Abstract
Dietary lipid peroxides (LOOHs) have been linked to gut pathologies including inflammatory bowel disease and cancer. As poorly differentiated (PDiff) intestinal epithelial (Caco-2) cells represent tumor cells and could model intestinal crypt cells, we investigated the cellular response of PDiff Caco-2 cells to the most common dietary LOOH, 13-hydroperoxyoctadecadienoic acid (13-HPODE), using RNA sequencing (RNA-seq). Further, we compared the results with the transcriptomic profiles of PDiff cells exposed to linoleic acid (LA) or hydrogen peroxide (H2O2). The results showed that 13-HPODE treatment induces expression of genes related to detoxification and several metabolic pathways including glycogen and amino acid metabolism, which may create a tumorigenic environment despite the downregulation of some proliferation-related genes. 13-HPODE also enhanced peroxisome proliferator-activated receptor signaling involved in lipid metabolism, homeostasis, and inflammation. Additionally, results indicated that 13-HPODE impacts ribosome biogenesis, phagosome, and mitochondrial function through disrupted electron transport chain, which may contribute to disease development or progression. RNA-seq results were validated using qRT-PCR. This study provides an understanding of PDiff Caco-2 cell response to 13-HPODE and the mechanisms by which 13-HPODE modulates cellular processes that may contribute to disease development or progression.
1. Introduction
The human intestinal epithelium is exposed to different chemicals that we ingest with food. Among those chemicals are lipid peroxides (LOOHs) that usually form in overheated oils (fried food). LOOHs, which are by-products of pre- or auto-oxidation of polyunsaturated fatty acids (PUFAs), have been implicated in various diseases including atherosclerosis [1,2], neurodegenerative disease [3,4], rheumatoid arthritis [5], inflammatory bowel disease [6,7], and colorectal cancer (CRC) [8]. Oxidation of linoleic acid (LA), the most abundant PUFA in mammals and plants, results in the formation of 13-hydroperoxyoctadecadienoic acid (13-HPODE), which is rapidly decomposed by cells into toxic aldehydes and carboxylic acids [9,10]. Lipid peroxidation products including malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE) have shown to modulate gene expression and contribute to CRC [11]. In addition, frying oil has shown to worsen the inflammatory condition and tumorigenesis in the colon [12]. Caco-2 cells, which are derived from human colorectal adenocarcinoma [13], have been widely used to study intestinal metabolism of molecules and drugs [14,15]. These cells, under standard culture conditions, have the ability to become differentiated (Diff; 14-Day) and acquire mature enterocyte-like structural and functional characteristics which makes it a suitable model to study lipid peroxidation [16,17,18,19,20,21,22]. On the other hand, undifferentiated or poorly differentiated (PDiff; 4-Day) Caco-2 cells mimic in vivo intestinal cancer cells and could model intestinal crypt cells, as they share the proliferative phenotype and lack of microvilli cells [23,24], which might be exposed to dietary LOOHs and contribute to disease.
It has been shown that the absorption of LA, the most common dietary PUFA, by PDiff and Diff Caco-2 cells was comparable; however, the absorption of peroxidized LA, 13-HPODE, by PDiff cells was less efficient compared to its absorption by Diff cells that possess brush borders [1]. It has been suggested that dietary oxidized fatty acids are absorbed by the intestine and are esterified into complex oxidized lipids that are transported in the plasma by lipoproteins [1]. Intestinal epithelial lipid transport by apolipoprotein B (ApoB) during differentiation depends on microsomal triglyceride transfer protein (MTTP), which is suppressed by nuclear receptor 2 family 1 (NR2F1) in PDiff Caco-2 cells [25]. It has been demonstrated that MTTP overexpression in PDiff Caco-2 cells increases triglyceride (TG) transfer activity and cholesterol esterification indicating the essential role of MTTP in lipid absorption [26]. Effects of linoleic acid on cells range from production of nitric oxide (NO) and reactive oxygen species (ROS) to protection against oxidative stress [27,28]. In addition, dietary PUFAs are highly susceptible to peroxidation, which has been implicated in the development of colon cancer [11]. Moreover, oral consumption of frying oil in mice has shown to aggravate colon tumorigenicity [12]. Our recent study demonstrated alterations in the expression of inflammatory genes and reduced cell viability on annexin V staining of PDiff Caco-2 cells when treated with peroxidized LA, 13-HPODE [6]. Despite these studies, the effects of dietary LOOHs on PDiff Caco-2 cells, which are sensitive to chemicals, have not been characterized fully.
