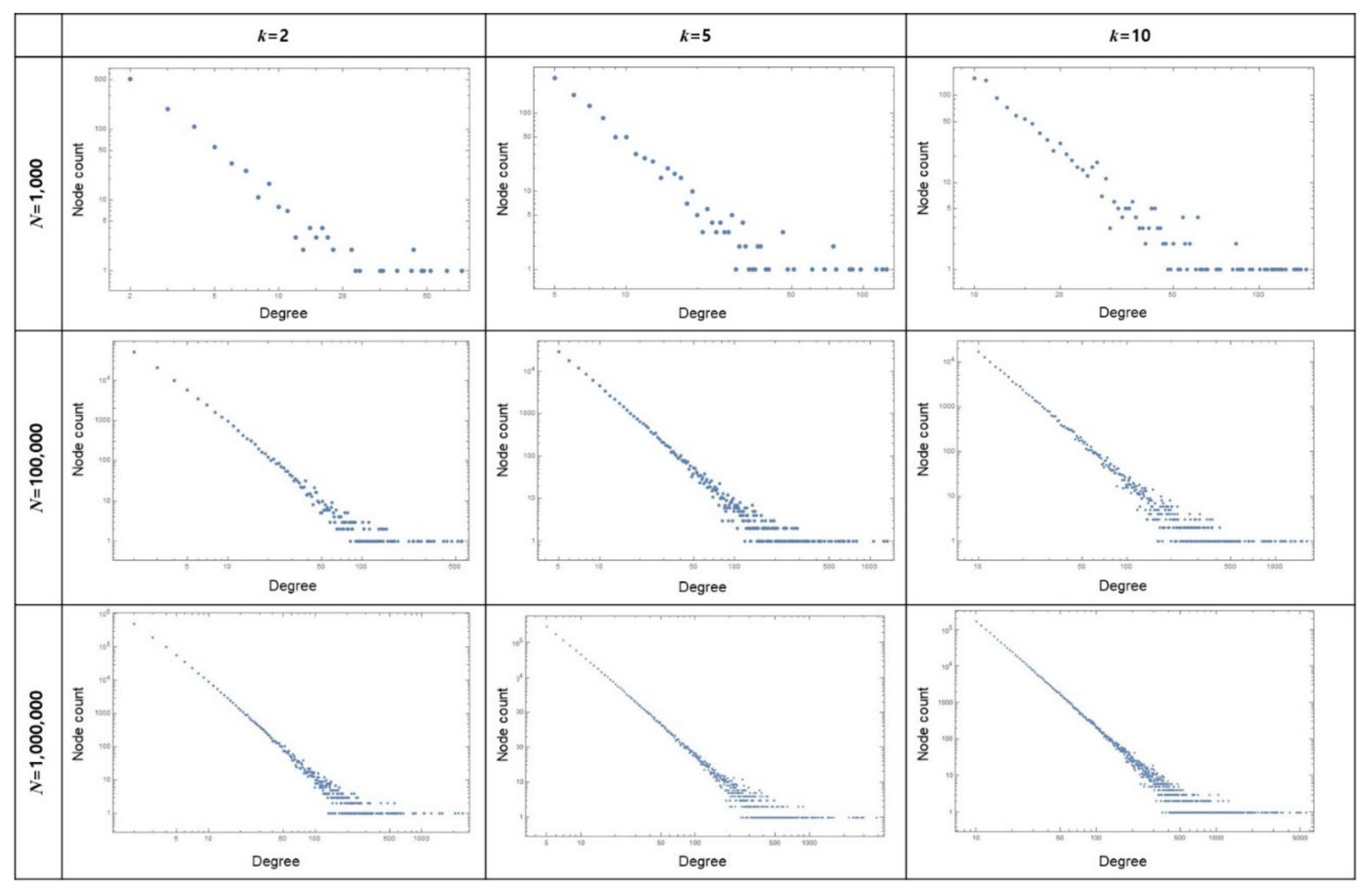
2.1. Network Generation Based on a Scale-Free Model
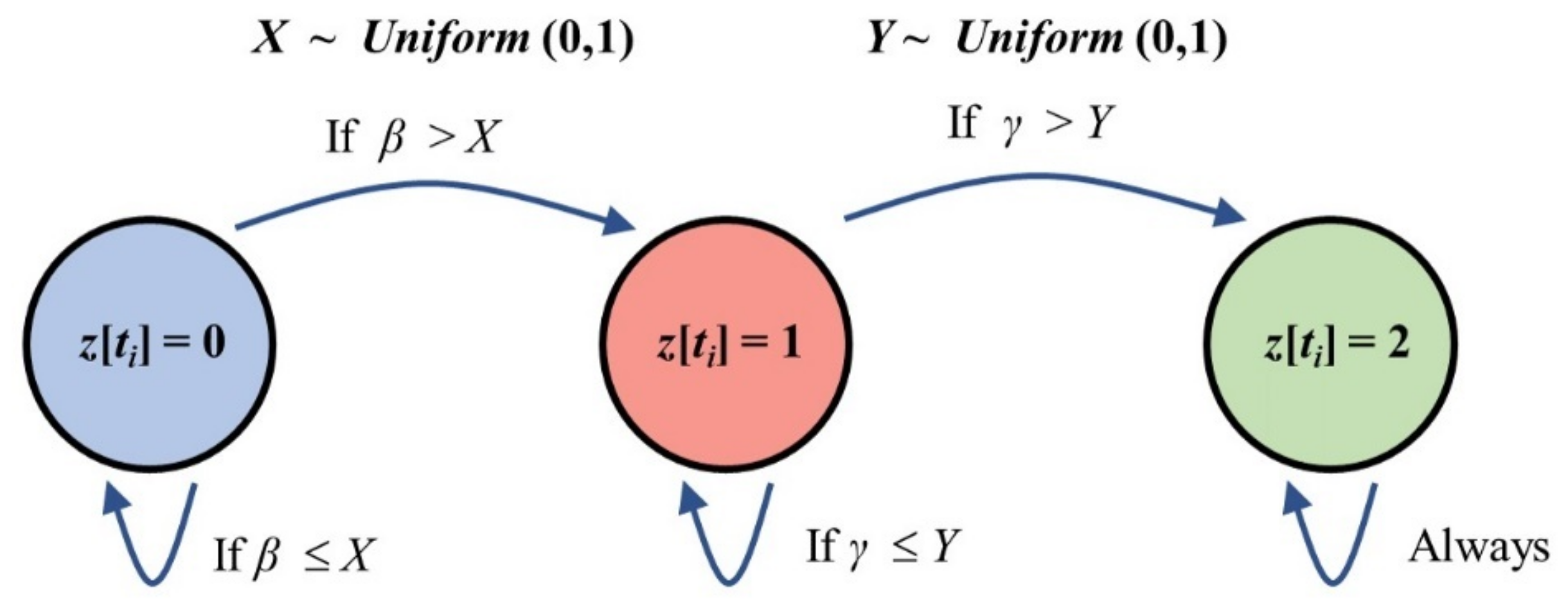
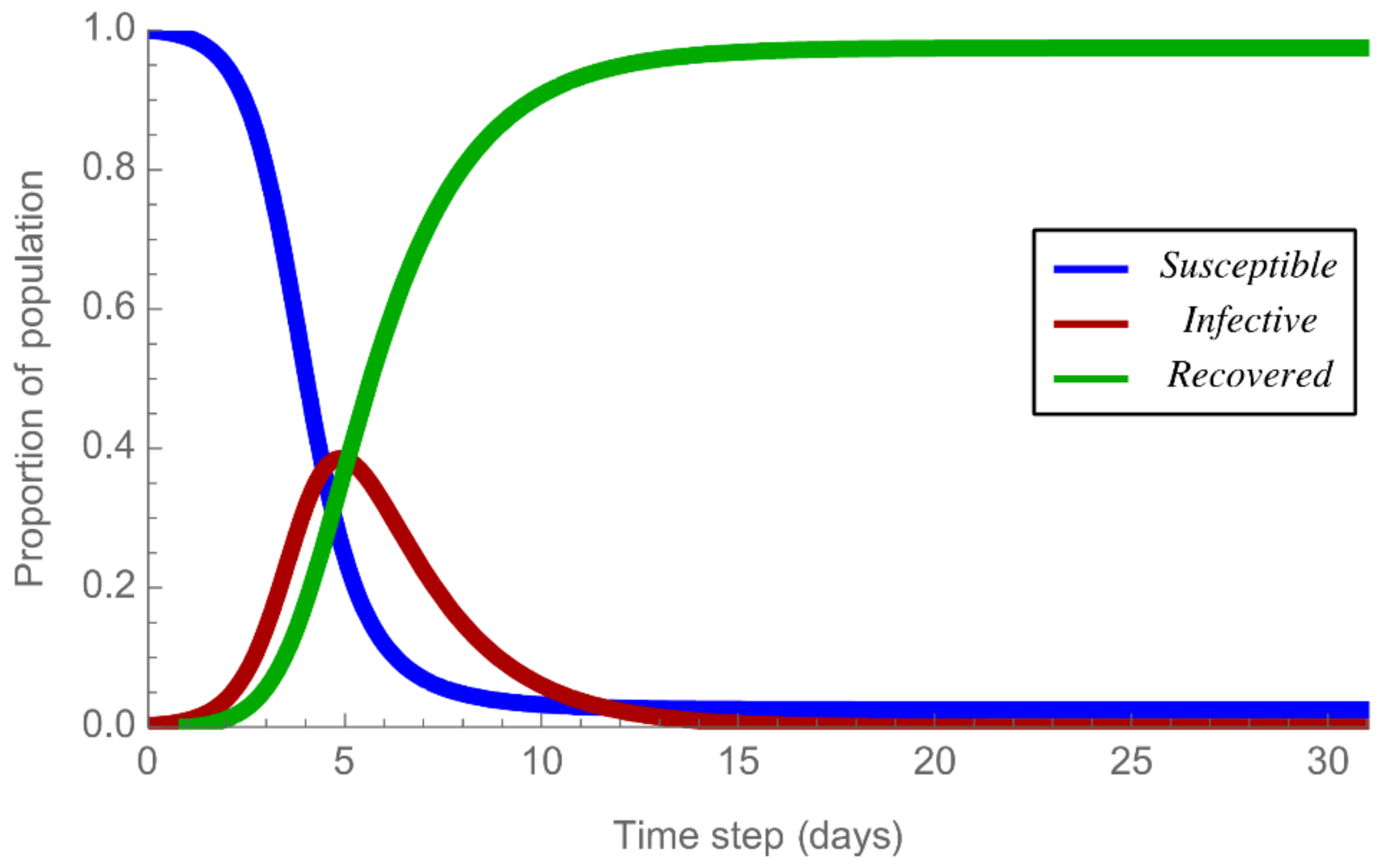
2.2. Epidemic Spreading Based on the SIR Model
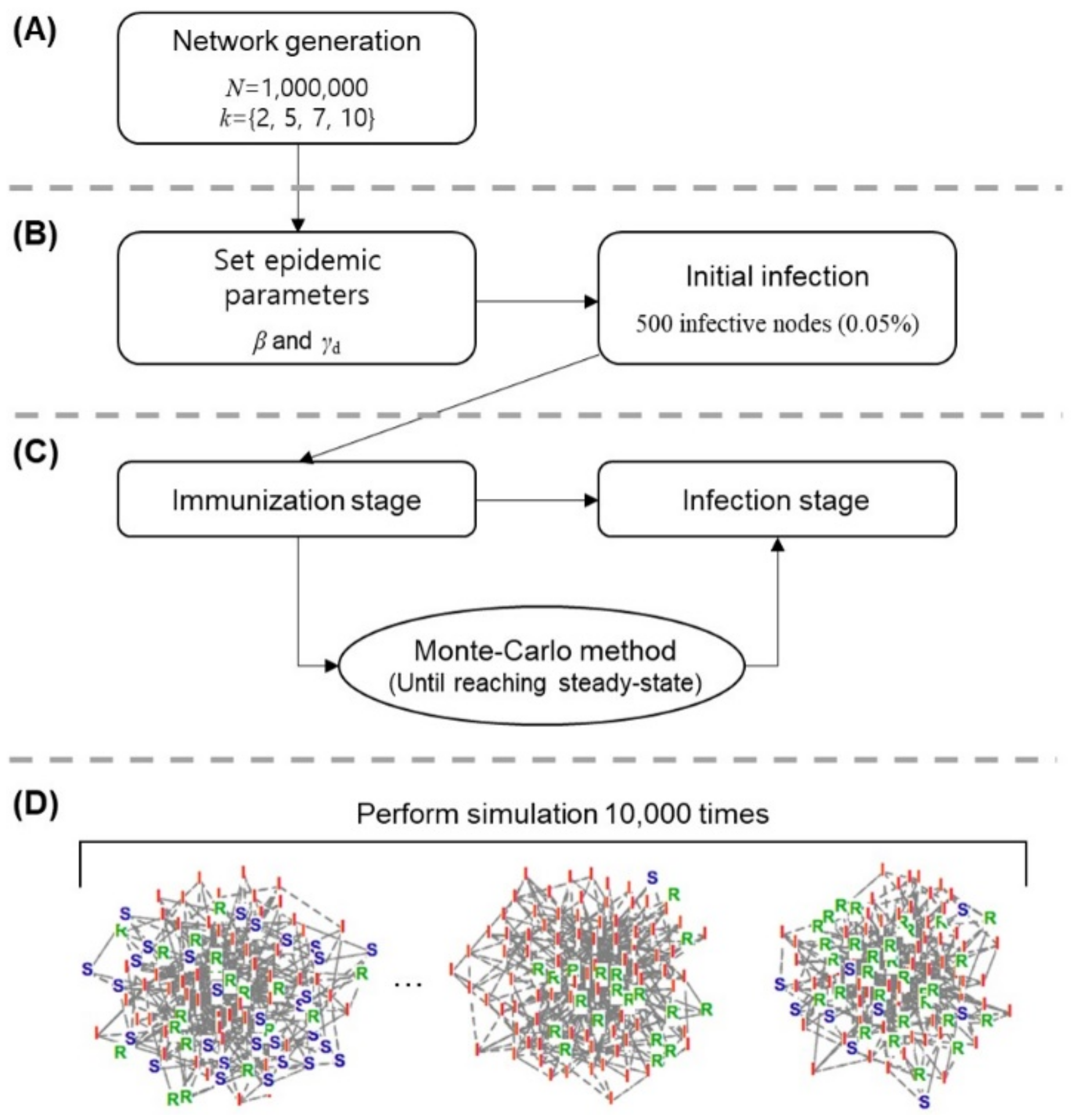
2.3. Investigation of Epidemic Status Based on the Monte Carlo Method
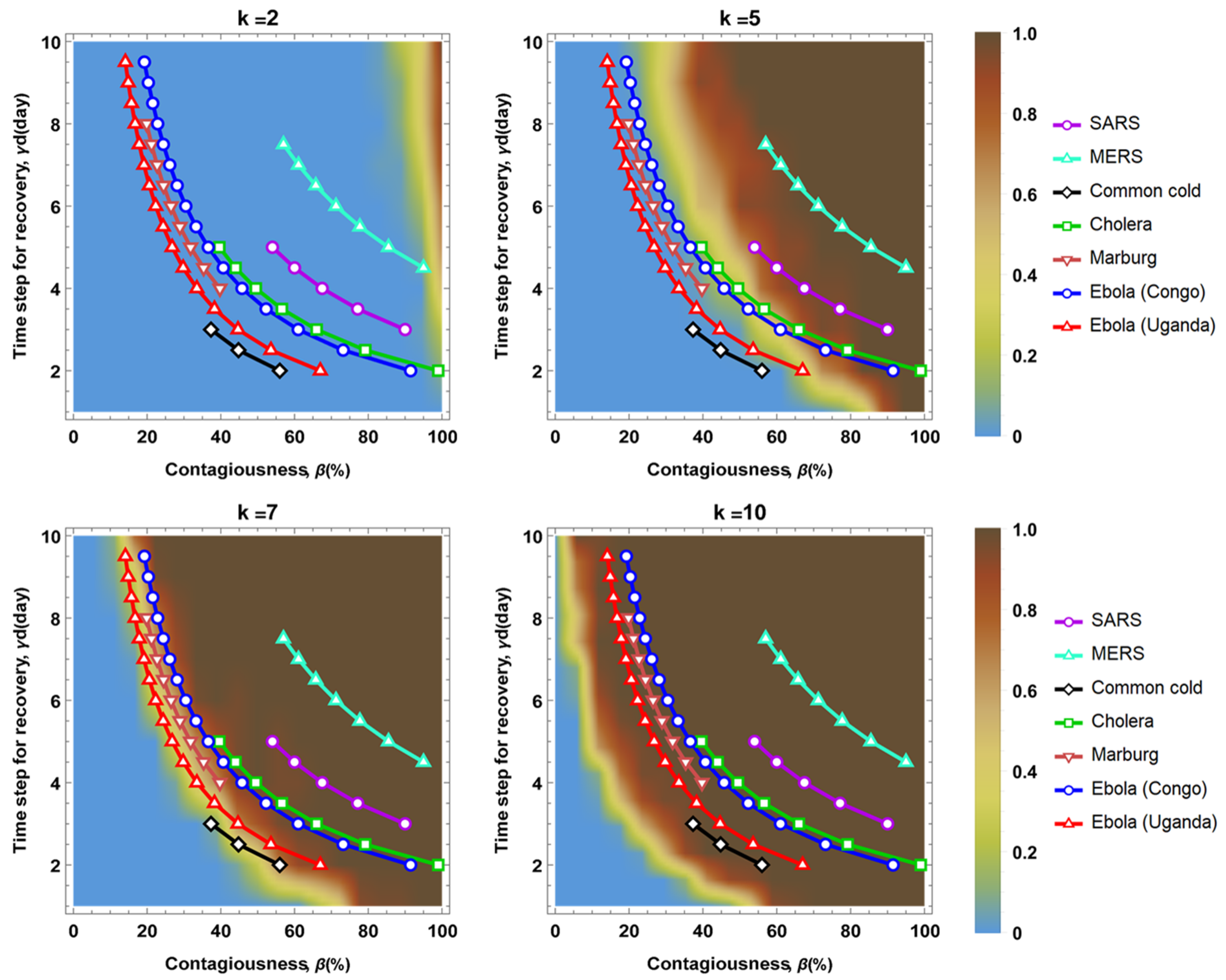
2.4. Simulation Parameters
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baum, S.D. The far future argument for confronting catastrophic threats to humanity: Practical significance and alternatives. Futures 2015, 72, 86–96. [Google Scholar] [CrossRef]
- Flahault, A.; Vergu, E.; Coudeville, L.; Grais, R.F. Strategies for containing a global influenza pandemic. Vaccine 2006, 24, 6751–6755. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.; Litt, J.; Moore, M.; Winzenberg, T.; Shaw, K. General practice: Professional preparation for a pandemic. Med. J. Aust. 2006, 185, S66–S69. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.L. A pest in the land: New World epidemics in a global perspective–Alchon, Suzanne Austin. J. R. Anthropol. Inst. 2006, 12, 219–220. [Google Scholar] [CrossRef]
- Benedictow, O.J. The Black Death, 1346–1353: The Complete History; Boydell & Brewer: Suffolk, UK, 2004. [Google Scholar]
