Nodule Detection with Convolutional Neural Network Using Apache Spark and GPU Frameworks
Abstract
1. Introduction
- The proposed NG-CNN model shows a great accuracy percentage in detecting the lung nodule even when the nodule’s size is less than 1.5 mm as we applied a noise filter as an extensive pre-processing technique. Measurement of correntropy is integrated with the autoencoder based deep neural network.
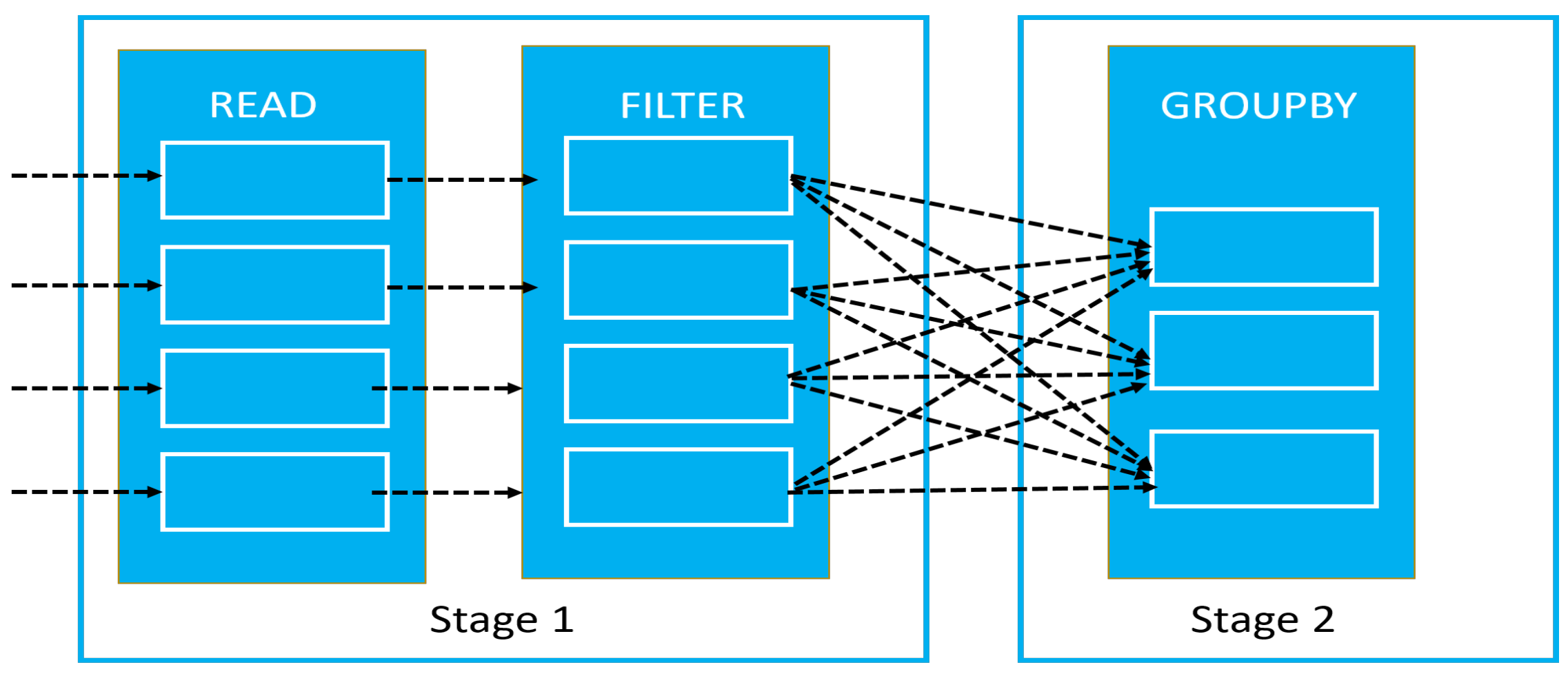
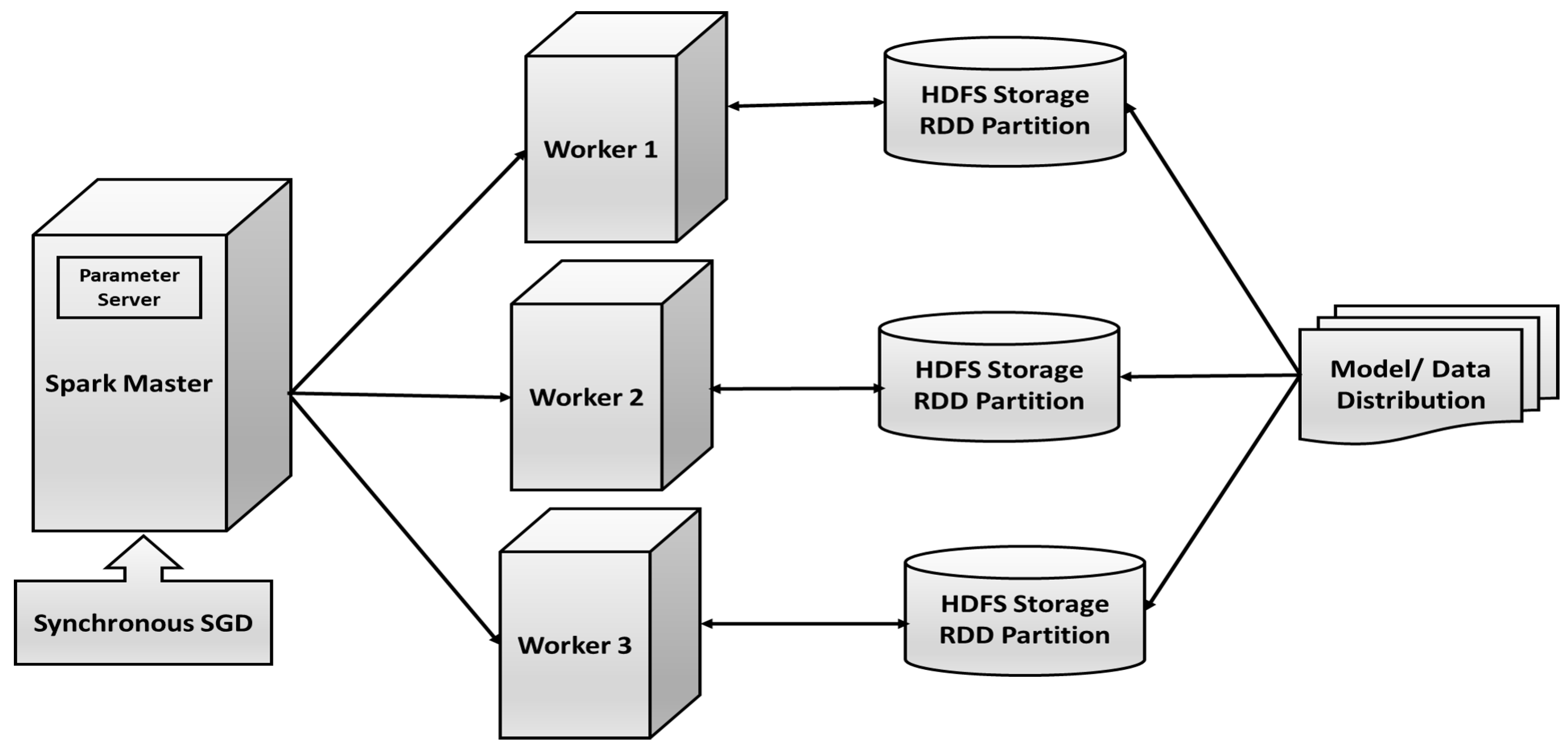
- We designed Apache Spark deep learning framework for our proposed NG-CNN model. Training time and performance are analyzed using detailed experimentation on various platforms, such as Apache Spark, GPU, and CPU.
- Large labeled lung CT scan dataset of size 150 GB has been used for evaluation, making it perfect for CNN training. The cumulative number of cases is more than 1600, with multiple slices per patient.
- Classification accuracy, sensitivity, specificity, and area under the ROC curve (AUC) of the model performance are compared with various combinations of CNN parameters and other deep neural networks.