PDiff Caco-2 cells are used in cancer studies because their gene expression profile resembles that of human colon cancer cells [23], and might also reflect other subpopulation cell types in the human colon [29]. Of note, intestinal proliferative crypt cells might undergo programmed cell death, under stressful conditions, to remove damaged cells and protect from tumorigenesis [30] suggesting that crypt cells could initiate gut pathology. In this study, we investigated the response of PDiff Caco-2 cells to 13-HPODE, in terms of differential gene expression, gene ontology, and pathway enrichment, using mRNA sequencing (RNA-seq) as this group of cells represents intestinal tumor cells and could also model intestinal crypt cells, which could be exposed to the stress of dietary LOOHs. In addition, studying PDiff Caco-2 cells allows us to look at a heterogeneous population of Caco-2 cells before differentiation, thus having broader plasticity in the cellular physiology and response to stress, as compared to Diff cells. We also compared our results to the transcriptomic changes of PDiff cells exposed to unoxidized LA or hydrogen peroxide (H2O2), which is another source of oxidative stress and similar antioxidant response between 13-HPODE and H2O2 treatments was observed in smooth muscle cells [31]. We validated our RNA-seq results using quantitative RT-PCR. The overall goal of this study was to understand how the transcriptomic profile of PDiff Caco-2 cells changes in response to the most common dietary lipid peroxide, 13-HPODE. These changes in gene expression and pathway regulation could provide mechanisms that might be implicated in the development or progression of disease in intestinal crypt cells or tumor cells exposed to dietary LOOHs.
2. Materials and Methods
2.1. Materials
Dulbecco’s modified eagle medium (DMEM), L-glutamine, penicillin/streptomycin, fetal bovine serum (FBS), phosphate-buffered saline (PBS), TRIzolTM reagent, and TURBO DNA-free kit were obtained from Invitrogen (Carlsbad, CA, USA). Trypsin-EDTA solution was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Linoleic acid and soybean lipoxydase were purchased from Sigma (St. Louis, MO, USA). Leucomethylene blue (LMB) was purchased from Alfa Aesar (Ward Hill, MA, USA). NEBNext Ultra II Directional RNA Library Prep kit for Illumina, indexed adaptors and NEBNext Library Quant Kit were purchased from New England Biolabs (NEB; Ipswich, MA, USA). High Sensitivity D1000 reagents were obtained from Agilent (Santa Clara, CA, USA). SsoAdvancedTM Universal SYBR® Green Supermix and PCR Primers were purchased from BioRad (Hercules, CA, USA) and Eurofins (Luxembourg), respectively.
2.2. Cell Culture
Colorectal adenocarcinoma (Caco-2) cells were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured in DMEM and supplemented with 2 mM L-glutamine, 1% penicillin/streptomycin, and 15% FBS. Trypsin-EDTA solution 0.25% was used to sub-culture confluent monolayers and 2.5 × 105–3 × 105 cells/well were seeded in 6-well plates and maintained for 4 days. All experiments were carried out on PDiff cells (Day-4 post-confluence) and run in triplicate.
2.3. Preparation of 13-Hydroperoxyoctadecadienoic Acid (13-HPODE)
13-HPODE was freshly prepared as previously described [22,32,33,34]. Briefly, linoleic acid (LA; 200 µM) in PBS was oxidized with the addition of 10 units soybean lipoxydase. Conjugated diene (HPODE) formation was monitored by scanning the absorption spectrophotometrically (Uvikon XL, Biotek 125 Instruments, El Cajon, CA, USA) at optical density 234 nm; PBS was used as a reference. Lipid peroxide formation was determined using LMB assay [35]. The freshly prepared 13-HPODE was filter sterilized to minimize the risk of contamination and used immediately for the culture experiments to reduce spontaneous peroxide decomposition. No significant difference was observed in peroxide content prior and after filter sterilization of 13-HPODE, which was confirmed by LMB assay as shown in Figure S1. 13-HPODE concentration was calculated using UV spectrophotometry.
2.4. Treatment of Cells with 13-HPODE, LA or H2O2
PDiff Caco-2 cells were starved in serum-free medium for 3 h prior to treatments. Cells were treated with 100 µM of either 13-HPODE, LA, or H2O2 for 24 h in serum-free medium. This concentration was used as it resulted in the significant differences between 13-HPODE-treated and untreated Caco-2 cells in our previous study [6]. Control (untreated) cells were maintained in serum-free medium with equal volume of PBS (500 µL). After 24 h incubation, cells were rinsed with PBS and immediately harvested into TRIzol for total RNA isolation.
2.5. Total RNA Extraction and Quantification
TRIzol reagent was used to lyse the cells directly in the culture plates. Chloroform was added to cell lysate and samples were incubated for 3 min at room temperature then centrifuged at 12,400× g for 15 min. The aqueous phase was transferred into new tubes from which total RNA was precipitated using isopropyl alcohol and resuspended in nuclease-free water. RNA concentration, quality, and purity were tested via NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) showing absorbance ratios of > 1.8 at 260 nm and 280 nm. The TURBO DNA-free kit was used to remove co-extracted DNA from the RNA samples following the manufacturer’s instructions.