- Johnson, N.P.; Mueller, J. Updating the accounts: Global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Riedel, S. Edward Jenner and the history of smallpox and vaccination. In Baylor University Medical Center Proceedings; Taylor & Francis: Oxfordshire, UK, 2005; pp. 21–25. [Google Scholar]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W. A novel coronavirus associated with severe acute respiratory syndrome. New Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef]
- Feldmann, H.; Geisbert, T.W. Ebola haemorrhagic fever. Lancet 2011, 377, 849–862. [Google Scholar] [CrossRef]
- Tatem, A.J.; Rogers, D.J.; Hay, S. Global transport networks and infectious disease spread. Adv. Parasitol. 2006, 62, 293–343. [Google Scholar]
- Morse, S.S. Factors in the emergence of infectious diseases. In Plagues and Politics; Springer: Berlin/Heidelberg, Germany, 2001; pp. 8–26. [Google Scholar]
- Balcan, D.; Colizza, V.; Gonçalves, B.; Hu, H.; Ramasco, J.J.; Vespignani, A. Multiscale mobility networks and the spatial spreading of infectious diseases. Proc. Natl. Acad. Sci. USA 2009, 106, 21484–21489. [Google Scholar] [CrossRef]
- Eubank, S. Network based models of infectious disease spread. Jpn. J. Infect. Dis. 2005, 58, S9–S13. [Google Scholar]
- Alvarez, F.; Crépey, P.; Barthélemy, M.; Valleron, A.-J. Sispread: A software to simulate infectious diseases spreading on contact networks. Methods Inf. Med. 2007, 46, 19–26. [Google Scholar] [PubMed]