2. Related Works
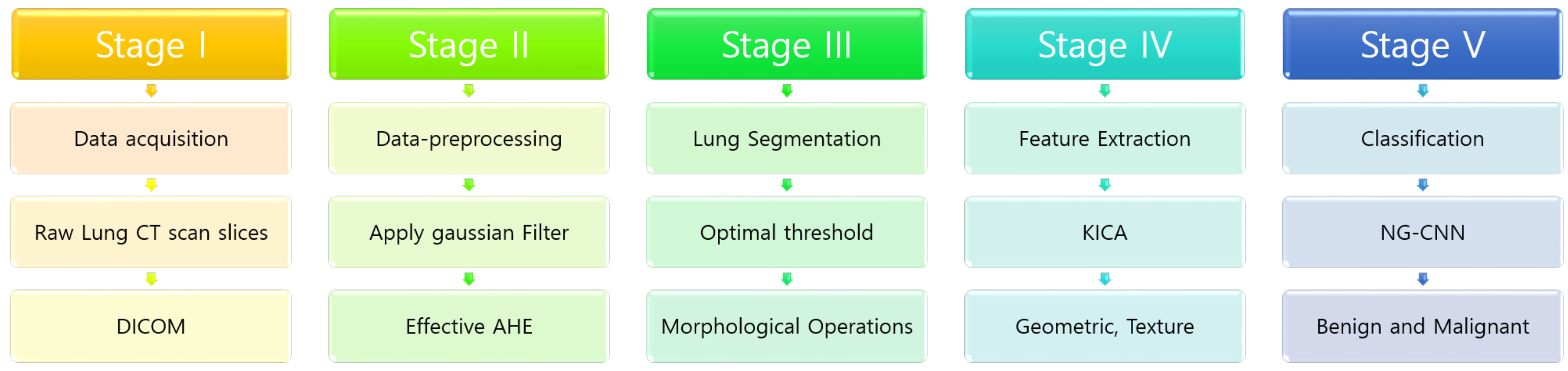
3. Our Proposed Methodology
3.1. Pre-Processing
| Algorithm 1 Pseudocode of pre-processing step in NG-CNN algorithm. |
INPUT: Set of lung CT images with ground truth labels OUTPUT: Binary classes 0 or 1 for cancerous and non-cancerous nodules.
|
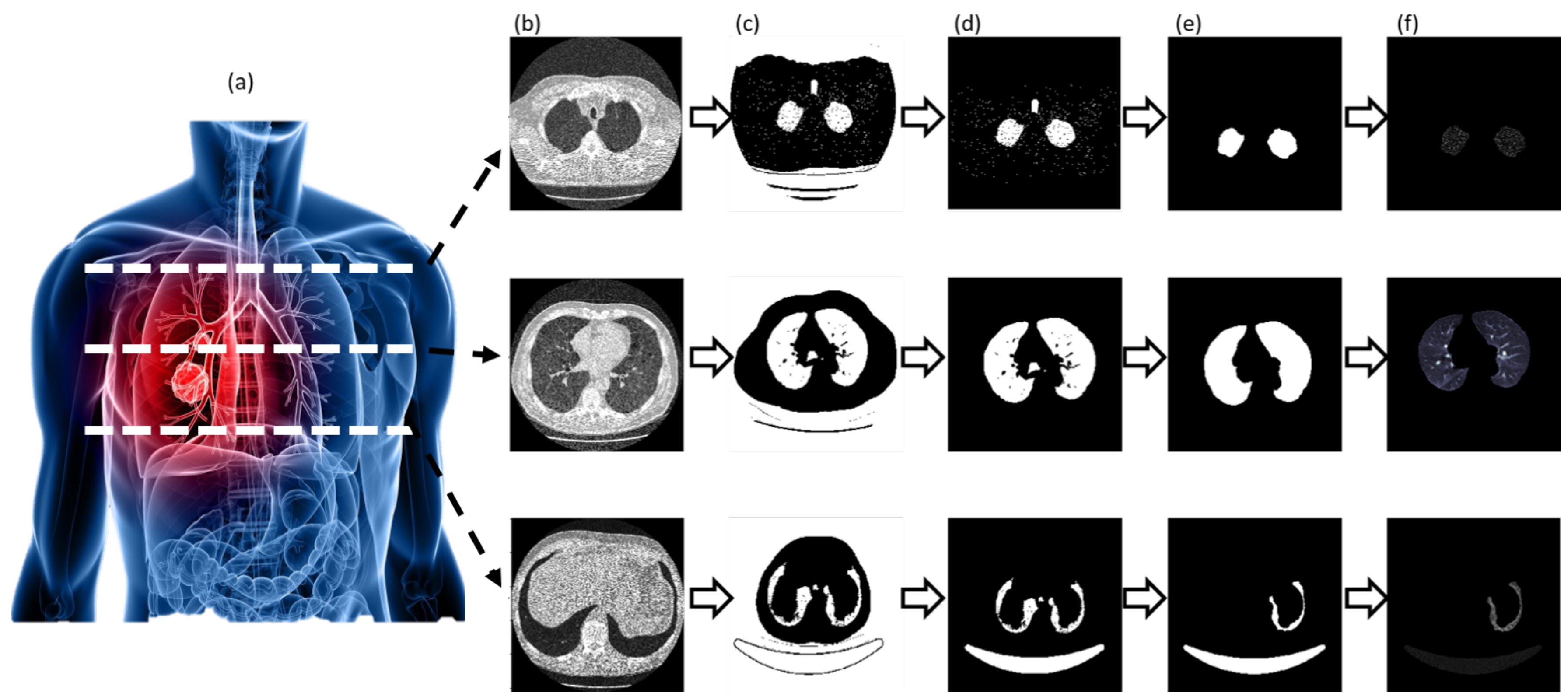
3.2. Segmentation and Candidate Nodules
3.3. Feature Extraction
3.4. Training the Neural Net
4. Results & Discussion
4.1. Dataset
4.2. Experimentation
4.3. Performance Analysis
- Parameters, such as weight, and biases are randomly initialized based on NG-CNN architecture.
- First copy of the parameters is distributed to all the Spark worker nodes.
- The lung image dataset is divided upon the worker nodes, and they train their subset of data.
- Update the calculated parameters after a certain number of iterations from the server node.
- Repeat from step 2 till the training converges.
5. Discussions & Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Paul. Key Statistics for Lung Cancer. Version 1.6.0. Available online: https://www.cancer.org/cancer/non-small-cell-lung-cancer/about/key-statistics.html (accessed on 15 May 2019).
- Zhou, Z.H.; Jiang, Y.; Yang, Y.B.; Chen, S.F. Lung cancer cell identification based on artificial neural network ensembles. Artif. Intell. Med. 2002, 24, 25–36. [Google Scholar] [CrossRef]
- Boroczky, L.; Zhao, L.; Lee, K.P. Feature subset selection for improving the performance of false positive reduction in lung nodule CAD. IEEE Trans. Inf. Technol. Biomed. 2006, 10, 504–511. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Suzuki, K. Comparing two classes of end-to-end machine-learning models in lung nodule detection and classification: MTANNs vs. CNNs. Pattern Recognit. 2017, 63, 476–486. [Google Scholar] [CrossRef]