2.6. RNA-seq Library Preparation and Sequencing
We used the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, E7490S) for mRNA isolation from 500 ng of total RNA input followed by the NEBNext Ultra II Directional RNA Library Prep kit for Illumina (NEB, E7760S) for library preparation according to the manufacturer’s instructions. mRNA was fragmented to a target size of 200 base pairs and ligated to indexed adaptors for Illumina sequencing (NEB, E7710, E7730). We used NEBNext Sample Purification Beads to remove unincorporated Illumina adapters. Quality and quantity of RNA-seq libraries were assessed by using High Sensitivity D1000 reagents on the TapeStation 2200 (Agilent, Santa Clara, CA, USA), and NEBNext Library Quant Kit (NEB, E7630S), respectively. Individual libraries were combined into an equimolar sequencing pool, which was checked on the TapeStation using High Sensitivity D1000 ScreenTape assay and showed good quality. The library pool was sent for HiSeq4000 sequencing (2 × 150 bp) to GENEWIZ (South Plainfield, NJ, USA).
2.7. Sequence Data Processing
Paired-end read quality was assessed using FastQC (version 0.11.7) [36]. Poor-quality bases were trimmed and adapters were removed using Trimmomatic (version 0.36) [37]. Retained reads were aligned to a human reference genome (Ensembl/Genome Reference Consortium Human Build 38, GRCh38) using Hisat2 (version 2.1.0) [38]. FeatureCounts (version 1.5.0) was used to count the fragments mapped to an exon/gene [39]. Differentially expressed genes (DEGs; adjusted p < 0.05) between different groups were determined using an R-package DESeq2 (version 1.30.0) [40]. One sample of the LA-treated group was excluded from the study as its differential gene expression results did not correlate with the other samples.
2.8. Gene Ontology and Pathway Enrichment Analyses
Gene Ontology (GO) analysis was performed on DEGs of adjusted p < 0.05 (from DESeq2) using clusterProfiler (version 3.18.0) R-package [41]. The function “enrichGO” of clusterProfiler was used to determine the enriched (adjusted p < 0.05) GO biological processes, molecular functions, and cellular components in treated cell groups compared to the untreated group. Pathway enrichment analysis was carried out using Generally Applicable Gene-set Enrichment (GAGE; version 2.40.0) R-package [42]. The function “gage” was used to determine the dysregulated pathways (p < 0.05) in each treatment group based on DEGs and their log2 fold change.
2.9. Validation Using qRT-PCR
Representative genes were chosen, based on differential gene expression and enrichment analysis, for validation of RNA-seq results using qRT-PCR. Eighteen genes were chosen for 13-HPODE-treated group, 12 genes for LA-treated group, and 11 genes for H2O2-treated group. Each Ten microliters of PCR reaction consisted of 5 μL of SsoAdvancedTM Universal SYBR® Green Supermix, 2.5 μM of forward and reverse primers, and 0.1 ng of cDNA. The thermal cycling protocol included an initial denaturation step at 95 °C for 30 s followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. GAPDH was used as the control gene. Relative mRNA expression was calculated using the Ct method, and statistical differences between treated and untreated groups were determined using the t-test. The data were presented as mean ± SD. Primer sequences used in qRT-PCR are provided in Table S1.
3. Results
We treated PDiff Caco-2 cells with 13-HPODE, LA, or H2O2 for 24 h. Untreated cell group was maintained in PBS and used as a control. RNA extraction and processing protocol is described in the Materials and Methods section. The sequencing reads ranged between 31 and 60 million reads (Table S2) with mean quality score > 37.2. Differential gene expression, gene ontology, and pathway analysis were conducted using DESeq2, clusterProfiler and GAGE R-packages, respectively. Principal component analysis (PCA; Figure S2) and sample-to-sample distance heatmap (Figure S3) showed that samples from each group were closely similar in gene expression with respect to treatment and that untreated and treated cell groups were well separated. Genes with adjusted p-value < 0.05 and large mean expression across all cell groups call for significance as demonstrated in log ratio vs. mean average (MA) plots (red dots; Figure S4).
3.1. 13-HPODE-Treated PDiff Caco-2 Cells
3.1.1. Differential Gene Expression
We identified 6648 differentially expressed genes (DEGs; adjusted p < 0.05) between untreated and 13-HPODE-treated PDiff Caco-2 cells using DESeq2 (Table S3); 3345 genes were upregulated and 3303 genes were downregulated upon treating the cells with 13-HPODE. Figure 1 demonstrated a heatmap and hierarchical clustering of the top 50 DEGs in 13-HPODE-treated cell groups.
