Abstract
Prions are misfolded, self-replicating, and transmissible proteins capable of causing different conditions that affect the brain and nervous system in humans and animals. Yeasts are the perfect model to study prion formation, dissemination, and the structure of protein aggregates. Yeast prions are related to stress resistance, cell fitness, and viability. Applying a pulsed electric field (PEF) as a factor capable of disintegrating the amyloid aggregates arises from the fact that the amyloid aggregates form via noncovalent bonds and stabilize via electrostatic interactions. In this research, we applied 2–26 kV/cm PEF delivered in sequences of 5 pulses of 1 ms duration to the Saccharomyces cerevisiae cell without prions and containing strong and weak variants of the [PSI+] prion (prion form of Sup35 translation termination factor). We determined that prions significantly increase cell survivability and resistance to PEF treatment. The application of PEF to the purified Sup35NM fibrils showed that the electric field causes significant reductions in the length of fibrils and the full disintegration of fibrils to Sup35 oligomers can be achieved in higher fields.
1. Introduction
Prions are misfolded, self-replicating, and transmissible proteins that are related to the loss of function of the native protein. Originally, the concept of the prion was used to describe transmissible spongiform encephalopathies: human neurodegenerative diseases Creutzfeldt-Jakob, scrapie in sheep, the chronic wasting disease in deer, and bovine spongiform encephalopathy in cattle (known as “mad cow disease”) [1]. Structurally, prions are amyloid-like aggregated state. Yeast prions have become important research objects in elucidating the underlying mechanisms of human amyloid disease [2]. Saccharomyces cerevisiae is common to model systems for prion research. Almost all prions in yeast are found in infectious amyloid conformation [3] and prions in yeast can appear spontaneously with the different variants [4]. Amyloid yeast prions are rare in wild-type strains. A single prion protein sequence may have several different prion variants, have a parallel β-fold amyloid structure, are harmful to their hosts by causing them stress, and are the subject of prion disease research [4,5]. Variants of [PSI+] yeast prion (an amyloid form of Sup35 translation termination factor) capable to manifest themselves as strong and weak forms that can be discriminated by their phenotypic intensity and stability during cell division [6]. It is shown that a full-length protein is not necessary for the formation of aggregate inside the cells. In SUP35 protein, the Sup35NM domain, located at 1–253 residues of the amino acid sequence, is capable of inducing and propagating the prion state of the [PSI+] prion [5,7]. Mutations in ADE1 and ADE2 mutant stains allow for the discrimination of different prion variants by color. In 2014, it was confirmed that the weak and strong variants of prions differ in the M middle domain, which is 40% more dynamic in weak prions than in strong. Those conformational changes have an impact on the interaction of the amyloid fibrils, chaperons, and fibril functions [8,9,10]. The functions and biological meaning of the prion proteins are still under discussion. How can the elements cause cell death and resist its evolutional elimination? Prions are related to various cell functions: stress resistance, copper homeostasis, cellular differentiation, proliferation, adhesion, control of cell morphology, and cell fitness, thus providing adaptation benefits in the changing environment [11,12,13,14,15]. In this research, we subjected yeast cells containing no prions and strong or weak prion variants to the pulsed electric field (PEF) of varied strength. PEF is proven to be an effective technique in drug delivery during electrochemotherapy [16,17,18], various microorganism transformation [19,20,21], electrofusion [22,23,24], as well as biocontrol of microorganisms in food protection and biotechnology [25,26,27]. It can provide a rapid and nonselective method for the elimination of various types of yeast. The PEF conditions can be easily optimized for each cell type for the most efficient effect.
Proteins in the amyloid conformation are highly resistant to various inactivation mechanisms and substances such as heat shock or proteases [14]. Electrostatic interactions, such as salt bridges (a combination of two non-covalent interactions: hydrogen and ionic bonding between the same amino acid residues) and self-energy, are key factors in stabilizing the secondary and tertiary structure-forming elements in prion proteins [28]. Therefore, in this research, it was investigated how the electric field can affect the polymeric form of the [PSI+] prion fibril. Ideally, the fibril could be cleaved to Sup35NM oligomers, which would enable an alternative method of prion treatment or a new tool for the amyloid disintegration in vitro. It is also important to note that the prion fibril does not contain any covalent bonds that would not be theoretically capable of breaking down by an electric field.