- Kitsak, M.; Gallos, L.K.; Havlin, S.; Liljeros, F.; Muchnik, L.; Stanley, H.E.; Makse, H.A. Identification of influential spreaders in complex networks. Nat. Phys. 2010, 6, 888. [Google Scholar] [CrossRef]
- Hethcote, H.W.; Yorke, J.A. Gonorrhea Transmission Dynamics and Control; Springer: Berlin/Heidelberg, Germany, 1984. [Google Scholar]
- Parshani, R.; Carmi, S.; Havlin, S. Epidemic threshold for the susceptible-infectious-susceptible model on random networks. Phys. Rev. Lett. 2010, 104, 258701. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, Z.; Yang, Z.; Zhang, Z.-K.; Zhou, T.; Sun, G.-Q. Global analysis of an SIS model with an infective vector on complex networks. Nonlinear Anal. Real World Appl. 2012, 13, 543–557. [Google Scholar] [CrossRef]
- Black, A.J.; McKane, A.J.; Nunes, A.; Parisi, A. Stochastic fluctuations in the susceptible-infective-recovered model with distributed infectious periods. Phys. Rev. E 2009, 80, 021922. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Infectious Diseases of Humans: Dynamics and Control; Oxford University Press: Oxford, UK, 1992. [Google Scholar]
- Kermack, W.O.; McKendrick, A.G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. A Contain. Pap. A Math. Phys. Character 1927, 115, 700–721. [Google Scholar]