- Sivakumar, S.; Chandrasekar, C. Lung nodule detection using fuzzy clustering and support vector machines. Int. J. Eng. Technol. 2013, 5, 179–185. [Google Scholar]
- Russell, S.J.; Norvig, P. Artificial Intelligence: A Modern Approach; Pearson Education Limited: Malaysia, 2016. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef]
- Vedaldi, A.; Lenc, K. Matconvnet: Convolutional neural networks for matlab. In Proceedings of the 23rd ACM international conference on Multimedia, Brisbane, Australia, 12 October 2015; ACM: New York, NY, USA, 2015; pp. 689–692. [Google Scholar]
- Polacin, A.; Kalender, W.A.; Marchal, G. Evaluation of section sensitivity profiles and image noise in spiral CT. Radiology 1992, 185, 29–35. [Google Scholar] [CrossRef]
- Huang, B.; Law, M.W.M.; Khong, P.L. Whole-body PET/CT scanning: Estimation of radiation dose and cancer risk. Radiology 2009, 251, 166–174. [Google Scholar] [CrossRef]
- Pearce, M.S.; Salotti, J.A.; Little, M.P.; McHugh, K.; Lee, C.; Kim, K.P.; Howe, N.L.; Ronckers, C.M.; Rajaraman, P.; Craft, A.W.; et al. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: A retrospective cohort study. Lancet 2012, 380, 499–505. [Google Scholar] [CrossRef]
- Ilango, G.; Gowri, B.S. Noise from CT–Images. International Journal of Applied Information Systems (IJAIS) ISSN: 2249-0868. Available online: https://www.techrepublic.com/resource-library/company/international-journal-of-applied-information-systems-ijais/ (accessed on 31 January 2021).