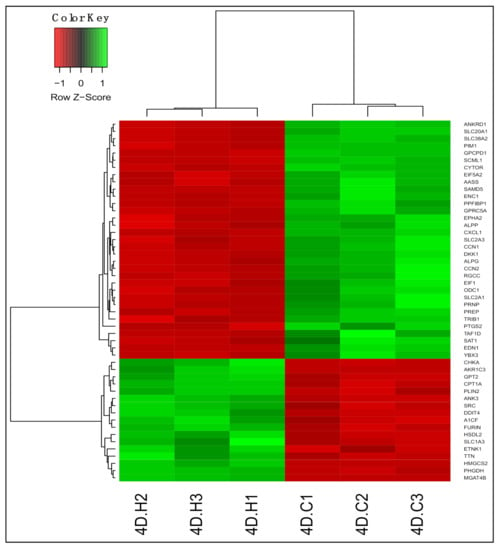
Figure 1.
Differential gene expression in 13-hydroperoxyoctadecadienoic acid (13-HPODE)-treated Caco-2 cells compared to untreated cells. Heatmap shows the top differentially expressed genes (DEGs; adjusted p < 0.05) in 13-HPODE-treated (4D.H1, 4D.H2, and 4D.H3) compared to untreated (control) cells (4D.C1, 4D.C2, and 4D.C3). Green, upregulated; red, downregulated.
Treating PDiff Caco-2 cells with 13-HPODE caused upregulation of genes involved in peroxisome proliferator-activated receptor (PPAR) signaling, which has been implicated in metabolic syndrome and cancer [43]. Among those genes were HMGCS2, which initiates the first step of ketogenesis, CPT1A, which facilitates fatty acid uptake for mitochondrial beta-oxidation, PLIN2, which coats the surface of intracellular lipid droplets, and FABP1, which is involved in fatty acid uptake and transport. Genes that play a role in biosynthesis of phospholipids such as CHKA and ETNK1 were upregulated indicating phospholipid metabolic alterations. Other genes involved in lipid metabolism and uptake were induced such as A1CF, a complementation factor of apolipoprotein B mRNA editing enzyme involved triglyceride uptake, CREB3L3, which is triggered by endoplasmic reticulum stress and involved in acute inflammatory response and triglyceride metabolism, and LRP2, which is endocytic glycoprotein receptor involved in uptake of sterols and lipoproteins. Genes involved in amino acid metabolism such as PHGDH and GPT2 were upregulated; the latter also plays a role in gluconeogenesis. These results suggest significant metabolic alterations in response to 13-HPODE treatment.
Genes involved in the regulation of cellular processes were upregulated such as CEBPA/PB, which inhibit proliferation, promote differentiation, and mediate acute phase response [44], ADM2, which regulates cardiovascular and intestinal activities and ameliorates intestinal inflammation and protects epithelial barrier [45], and ANK3, which links integral membrane proteins to cytoskeleton and regulates cellular activities. The latter’s expression has shown to be increased during Caco-2 polarization [46]. In addition, TTN, which is a key component of stress fibers and brush borders [47], was upregulated. On the other hand, CCN1/N2, CYTOR, and PIM1, which promote cell proliferation, and ODC1, which is important for polyamine biosynthesis, were downregulated. Alterations of the previously mentioned genes indicate low proliferative potential and possible shift towards differentiation. Expression of DDIT4 was increased upon 13-HPODE treatment, which inhibits mTORC1 and regulates cellular processes in response to stress. Upregulated expression of AKR1C1/C2/C3 genes, which have detoxifying activity and reduce aldehydes to alcohols, may indicate conversion of 13-HPODE to aldehydes in the cells. IDH1, which generates NADPH, which is an important cofactor in drug metabolism, antioxidant response, and inhibition of ROS production, was also upregulated. Among downregulated genes were transporters of small molecules such as glucose transporters-SLC2A1/A3, which suggest altered transport of substances across biological membranes by 13-HPODE. Genes involved in inflammation such as CXCL1/L3, chemotactic factors, and CCL20, inflammatory chemokine, were downregulated, which may indicate an anti-inflammatory effect of 13-HPODE.
3.1.2. Gene Ontology
Biological processes that were enriched upon 13-HPODE treatment included ribosomal biogenesis, translation, RNA processing and transport, and protein targeting. In addition, DNA metabolic process, response to endoplasmic reticulum stress, and response to unfolded protein were also enriched. Intrinsic apoptotic signaling pathway and response to oxidative stress were also enriched. This suggests that a significant stress was brought to the cells by 13-HPODE. Biological processes involved in metabolism were affected as well, such as lipid catabolic process, steroid metabolic processes, regulation of carbohydrate metabolism, and cellular aldehyde metabolic process. Peroxisome organization and establishment of cell polarity were also enriched (Figure 2a; Table S4).