- Dodds, P.S.; Watts, D.J. Universal behavior in a generalized model of contagion. Phys. Rev. Lett. 2004, 92, 218701. [Google Scholar] [CrossRef] [PubMed]
- Wearing, H.J.; Rohani, P.; Keeling, M.J. Appropriate models for the management of infectious diseases. PLoS Med. 2005, 2, e174. [Google Scholar] [CrossRef]
- Sartwell, P.E. The Distribution of Incubation Periods of Infectious Diseases. Am. J. Hyg. 1950, 51, 310–318. [Google Scholar]
- Simpson, R.H. Infectiousness of communicable diseases in the household (measles, chickenpox, and mumps). Lancet 1952, 549–554. [Google Scholar] [CrossRef]
- Bailey, N.T. The Mathematical Theory of Infectious Diseases and Its Applications; Charles Griffin & Company Ltd: High Wycombe, UK, 1975. [Google Scholar]
- Barabási, A.-L.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509–512. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Albert, R.; Jeong, H. Mean-field theory for scale-free random networks. Phys. A Stat. Mech. Its Appl. 1999, 272, 173–187. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Bonabeau, E. Scale-free networks. Sci. Am. 2003, 288, 60–69. [Google Scholar] [CrossRef]
- Albert, R.; Barabási, A.-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002, 74, 47. [Google Scholar] [CrossRef]
- Diekmann, O.; Heesterbeek, J.A.P. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation; John Wiley & Sons: Hoboken, NJ, USA, 2000; Volume 5. [Google Scholar]