- Kijewski, M.F.; Judy, P.F. The noise power spectrum of CT images. Phys. Med. Biol. 1987, 32, 565. [Google Scholar] [CrossRef]
- Lehtinen, J.; Munkberg, J.; Hasselgren, J.; Laine, S.; Karras, T.; Aittala, M.; Aila, T. Noise2Noise: Learning Image Restoration without Clean Data. arXiv 2018, arXiv:1803.04189. [Google Scholar]
- Eigen, D.; Rolfe, J.; Fergus, R.; LeCun, Y. Understanding deep architectures using a recursive convolutional network. arXiv 2013, arXiv:1312.1847. [Google Scholar]
- Johnsirani Venkatesan, N.; Nam, C.; Ryeol Shin, D. Lung Nodule Classification on CT Images Using Deep Convolutional Neural Network Based on Geometric Feature Extraction. J. Med. Imaging Health Inform. 2020, 10, 2042–2052. [Google Scholar] [CrossRef]
- Yang, Y.; Xiang, P.; Kong, J.; Zhou, H. A GPGPU compiler for memory optimization and parallelism management. ACM Sigplan Not. 2010, 45, 86–97. [Google Scholar] [CrossRef]
- Fung, J.; Mann, S. OpenVIDIA: Parallel GPU computer vision. In Proceedings of the 13th Annual ACM International Conference on Multimedia, Singapore, 6–11 November 2005; ACM: New York, NY, USA, 2005; pp. 849–852. [Google Scholar]
- Zaharia, M.; Xin, R.S.; Wendell, P.; Das, T.; Armbrust, M.; Dave, A.; Meng, X.; Rosen, J.; Venkataraman, S.; Franklin, M.J.; et al. Apache spark: A unified engine for big data processing. Commun. ACM 2016, 59, 56–65. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Kalinovsky, A.; Kovalev, V. Lung Image Segmentation Using Deep Learning Methods and Convolutional Neural Networks; Center of Ball State University: Muncie, Indiana, 2016. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Handa, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for robust semantic pixel-wise labelling. arXiv 2015, arXiv:1505.07293. [Google Scholar]
- Yang, H.; Yu, H.; Wang, G. Deep learning for the classification of lung nodules. arXiv 2016, arXiv:1611.06651. [Google Scholar]
- Romero, A.; Gatta, C.; Camps-Valls, G. Unsupervised deep feature extraction for remote sensing image classification. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1349–1362. [Google Scholar] [CrossRef]
- Gruetzemacher, R.; Gupta, A. Using Deep Learning for Pulmonary Nodule Detection & Diagnosis. 2016. Available online: https://aisel.aisnet.org/amcis2016/Intel/Presentations/3/ (accessed on 31 January 2021).
- Setio, A.A.A.; Traverso, A.; De Bel, T.; Berens, M.S.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
- Song, Q.; Zhao, L.; Luo, X.; Dou, X. Using Deep Learning for Classification of Lung Nodules on Computed Tomography Images. J. Healthc. Eng. 2017, 2017, 8314740. [Google Scholar] [CrossRef]
- Ciompi, F.; Chung, K.; Van Riel, S.J.; Setio, A.A.A.; Gerke, P.K.; Jacobs, C.; Scholten, E.T.; Schaefer-Prokop, C.; Wille, M.M.; Marchianò, A.; et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci. Rep. 2017, 7, 46479. [Google Scholar] [CrossRef]
- Lo, S.C.; Lou, S.L.; Lin, J.S.; Freedman, M.T.; Chien, M.V.; Mun, S.K. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans. Med Imaging 1995, 14, 711–718. [Google Scholar] [CrossRef]
- Anirudh, R.; Thiagarajan, J.J.; Bremer, T.; Kim, H. Lung nodule detection using 3D convolutional neural networks trained on weakly labeled data. In Medical Imaging 2016: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2016; Volume 9785, p. 978532. Available online: https://accucoms.com/publishers/international-society-for-optics-and-photonics/ (accessed on 31 January 2021).
- Gupta, A.; Thakur, H.K.; Shrivastava, R.; Kumar, P.; Nag, S. A Big Data Analysis Framework Using Apache Spark and Deep Learning. In Proceedings of the 2017 IEEE International Conference on Data Mining Workshops (ICDMW), New Orleans, LA, USA, 18–21 November 2017; pp. 9–16. [Google Scholar]
- Team, D. Deeplearning4j: Open-source distributed deep learning for the jvm. Apache Softw. Found. Licens. 2016, 2. Available online: https://mgubaidullin.github.io/deeplearning4j-docs/ (accessed on 31 January 2021).