2. Materials and Methods
2.1. Yeast Strains. Saccharomyces Cerevisiae 74-D694
The genotype of this strain (MATa, ade1-14 (UGA), trp1-289, his3-Δ200, ura3-52, leu2-3,112) (Chernoff et al., 1995). Thus, this strain cannot produce adenine, tryptophan, histidine, uracil, and leucine de novo—these amino acids must be present in the culture medium. ADE1 and ADE2 mutant phenotypes allow the discrimination of the cells without prions or containing strong or weak prions by the color of the colonies, while grown under adenine limitation conditions (Figure 1). The following were used in this research: S. cerevisiae 74-D694 [psi−][PIN+] strain with [PIN+] prion but without Sup35 prion forms; S. cerevisiae 74-D694 [PSI+][PIN+]—strain with [PIN+] prion and strong Sup35 prion variant; S. cerevisiae 74-D694 [PSI+][PIN+]—strain with [PIN+] prion and weak Sup35 prion variant.
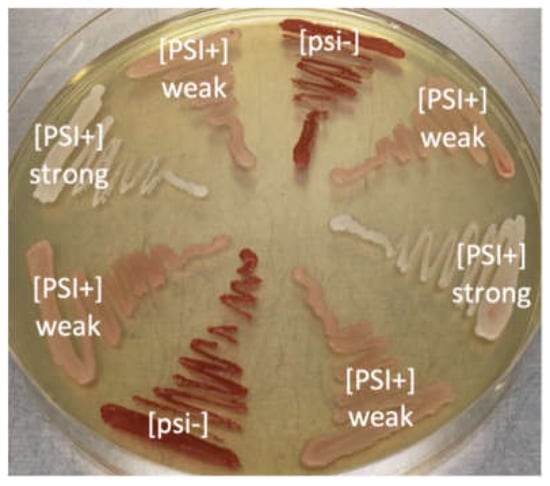
Figure 1.
Phenotype of ADE1 mutants in the absence or presence of different variants of [PSI] prions in an synthetic complete (SC) medium. White—cells containing strong [PSI+] prion, pink—cells containing weak [PSI+] prion, and red—cells without prion ([psi−]).
The literature states that [PIN+] facilitates the bypass of amyloid regulatory mechanisms by other proteins in vivo and thus promotes the formation of other amyloids [6,29,30,31]. The functions of the native protein are unknown, but it is not vital for cell survival. Cells without the [PIN+] prion were not used in the study, so the name of the strains were abbreviated according to the presence of [PSI+] prion: S. cerevisiae [PSI+] or S. cerevisiae [psi−], respectively.
The strain and its different variants were obtained from Dr. Young-Jun Choe at the Max–Planck Institute of Biochemistry, Germany.
2.2. Cell Growth Dynamics
S. cerevisiae cells with different prion variants of [PSI+] are plated on agarized YPD (rich medium) medium and grown for 48 h at 30 °C. One colony of the respective culture is then transferred to 5 mL of fresh YPD medium and grown overnight (~12 h) at 30 °C with aeration, at 130 rpm. The overnight culture is transferred to 50 mL of fresh synthetic complete (SC, medium, containing 2% glucose, 2% yeast nitrogen base without amino acids, and supplemented with the appropriate amino acids according to the genotype of the strains) or the YPD medium to the final optic density (OD) of 0.04 to 0.06 AU at a 600 nm wavelength. Culture samples are taken every 3 h and cell growth was measured with a biophotometer (Eppendorf, Germany). Growth dynamics were observed until culture reached the stationary growth phase. All the experiments were repeated independently 3 times.
2.3. PEF Permeabilization Assay
A unique square pulse wave PEF generator was developed at the Institute of High Magnetic Fields (Vilnius Gediminas Technical University Vilnius, Lithuania), generating 100 ns–1 ms pulses. Up to 30 kV was used in this research [32]. PEF experiments were performed in 1 M sorbitol. The impedance of the cuvette was in the range of 9.1 ± 0.1 kΩ, the highest expected current was in the range 0.02 to 0.3 A, corresponding to 2 and 26 kV/cm treatments. Temperature rise was measured using a Pt1000 sensor (InnovativeSensor Technology, Wattwil, Switzerland).
The summary of PEF treatments, including the energy of each protocol, is presented in Table 1.