- Keeling, M.J. The effects of local spatial structure on epidemiological invasions. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1999, 266, 859–867. [Google Scholar] [CrossRef]
- Heesterbeek, J.A.P. A brief history of R 0 and a recipe for its calculation. Acta Biotheoretica 2002, 50, 189–204. [Google Scholar] [CrossRef]
- Hethcote, H.W. The mathematics of infectious diseases. Siam Rev. 2000, 42, 599–653. [Google Scholar] [CrossRef]
- McCracken, D.D. The monte carlo method. Sci. Am. 1955, 192, 90–97. [Google Scholar] [CrossRef]
- Rubinstein, R.Y.; Kroese, D.P. Simulation and the Monte Carlo Method; John Wiley & Sons: Hoboken, NJ, USA, 2016; Volume 10. [Google Scholar]
- Porta, M. A Dictionary of Epidemiology; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Li, F.; Choi, B.; Sly, T.; Pak, A. Finding the real case-fatality rate of H5N1 avian influenza. J. Epidemiol. Community Health 2008, 62, 555–559. [Google Scholar] [CrossRef]
- Last, J.M.; Harris, S.S.; Thuriaux, M.C.; Spasoff, R.A. A Dictionary of Epidemiology; International Epidemiological Association, Inc.: Chicago, IL, USA, 2001. [Google Scholar]
- Mikler, A.R.; Venkatachalam, S.; Abbas, K. Modeling infectious diseases using global stochastic cellular automata. J. Biol. Syst. 2005, 13, 421–439. [Google Scholar] [CrossRef]