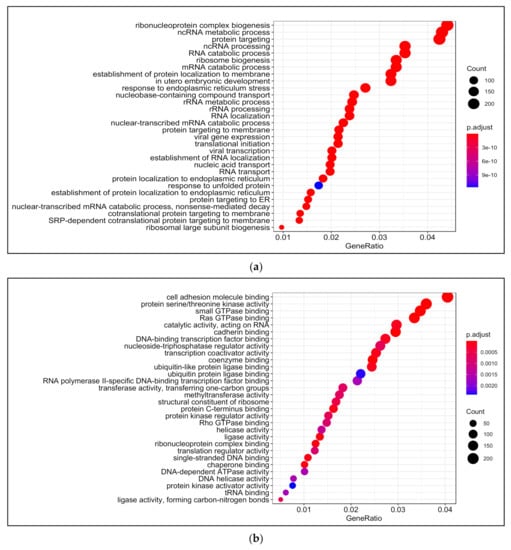
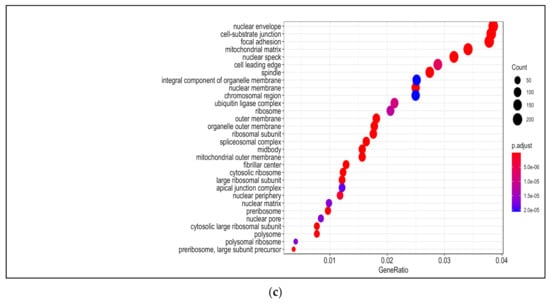
Figure 2.
Gene ontology (GO) analysis in 13-HPODE-treated poorly differentiated (PDiff) Caco-2 cells. Top enriched biological processes (a), molecular function (b), and cellular components (c) in 13-HPODE-treated cells relative to untreated (control) cells (adjusted p < 0.05).
Among molecular functions enriched in 13-HPODE-treated cells were cadherin binding, cell adhesion molecule binding, protein serine/threonine kinase activity, coenzyme binding, and GTPase binding. In addition, ligase activity, ribonucleoprotein complex binding, transcription coactivator activity, protein kinase regulator activity, methyltransferase activity, and helicase activity were enriched (Figure 2b; Table S5).
Among enriched cellular components in 13-HPODE-treated Caco-2 cells were focal adhesion, cell–substrate junction, cell–cell junction, spindle, and cell-leading edge. Furthermore, ribosomal subunit, nuclear envelop/membrane/speck, and spliceosomal complex were enriched. In addition, outer membrane, organelle outer membrane, mitochondrial outer membrane, mitochondrial matrix, and ubiquitin ligase complex. Brush border and peroxisome were enriched as well (Figure 2c; Table S6).
3.1.3. Pathway Analysis
We found that treating PDiff Caco-2 cells with 13-HPODE led to upregulation of retinol metabolism, which could be a counter-regulatory defense mechanism against 13-HPODE, which is considered a xenobiotic. Consistent with previous reports [48], 13-HPODE induced steroid hormone biosynthesis, which has a role in maintaining intestinal barrier [49]. Additionally, starch and sucrose metabolism as well as amino acid metabolism were upregulated. Other pathways that were enriched with slightly higher p values included PPAR signaling as well as peroxisome pathway and metabolism of xenobiotics by cytochrome P450 suggesting that 13-HPODE could trigger detoxification mechanisms. On the other hand, ribosome, RNA polymerase, spliceosome, and pyrimidine metabolic pathways were suppressed due to 13-HPODE treatment. Phagosome degradation pathway was also downregulated. Moreover, 13-HPODE treatment caused downregulation of oxidative phosphorylation indicating possible impaired mitochondrial function (Table 1).

Table 1.
Pathway enrichment analysis in 13-HPODE-treated PDiff Caco-2 cells. Pathway analysis demonstrates upregulated and downregulated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in 13-HPODE-treated cells relative to untreated (control) cells.