Table 1.
The summary of pulsed electric field (PEF) treatments.
Different prion variants of S. cerevisiae PSI+ and S. cerevisiae psi− prion-free strains were plated on agarized YPD mediums and grown for 48 h at 30 °C. One sterile colony was transferred to a tube with 5 mL of a YPD medium and grown in a shaker with aeration for 12 h at 30 °C, 130 rpm. The overnight culture was transferred again to a sterile 5 mL YPD tube and diluted to the final OD 0.2 and grown for 3–4 h until 0.5–0.6 OD was reached. This indicated a transition of the culture to an exponential growth phase. Then, yeast cells were harvested via centrifugation at 9000× g for 90 s. After centrifugation, all of the following steps were performed by keeping the cells cold, on ice, to slow down the metabolic reactions and cell division. To remove unnecessary salts and ions adhering to the cell wall that could increase the sample’s electrical conductivity, the cells were washed 3 times with 1.5 mL of cold 1 M sorbitol solution. The washed cells were suspended in 1 mL of 1 M sorbitol to the final concentration of 109. After cell suspension was transferred to the 1 mm gap electroporation cuvette and subjected to the PEF ranging from 2 kV/cm to 26 kV/cm, 5 pulses with the duration of 1 ms. After PEF treatment, cells were plated on a fresh medium and grown for 48–72 h, wherein colonies were counted and analyzed for the appearing prions, indicated by the switch of the colony’s color. In total, two controls were used in this research: the first control was prepared and plated before the PEF experiments, showing the number of the viable cells in the suspension; the second control had cells that were prepared and stored on ice throughout the duration of the experiment, showing a general survivability during the experimental procedure.
2.4. Expression and Purification
Sup35NM was performed similarly to the previously described method [33]. In brief, plasmid encoding C-terminally tagged 8x-histidine Sup35NM protein was transformed into BL21(DE3) one star Escherichia coli cells. Overnight culture of transformed cells was used for protein expression, which was carried out by inoculating 2.4 L of a auto-inductive Zym-5052 medium [34] containing ampicillin (100 µg/mL) and grown at 37 °C for 12 h. To collect the cell pellet, the cell suspension was centrifuged at 4000 g at 4 °C for 30 min. Then, the cell pellet was homogenized with a Potter–Elvehjem homogenizer in a 100 mL buffer (20 mM sodium phosphate, 6 M GuHCl, pH 8.0) and sonicated for 15 min on ice using Sonopuls 3100 (Bandelin) ultrasonic homogenizer equipped with a VS70/T tip (using 70% power, 30 s/30 s sonication and rest intervals). The cell suspension lysate was centrifuged at 18,000 g at 4 °C for 20 min and then the supernatant was filtered using 0.22 µm mixed cellulose syringe filters. Finally, the protein mixture was loaded on an immobilized metal affinity chromatography nickel column, which was equilibrated using a loading buffer (20 mM sodium phosphate, 6 M GuHCl, pH 8.0). The protein purification was performed using an imidazole step-gradient where protein eluted at 350 mM of imidazole. Imidazole was then removed from the solution by exchanging the buffer while concentrating the protein. Purified protein was concentrated to 250 µM in buffer (20 mM potassium phosphate, 50 mM NaCl, 6 M GuHCl, pH 7.4), filtered, and stored at –80 °C for further use.
2.5. Preparation of Sup35 Aggregates
Protein stock solution was mixed with aggregation buffer (20 mM KH2PO4, 50 mM NaCl, pH 7.4) to the final 10 µM protein concentration, stored in low-binding tubes, and incubated at 37 °C with constant mixing at 10 RPM for 48 h.
2.6. Atomic Force Microscopy
The following is how we prepared the microscopy: 50 μL of 10 μM purified Sup35NM in a buffer (20 mM KH2PO4, 50 mM NaCl, 0.24 M GdnHCl, pH 7.4) was transferred to a sterile 1 mm electroporation cuvette and subjected to 10 kV and 20 kV (±0.05 kV/cm) for 10 impulses of 1000 μs or 50 μs duration. Imaging of the sample topography via atomic force microscopy then occurred, where the sample was transfer to a 1 × 1 cm cleaved mica substrate (V grade, SPI supplies, West Chester, PA, USA) and incubated for 5–10 min until the proteins adhered to it. The plate was then washed with distilled H2O for 10 s and dried with nitrogen gas for 55 s, so the sample became fixed. Imaging of the samples was performed with a DimensionIcon (Bruker, MA, USA) atomic force microscope using a tapping mode in the air. The manufacturer′s (Bruker, MA, USA) probes were used: FESP (nominal elastic modulus 5 N/m) and TESP (nominal elastic modulus 40 N/m). Standard imaging parameters were used 10 µm × 10 μm area; 0.4 Hz speed; 512-pixel resolution. The obtained images were processed with the NanoScope Analysis (version 1.9) software package.