- Eggo, R.M.; Scott, J.G.; Galvani, A.P.; Meyers, L.A. Respiratory virus transmission dynamics determine timing of asthma exacerbation peaks: Evidence from a population-level model. Proc. Natl. Acad. Sci. USA 2016, 113, 2194–2199. [Google Scholar] [CrossRef]
- Tien, J.H.; Poinar, H.N.; Fisman, D.N.; Earn, D.J. Herald waves of cholera in nineteenth century London. J. R. Soc. Interface 2011, 8, 756–760. [Google Scholar] [CrossRef][Green Version]
- Pourabbas, E.; d’Onofrio, A.; Rafanelli, M. A method to estimate the incidence of communicable diseases under seasonal fluctuations with application to cholera. Appl. Math. Comput. 2001, 118, 161–174. [Google Scholar] [CrossRef]
- Bettencourt, L.M. An ensemble trajectory method for real-time modeling and prediction of unfolding epidemics: Analysis of the 2005 Marburg Fever outbreak in Angola. In Mathematical and Statistical Estimation Approaches in Epidemiology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 143–161. [Google Scholar]
- Ajelli, M.; Merler, S. Transmission potential and design of adequate control measures for Marburg hemorrhagic fever. PLoS ONE 2012, 7, e50948. [Google Scholar] [CrossRef]
- Camacho, A.; Kucharski, A.; Funk, S.; Breman, J.; Piot, P.; Edmunds, W. Potential for large outbreaks of Ebola virus disease. Epidemics 2014, 9, 70–78. [Google Scholar] [CrossRef]