3.2. LA-Treated PDiff Caco-2 Cells
3.2.1. Differential Gene Expression
We identified 4225 differentially expressed genes (adjusted p < 0.05) between untreated and LA-treated PDiff Caco-2 cells. Of these, 2080 genes were upregulated and 2145 genes were downregulated in LA-treated cells compared to untreated cells (Table S7). As in 13-HPODE-treated cells, genes involved in lipid uptake and metabolism such as FABP1 and HMGCS2 were also induced upon LA treatment. APOL6, which has a role in lipid transport in the cytosol, was also upregulated. On the contrary, other genes of lipid metabolism were downregulated including MGLL, which hydrolyzes monoacylglycerol to fatty acid and glycerol, and OLR1, which promotes internalization of oxidized LDL. PCK2 involved in glucose production was upregulated as well. DDIT4 and CHAC1, which have a role in stress and unfolded protein response, were upregulated. CEBPB, which have shown to inhibit proliferation, promote differentiation, and mediate acute phase response [44], and IHH, that regulates proliferation and promotes differentiation [50], were also upregulated. On the other hand, similar to 13-HPODE-treated cells, RGCC, CCN1/N2, PIM1, and CYTOR were downregulated in LA-treated cells, which may indicate reduced proliferative potential and enhanced differentiation of PDiff cells by both treatments. While some small molecule transporters such as SLC7A11, an anionic amino acid transporter, were upregulated; others were downregulated such as SLC20A1, a sodium-phosphate symporter, suggesting altered membrane transport of substrates by LA. Reduced expression of CXCL1 and CCL20 chemotactic factors was also seen indicating anti-inflammatory effect of LA. The top 50 DEGs upon LA treatment are shown in Figure 3.
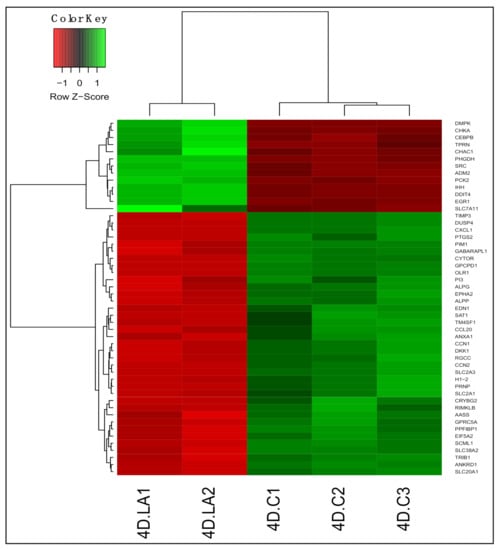
Figure 3.
Differential gene expression in LA-treated Caco-2 cells compared to untreated cells. Heatmap shows the top differentially expressed genes (DEGs; adjusted p < 0.05) in LA-treated (4D.LA1 and 4D.LA2) compared to untreated (control) cells (4D.C1, 4D.C2, and 4D.C3). Green, upregulated; red, downregulated.
3.2.2. Gene Ontology
Among enriched biological processes upon treating PDiff Caco-2 cells with LA were coenzyme metabolic process, regulation of catabolic process, response to insulin, and autophagy. Nucleobase-containing compound transport, ribonucleotide, and nucleotide biosynthetic processes were also enriched. Similar to 13-HPODE-treated cells, we observed enrichment of response to endoplasmic reticulum stress and intrinsic apoptotic signaling pathway in LA-treated cells. Furthermore, alcohol, lipid, and glucose metabolic processes were enriched indicating significant impact of LA on cellular metabolism (Figure 4a; Table S8).
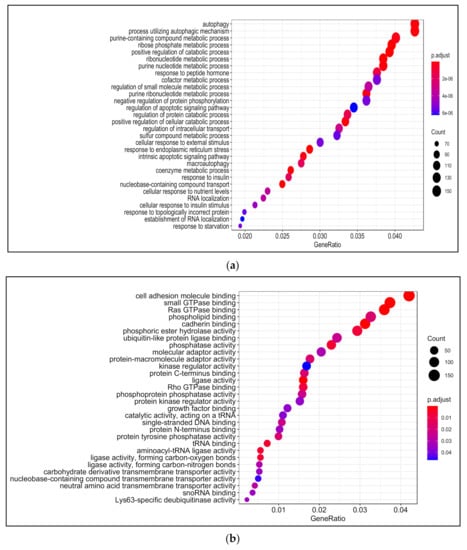
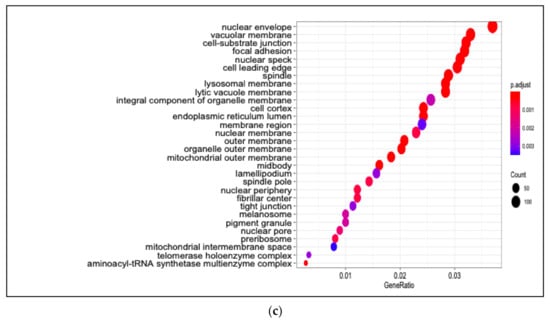
Figure 4.
Gene Ontology (GO) analysis in LA-treated PDiff Caco-2 cells. Top enriched biological processes (a) molecular functions (b) and cellular components (c) in LA-treated cells relative to untreated (control) cells (adjusted p < 0.05).