2.7. Statistical Analysis
At least 3 independent experimental sessions were performed for each unique set of parameters. Atomic force microscopy analysis represented the data from 3 independent experiments. Each time, the length of the fibrils was measured in 10 independent images and represented as an average. Statistical analysis was performed for all acquired data (OriginPro 8.5, OriginLab, Northhampton, MA, USA). The paired t-test was used to estimate the statistical significance. The results were considered statistically significant at p < 0.05.
3. Results
Yeast cells were grown until the culture reached the stationary phase (Figure 2). The results showed that yeast prions significantly affected cell growth dynamics. Psi− in the SC medium reached the stationary phase after 48 h of growth. The growth dynamics of the [PSI+] cells, with a weak variant of the prion, exhibited the same growth dynamics in the Lag and Log phase, but the Log phase was prolonged when compared to the [psi−] strain. The stationary phase reached 82 h of growth. A strong [PSI+] variant significantly altered the cell growth dynamics. The Lag phase was unusually extended, showing that prions affected cell adaptations in a new environment. The Log phase started after 48 h of growth; the stationary phase started after 78 h. S. cerevisiae strain containing a strong [PSI+] variant reached about 3 times higher cell density in the SC medium as compared to the weak prion and 6 times higher as compared to the [psi−] strain.
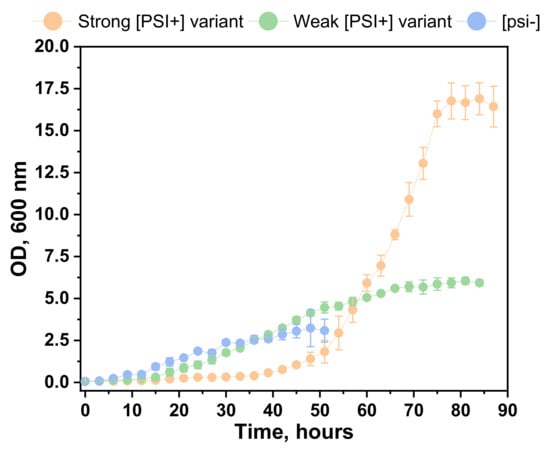
Figure 2.
Growth dynamics of S. cerevisiae cells in the SC medium. The orange line represents the growth of S. cerevisiae with a strong [PSI+] variant; the green line represents the growth of S. cerevisiae with a weak [PSI+] variant; the blue line represents growth of S. cerevisiae without [PSI+]. Data presented as average ±SE.
The PEF treatment results are presented in Figure 3. The presence of the prions in the yeast cells significantly increased cell resistance to PEF treatment. Further, [psi−] cells viability decreased to 40.55 ± 9.59% after applying 2 kV/cm 1000 μs pulse duration, 5 pulses; therefore, cells with strong and weak prions were not affected. Significant differences in cell viability between different prion variants were observed after applying 6 kV/cm PEF. Weak prions were less resistant to PEF as compared to strong ones. Even after 22 kV/cm PEF treatment, 9.58 ± 0.69% viability of strong [PSI+] variant was observed.
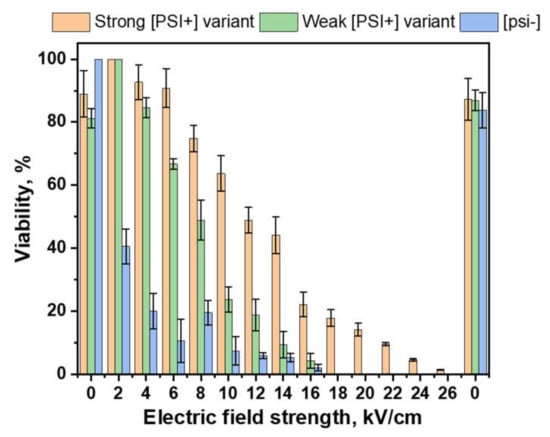
Figure 3.