- Chowell, G.; Hengartner, N.W.; Castillo-Chavez, C.; Fenimore, P.W.; Hyman, J.M. The basic reproductive number of Ebola and the effects of public health measures: The cases of Congo and Uganda. J. Theor. Biol. 2004, 229, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Lekone, P.E.; Finkenstädt, B.F. Statistical inference in a stochastic epidemic SEIR model with control intervention: Ebola as a case study. Biometrics 2006, 62, 1170–1177. [Google Scholar] [CrossRef]
- Chowell, G.; Nishiura, H. Transmission dynamics and control of Ebola virus disease (EVD): A review. BMC Med. 2014, 12, 196. [Google Scholar] [CrossRef]
- Chan-Yeung, M.; Xu, R.H. SARS: Epidemiology. Respirology 2003, 8, S9–S14. [Google Scholar] [CrossRef]
- Park, J.-E.; Jung, S.; Kim, A. MERS transmission and risk factors: A systematic review. BMC Public Health 2018, 18, 574. [Google Scholar] [CrossRef]
- Liao, S.; Wang, J. Stability analysis and application of a mathematical cholera model. Math. Biosci. Eng. 2011, 8. [Google Scholar] [CrossRef]
- Chen, Q. Application of SIR model in forecasting and analyzing for SARS. Beijing Da Xue Xue Bao Yi Xue Ban J. Peking Univ. Health Sci. 2003, 35, 75–80. [Google Scholar]
- Berge, T.; Lubuma, J.-S.; Moremedi, G.; Morris, N.; Kondera-Shava, R. A simple mathematical model for Ebola in Africa. J. Biol. Dyn. 2017, 11, 42–74. [Google Scholar] [CrossRef] [PubMed]