LA caused enrichment of molecular functions such as cadherin binding, cell adhesion molecule binding, ligase activity, and GTPase binding, which were also enriched upon 13-HPODE treatment. In addition, phosphatase activity, tRNA binding, and phospholipid binding were enriched in LA-treated cells (Figure 4b; Table S9).
As in 13-HPODE-treated cells, cellular, organelle, and mitochondrial outer membranes, nuclear envelop/speck/pore, focal adhesion, cell–substrate junction, cell leading edge, and spindle were among the enriched cellular components in LA-treated cells. Endoplasmic reticulum lumen, vacuolar membrane—that stores nutrients or waste products—and lysosomal membrane were enriched as well as the peroxisomal part (Figure 4c; Table S10).
3.2.3. Pathway Analysis
Pathway analysis revealed that most significantly enriched pathways in LA-treated cells were downregulated. Although ribosome biogenesis was downregulated, the aminoacyl-tRNA biosynthesis pathway, which is essential for protein synthesis, was marginally upregulated in LA-treated cells. The starch and sucrose metabolism and peroxisome pathway were also upregulated, with slightly higher p values, upon treating cells with LA. On the other hand, phagosome and lysosome pathways were downregulated. In addition, spliceosome and pyrimidine metabolism were downregulated. NOD-like receptor signaling involved in immune response was also suppressed (Table 2).

Table 2.
Pathway enrichment analysis in LA-treated PDiff Caco-2 cells. Pathway analysis demonstrates upregulated and downregulated KEGG pathways in LA-treated cells relative to untreated (control) cells.
3.3. H2O2-Treated PDiff Caco-2 Cells
3.3.1. Differential Gene Expression
We identified 5639 DEGs (adjusted p < 0.05) between untreated and H2O2-treated PDiff Caco-2 cells. Of these, 2743 genes were upregulated and 2896 were downregulated upon treatment of cells with H2O2 (Table S11). Similar to 13-HPODE-treated cells, we observed downregulation of PIM1, CCN1/N2, and CYTOR in H2O2-treated cells suggesting reduced cellular proliferation. SLC38A2, an amino acid transporter, SLC20A1, a phosphate transporter, and EPHA2 (ECK), which has a role in intestinal homeostasis and barrier function [51], were suppressed which might suggest impairment of molecule transport and intestinal barrier upon treating the cells with H2O2. In addition, DKK1, a Wnt signaling inhibitor, DUSP6/P4, which dephosphorylate and inactivate MAP/ERK kinase, FAT1, which has a role in cell polarization, and JAG1, which is involved in notch signaling, were downregulated, which may also suggest disrupted intestinal homeostasis. Tumor suppressor genes such as KLF6 and MAFF which also has a role in stress response [52], were downregulated. H2O2 may affect lipid metabolism as it induced the expression of HMGCS2, A1CF and HSDL2, which may have a role in fatty acid metabolism [53], while it suppressed other genes such as PTGS2 and OLR1. H2O2 treatment caused upregulation of antioxidant genes such as BPNT1, which has a detoxifying effect by preventing accumulation of phosphoadenosine phosphate (PAP) [54], CAT, which has a protective effect against H2O2, and IDH1, which is involved in NADPH production and reduces the production of ROS [55]. The top 50 DEGs upon treating PDiff cells with H2O2 are shown in Figure 5.
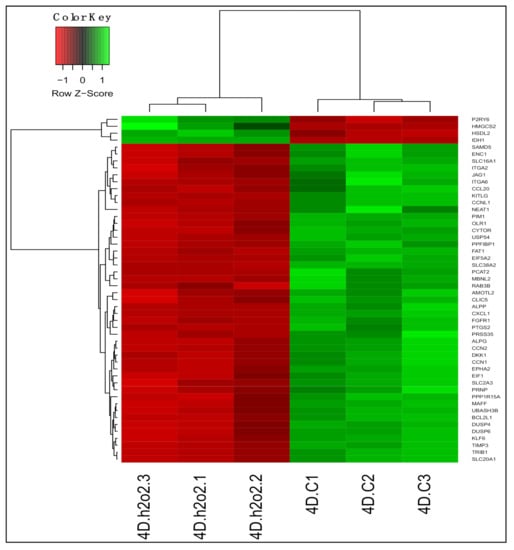
Figure 5.
Differential gene expression in H2O2-treated Caco-2 cells compared to untreated cells. Heatmap shows the top differentially expressed genes (DEGs; adjusted p < 0.05) in H2O2-treated (4D.h2o2.1, 4D.h2o2.2, and 4D.h2o2.3) compared to untreated (control) cells (4D.C1, 4D.C2, and 4D.C3). Green, upregulated; red, downregulated.