Viability (%) of the S. cerevisiae cells after exposure to a pulsed electric field (PEF) with different strength of electric field (kV/cm), 1 ms pulse duration, 5 pulses. Orange bars represent the viability of S. cerevisiae with a strong [PSI+] variant; green bars represent the viability of S. cerevisiae with a weak [PSI+] variant; blue bars represent the viability of S. cerevisiae without [PSI+]. Data presented as average ±SE.
The stress-induced prionization analyses revealed that our conditions of PEF treatment did not induce the appearance of the [PSI+] prions in yeast cells or switches from the weak to the strong variant of the prion.
Different electric field strength and pulse duration were tested in the disintegration of the Sup35NM amyloids (Figure 4). Figure 4A presents the control sample of the purified Sup35NM fibrils. The concentration of 10 µM was suitable for atomic force analysis. Fibrils were visible and single amyloid are present in the field without interfering with each other.
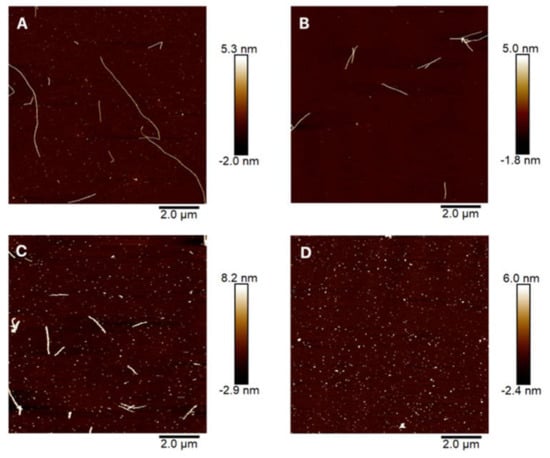
Figure 4.
Atomic force microscopy images of Sup35NM amyloid fibrils before (A) and after exposure to a pulsed electric field (PEF) using different parameters: (B) 10 kV/cm, 50 μs pulse duration, 10 pulses; (C) 10 kV/cm, 1000 μs pulse duration, 10 pulses; (D) 20 kV/cm, 1000 μs pulse duration, 10 pulses. The adjacent color scale indicates the height of the fibrils found in the samples.
The length of the PEF non-treated fibrils varies from 170 nm to 8.1 µm. Small white dots on the sample were the oligomers of Sup35NM. The diameter of the fibrils was 8.3 ± 2.19 nm. Application of 10 kV/cm, 50 μs pulse duration, and 10 pulses resulted in a length reduction in the amyloid fibrils when compared to the control (Figure 4B). The longest fibril found in the samples was 1.45 µm long and this was around four times less than the amyloid from the control samples. An increase in PEF pulse duration to 1000 µs resulted in higher fibril disintegration (Figure 4C). Fewer long amyloids and more oligomers were observed in the sample. Finally, 20 kV/cm, 1000 μs pulse duration, and 10 pulses were responsible for the full disintegration of the fibrils to oligomers (Figure 4D). It is important to point out that the diameter of the Sup35 fibrils increased according to the applied PEF duration. The diameter of 50 µs treated fibrils was 11.1 ± 3 nm and 1000 µs 20.4 ± 5 nm.
4. Discussion
The biological meaning and functions of prions are still under intensive discussion in the scientific community. [PSI+] yeast prion is a perfect model system to analyze the behavior and functions of the amyloid inside the cell. [PSI+] prion has mild pathogenicity, so the cell can survive longer and divide without lethal disturbances. Native Sup35 protein is a translation termination factor and [PSI+] prion helps to overcome STOP codon and prevents translation termination [35,36,37,38]. Strong and weak [PSI+] variants have different abilities to overcome translation termination. It is proven that strong prions form smaller aggregates as compared to weak ones, therefore giving more polymerization ends. Strong prions are more stable and better transmitted in the cell population [6,39].
In this research, we tried to answer three main scientific questions.