- Alshakhoury, N.S. Mathematical Modeling and Control of MERS-COV Epidemics. Ph.D. Thesis, College of Arts and Sciences, Denton, TX, USA, December 2017. [Google Scholar]

| Epidemic Disease | Infectious Period (γd) | Basic Reproduction Number (R0) | Transmission Rate (β) |
|---|---|---|---|
| Common cold | 3~7 | 1.12 | 0.16~0.37 |
| Cholera | 1~5 | 1.98 | 0.39~1.98 |
| Marburg | 3 | 1.59 | 0.53 |
| Ebola (Congo) | 6.5 | 1.83 | 0.28 |
| Ebola (Uganda) | 6.5 | 1.34 | 0.20 |
| SARS | 3~5 | 2.70 | 0.54~0.90 |
| MERS | 4.5~7.8 | 4.275 | 0.548~0.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Yoo, S.; Lee, S.; Lee, D.; Lee, K.-H. Network Analysis to Identify the Risk of Epidemic Spreading. Appl. Sci. 2021, 11, 2997. https://doi.org/10.3390/app11072997
Kim K, Yoo S, Lee S, Lee D, Lee K-H. Network Analysis to Identify the Risk of Epidemic Spreading. Applied Sciences. 2021; 11(7):2997. https://doi.org/10.3390/app11072997
Chicago/Turabian StyleKim, Kiseong, Sunyong Yoo, Sangyeon Lee, Doheon Lee, and Kwang-Hyung Lee. 2021. "Network Analysis to Identify the Risk of Epidemic Spreading" Applied Sciences 11, no. 7: 2997. https://doi.org/10.3390/app11072997
APA StyleKim, K., Yoo, S., Lee, S., Lee, D., & Lee, K.-H. (2021). Network Analysis to Identify the Risk of Epidemic Spreading. Applied Sciences, 11(7), 2997. https://doi.org/10.3390/app11072997