- Li, P.; Luo, Y.; Zhang, N.; Cao, Y. HeteroSpark: A heterogeneous CPU/GPU Spark platform for machine learning algorithms. In Proceedings of the 2015 IEEE International Conference on Networking, Architecture and Storage (NAS), Boston, MA, USA, 6–7 August 2015; pp. 347–348. [Google Scholar]
- Moritz, P.; Nishihara, R.; Stoica, I.; Jordan, M.I. SparkNet: Training deep networks in Spark. arXiv 2015, arXiv:1511.06051. [Google Scholar]
- Zhong, G.; Wang, L.N.; Ling, X.; Dong, J. An overview on data representation learning: From traditional feature learning to recent deep learning. J. Financ. Data Sci. 2016, 2, 265–278. [Google Scholar] [CrossRef]
- Messay, T.; Hardie, R.C.; Rogers, S.K. A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med. Image Anal. 2010, 14, 390–406. [Google Scholar] [CrossRef]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep sparse rectifier neural networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; pp. 315–323. [Google Scholar]
- Davis, J.; Goadrich, M. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on MACHINE Learning, Pittsburgh, PA, USA, 1 January 2016; ACM: New York, NY, USA, 2006; pp. 233–240. [Google Scholar]
- Riccardi, A.; Petkov, T.S.; Ferri, G.; Masotti, M.; Campanini, R. Computer-aided detection of lung nodules via 3D fast radial transform, scale space representation, and Zernike MIP classification. Med. Phys. 2011, 38, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Li, Q. High performance lung nodule detection schemes in CT using local and global information. Med. Phys. 2012, 39, 5157–5168. [Google Scholar] [CrossRef]
- Cascio, D.; Magro, R.; Fauci, F.; Iacomi, M.; Raso, G. Automatic detection of lung nodules in CT datasets based on stable 3D mass–spring models. Comput. Biol. Med. 2012, 42, 1098–1109. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Setio, A.A.; Jacobs, C.; Ciompi, F. Off-the-shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 286–289. [Google Scholar]





| ep | NG-CNN | CAE-DLF | DBN | CNN |
|---|---|---|---|---|
| 50 | 0.5879 | 0.5124 | 0.5469 | 0.6789 |
| 100 | 0.78632 | 0.6645 | 0.6214 | 0.7356 |
| 150 | 0.8021 | 0.6987 | 0.6987 | 0.78541 |
| 200 | 0.8432 | 0.7421 | 0.7548 | 0.8632 |
| 250 | 0.9584 | 0.8952 | 0.79215 | 0.9452 |
| ep | NG-CNN | CAE-DLF | DBN | CNN |
|---|---|---|---|---|
| 50 | 93.3150 | 90.3701 | 88.0467 | 95.7059 |
| 100 | 92.1970 | 89.1087 | 96.1765 | 96.5224 |
| 150 | 95.7167 | 95.9751 | 87.0868 | 99.7587 |
| 200 | 95.0442 | 91.4021 | 87.1534 | 98.6222 |
| 250 | 97.4947 | 89.9001 | 93.8550 | 97.5653 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkatesan, N.J.; Shin, D.R.; Nam, C.S. Nodule Detection with Convolutional Neural Network Using Apache Spark and GPU Frameworks. Appl. Sci. 2021, 11, 2838. https://doi.org/10.3390/app11062838
Venkatesan NJ, Shin DR, Nam CS. Nodule Detection with Convolutional Neural Network Using Apache Spark and GPU Frameworks. Applied Sciences. 2021; 11(6):2838. https://doi.org/10.3390/app11062838
Chicago/Turabian StyleVenkatesan, Nikitha Johnsirani, Dong Ryeol Shin, and Choon Sung Nam. 2021. "Nodule Detection with Convolutional Neural Network Using Apache Spark and GPU Frameworks" Applied Sciences 11, no. 6: 2838. https://doi.org/10.3390/app11062838
APA StyleVenkatesan, N. J., Shin, D. R., & Nam, C. S. (2021). Nodule Detection with Convolutional Neural Network Using Apache Spark and GPU Frameworks. Applied Sciences, 11(6), 2838. https://doi.org/10.3390/app11062838