- Do [PSI+] prions increase PEF resistance in S. cerevisiae cells and how do different variants change this resistance? Our results show that S. cerevisiae cells containing [PSI+] prions exhibited extremely increased resistance to PEF treatment even in the case of weak prion variants (Figure 3). There is also a significant difference between survivability after PEF between strong and weak variants. Almost 10% viability was observed in the case of strong prions, even after applying 22 kV/cm, 1000 μs pulse duration for 5 pulses. Prions provided cells the ability to survive in such harsh treatment. Other authors agree that prions are related to stress resistance and improve cell viability in chronological aging [40,41]. Prions in the amyloid conformation are resistant to proteases, heat shock, and other inactivation methods [14]. Franzmann et al. showed that [PSI+] prion can regulate the phase separation of native Sup35 protein during stress [15]. In cell growth, Sup35 acts as a translation terminator. In the stress conditions, the prion and sensor domains act together to promote phase separation and to establish protective and reversible biomolecular condensates.
- Can PEF induce the appearance of prions in the cell population de novo or change the type of prion from weak to strong by producing free amyloid ends for protein polymerization? There is a hypothesis that prions in the cell population can appear de novo in cells experiencing stress conditions [41,42] and prion variants can modulate prion-dependent proteomes in response to environmental stress [43], producing a facilitated adaptation of the cells by enhancing survival in rapidly changing environments [44,45,46]. We hypothesized that PEF could act as a factor capable of disrupting cell homeostasis and lead to prion induction. In the case of the cells already containing prions, it could serve as an agent that generates free amyloid ends for the more effective intake of the Sup35 to the amyloids. Analysis of the [psi−] and weak [PSI+] colonies after PEF treatment showed no signs of showing new prions or shift from weak to strong variants. This is the possibility that the PEF itself is not capable of inducing prionization in yeast cells, which makes PEF safe for the biocontrol of yeasts without the opportunity to induce prionization.
- Is it possible to disintegrate [PSI+] amyloids by applying PEF to the purified proteins? There is no covalent bond in the formation of the secondary and tertiary structure between the fibril assembling prion proteins. Stabilizing factors are electrostatic interactions, such as salt bridges and self-energy [28]. Therefore, we hypothesized that those interactions can easily be broken by applying PEF. The ability to use PEF for fibril disintegration could provide universal methods to disintegrate [PSI+] amyloid aggregates. In our research, atomic force microscopy revealed that amyloids of Sup35NM can be successfully disintegrated by using PEF (Figure 4). Even after applying 10 kV/cm, 50 μs pulse duration, and 10 pulses, we observed a length reduction in the amyloid fibrils as compared to the untreated ones. The longest fibril found in the samples was 1.45 µm long, which was around four times less than the amyloid from the control samples. This shows that, for the disintegration of the long fibrils into the smaller aggregates 10 kV/cm, 50 µs, and 10 pulses were enough. The diameter of the fibrils indicates that they belong to a strong prion variant. More than 80% of the strong prion amyloids are thin (~11.5 nm) and around 68% of the weak prions produce thick ~16 nm fibrils [47]. Our research showed that treating the fibrils with longer pulse results not only disintegrates fibrils but increases their diameter as well. The shift from 50 µs to 1000 µs resulted in the doubling of the fibril diameter. An electric field affects the charged groups of amino acids in the molecule, causing a change of the conformation for the fibril itself.
It can be concluded that prions providing yeast cells significantly increase resistance, but prions containing cells can be successfully eliminated using 26 kV/cm and long duration pulses. PEF itself is not capable of inducing prion appearance in yeast cells. Finally, PEF can be used as a fast and safe method in the disintegration of prion aggregates in vitro.
Author Contributions
J.J. and N.B. conducted the experiment, cell growth dynamics, pulsed electric field (PEF) treatment, and protein electroporation; V.N. constructed the PEF generator and optimized PEF conditions; V.S. and A.S. conducted Sup35NM amyloid expression and purification; E.L. supervised the research, resource acquisition, methodology, experiment adaptation, and the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors would like to thank the Marija Jankunec Institute of Biochemistry at the Life Sciences Center of Vilnius University for her valuable support with atomic force microscopy imaging.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Prusiner, S.B. Molecular biology of prion diseases. Science 1991, 252, 1515–1522. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Son, M.; Edskes, H.K. Prion variants of yeast are numerous, mutable, and segregate on growth, affecting prion pathogenesis, transmission barriers, and sensitivity to anti-prion systems. Viruses 2019, 11, 238. [Google Scholar] [CrossRef]
- Shewmaker, F.; McGlinchey, R.P.; Wickner, R.B. Structural insights into functional and pathological amyloid. J. Biol. Chem. 2011, 286, 16533–16540. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Son, M.; Bezsonov, E.E.; DeWilde, M.; Ducatez, M. Yeast Prions Compared to Functional Prions and Amyloids. J. Mol. Biol. 2018, 430, 3707–3719. [Google Scholar] [CrossRef] [PubMed]
- Bradley, M.E.; Liebman, S.W. The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol. Microbiol. 2004, 51, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Chernoff, Y.O.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Sant’anna, R.; Fernández, M.R.; Batlle, C.; Navarro, S.; De Groot, N.S.; Serpell, L.; Ventura, S. Characterization of amyloid cores in prion domains. Sci. Rep. 2016, 6, srep34274. [Google Scholar] [CrossRef]
- Tuite, M.F.; Howard, M.J.; Xue, W.F. Dynamic prions revealed by magic. Chem. Biol. 2014, 21, 172–173. [Google Scholar] [CrossRef][Green Version]
- Vos, T.; Flaxman, A.D.; Naghavi, M.; Lozano, R.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; Aboyans, V.; et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef]
- Frederick, K.K.; Debelouchina, G.T.; Kayatekin, C.; Dorminy, T.; Jacavone, A.C.; Griffin, R.G.; Lindquist, S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem. Biol. 2014, 21, 295–305. [Google Scholar] [CrossRef]
- Linden, R.; Martins, V.R.; Prado, M.A.M.; Cammarota, M.; Izquierdo, I.; Brentani, R.R. Physiology of the prion protein. Physiol. Rev. 2008, 88, 673–728. [Google Scholar] [CrossRef] [PubMed]
- Castle, A.R.; Gill, A.C. Physiological functions of the cellular prion protein. Front. Mol. Biosci. 2017, 4, 19. [Google Scholar] [CrossRef]
- Wulf, M.A.; Senatore, A.; Aguzzi, A. The biological function of the cellular prion protein: An update. BMC Biol. 2017, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Scheckel, C.; Aguzzi, A. Prions, prionoids and protein misfolding disorders. Nat. Rev. Genet. 2018, 19, 405–418. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nüske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359, eaao5654. [Google Scholar] [CrossRef] [PubMed]
- Miklavčič, D.; Mali, B.; Kos, B.; Heller, R.; Serša, G. Electrochemotherapy: From the drawing board into medical practice. Biomed. Eng. Online 2014, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Mir, L.M. Therapeutic perspectives of in vivo cell electropermeabilization. Bioelectrochemistry 2001, 53, 1–10. [Google Scholar] [CrossRef]
- Jakštys, B.; Ruzgys, P.; Tamošiūnas, M.; Šatkauskas, S. Different Cell Viability Assays Reveal Inconsistent Results After Bleomycin Electrotransfer In Vitro. J. Membr. Biol. 2015, 248, 857–863. [Google Scholar] [CrossRef] [PubMed]
- De Backer, M.D.; Maes, D.; Vandoninck, S.; Logghe, M.; Contreras, R.; Luyten, W.H.M.L. Transformation of Candida albicans by electroporation. Yeast 1999, 15, 1609–1618. [Google Scholar] [CrossRef]
- Tee, K.L.; Grinham, J.; Othusitse, A.M.; González-Villanueva, M.; Johnson, A.O.; Wong, T.S. An Efficient Transformation Method for the Bioplastic-Producing “Knallgas” Bacterium Ralstonia eutropha H16. Biotechnol. J. 2017, 12, 1700081. [Google Scholar] [CrossRef]
- Holo, H.; Nes, I.F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 1989, 55, 3119–3123. [Google Scholar] [CrossRef]
- Chang, D.C.; Saunders, J.A.; Chassy, B.M.; Sowers, A.E. Guide to Electroporation and Electrofusion; Academic Press: Cambridge, MA, USA, 2012; ISBN 9780121680411. [Google Scholar]
- Mekid, H.; Mir, L.M. In vivo cell electrofusion. Biochim. Biophys. Acta-Gen. Subj. 2000, 1524, 118–130. [Google Scholar] [CrossRef]
- Rems, L.; Ušaj, M.; Kandušer, M.; Reberšek, M.; Miklavčič, D.; Pucihar, G. Cell electrofusion using nanosecond electric pulses. Sci. Rep. 2013, 3, 3382. [Google Scholar] [CrossRef]
- Kotnik, T.; Frey, W.; Sack, M.; Haberl Meglič, S.; Peterka, M.; Miklavčič, D. Electroporation-based applications in biotechnology. Trends Biotechnol. 2015, 33, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Toepfl, S.; Siemer, C.; Saldaña-Navarro, G.; Heinz, V. Overview of Pulsed Electric Fields Processing for Food. In Emerging Technologies for Food Processing; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Robin, A.; Golberg, A. Pulsed Electric Fields and Electroporation Technologies in Marine Macroalgae Biorefineries. In Handbook of Electroporation; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Guest, W.C.; Cashman, N.R.; Plotkin, S.S. Electrostatics in the stability and misfolding of the prion protein: Salt bridges, self energy, and solvation. Biochem. Cell Biol. 2010, 88, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Serio, T.R. [PIN + ]ing down the mechanism of prion appearance. FEMS Yeast Res. 2018, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Dyda, F.; Tycko, R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. USA 2008, 105, 2403–2408. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions Affect the Appearance of Other Prions: The Story of [PIN] University of Illinois at Chicago. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Novickij, V.; Grainys, A.; Butkus, P.; Tolvaišienė, S.; Švedienė, J.; Paškevičius, A.; Novickij, J. High-frequency submicrosecond electroporator. Biotechnol. Biotechnol. Equip. 2016, 30, 607–613. [Google Scholar] [CrossRef]
- Espargaró, A.; Villar-Piqué, A.; Sabaté, R.; Ventura, S. Yeast prions form infectious amyloid inclusion bodies in bacteria. Microb. Cell Fact. 2012, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Tuite, M.F.; Cox, B.S. Propagation of yeast prions. Nat. Rev. Mol. Cell Biol. 2003, 4, 878–890. [Google Scholar] [CrossRef] [PubMed]
- Shorter, J.; Lindquist, S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 2005, 6, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.; Weissman, J.S.; DePace, A.H. Emerging principles of conformation-based prion inheritance. Annu. Rev. Biochem. 2004, 73, 617–656. [Google Scholar] [CrossRef]
- Chernoff, Y.O. Amyloidogenic domains, prions and structural inheritance: Rudiments of early life or recent acquisition? Curr. Opin. Chem. Biol. 2004, 8, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Uptain, S.M.; Sawicki, G.J.; Caughey, B.; Lindquist, S. Strains of [PSI+] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001, 20, 6236–6245. [Google Scholar] [CrossRef]
- Speldewinde, S.H.; Grant, C.M. The frequency of yeast [PSI+] prion formation is increased during chronological ageing. Microb. Cell 2017, 4, 127–132. [Google Scholar] [CrossRef]
- Tyedmers, J.; Madariaga, M.L.; Lindquist, S. Prion switching in response to environmental stress. PLoS Biol. 2008, 6, e294. [Google Scholar] [CrossRef]
- Chernova, T.A.; Chernoff, Y.O.; Wilkinson, K.D. Prion-based memory of heat stress in yeast. Prion 2017, 11, 151–161. [Google Scholar] [CrossRef]
- Allwein, B.; Kelly, C.; Kammoonah, S.; Mayor, T.; Cameron, D.M. Prion-dependent proteome remodeling in response to environmental stress is modulated by prion variant and genetic background. Prion 2019, 13, 53–64. [Google Scholar] [CrossRef]
- Halfmann, R.; Jarosz, D.F.; Jones, S.K.; Chang, A.; Lancaster, A.K.; Lindquist, S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 2012, 482, 363–368. [Google Scholar] [CrossRef]
- True, H.L.; Lindquist, S.L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 2000, 407, 477–483. [Google Scholar] [CrossRef] [PubMed]
- True, H.L.; Bedin, I.; Lindquist, S.L. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 2004, 431, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Dong, J.; Wall, J.; Frederick, K.K. Amyloid fibrils embodying distinctive yeast prion phenotypes exhibit diverse morphologies. FEMS Yeast Res. 2018, 18, foy059. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).