3.3.2. Gene Ontology
H2O2 treatment caused enrichment of mRNA catabolic process and rRNA processing, ribosome biogenesis, and translation initiation as well as processes involved in protein localization and targeting to membrane and ER, which were also enriched by 13-HPODE treatment. Consistent with previous studies [56,57,58], H2O2 led to enrichment of double-strand break repair. Similar to 13-HPODE, we also observed enrichment of apoptotic signaling pathway and response to oxidative stress due to H2O2 treatment. Among other enriched biological processes were autophagy, carboxylic acid catabolic process, and cholesterol biosynthetic process (Figure 6a; Table S12).
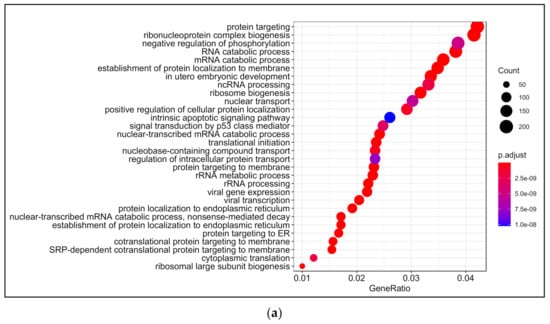
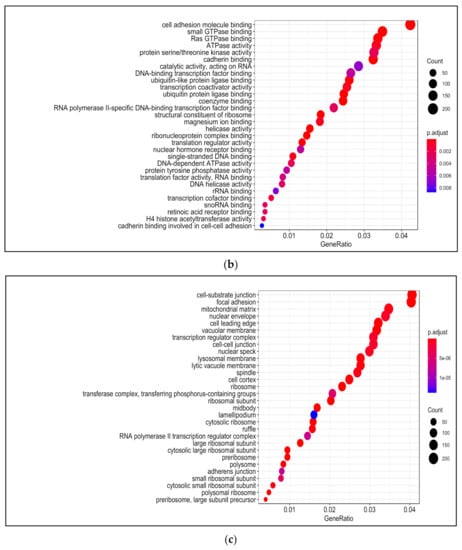
Figure 6.
Gene Ontology (GO) analysis in H2O2-treated PDiff Caco-2 cells. Top enriched biological processes (a) molecular functions (b) and cellular components (c) in H2O2-treated cells relative to untreated (control) cells (adjusted p < 0.05).
Among enriched molecular functions upon H2O2 treatment were cadherin binding, cell adhesion molecule binding, and GTPase binding, which was also seen in 13-HPODE treatment. Ribonucleoprotein complex binding, RNA binding, nuclear hormone receptor binding, and ubiquitin protein ligase binding were also enriched upon H2O2 treatment. Enriched activities included helicase, transcription coactivator, translation factor, protein serine/threonine kinase, and protein tyrosine phosphatase. In addition, coenzyme binding and retinoic acid receptor binding were enriched by H2O2 (Figure 6b; Table S13).
There was a relative similarity in the enriched cellular components between 13-HPODE and H2O2. Among enriched cellular components upon H2O2 treatment were focal adhesion, cell–cell junction, cell leading edge, cell cortex, and ruffle. Components involved in cell division such as midbody and spindle, nuclear components such as nuclear envelope, speck, and transcription factor complex were also enriched. Other enriched components included vacuolar membrane, lysosomal membrane, and mitochondrial matrix. H2O2 also caused enrichment of peroxisome, chromosome, and kinetochore (Figure 6c; Table S14).
3.3.3. Pathway Analysis
H2O2 treatment of PDiff Caco-2 cells led to upregulation of steroid and steroid hormone biosynthesis. It also promoted detoxifying pathways including peroxisome and cytochrome P450, which were also upregulated in 13-HPODE-treated cells, as well as glutathione metabolism. Consistent with a previous report that showed increased lipid uptake by ROS [59], fat digestion and absorption as well as propanoate metabolic pathways appeared to be upregulated by H2O2. Metabolism of amino acids was also affected by H2O2.
Similar to what we observed in 13-HPODE-treated cells, pathways involved in RNA processing and translation such as ribosome, RNA transport, and spliceosome were downregulated in upon treatment with H2O2. Pathways involved in cell adhesion were also downregulated including cell adhesion molecules (CAMs), adherens junction, focal adhesion, and tight junction suggesting significant disruption of epithelial barrier. In addition, Wnt signaling pathway was suppressed, which affects the regulation of physiological processes such as proliferation and differentiation, and immune response (Table 3).

Table 3.
Pathway enrichment analysis in H2O2-treated PDiff Caco-2 cells. Pathway analysis demonstrates upregulated and downregulated KEGG pathways in H2O2-treated cells relative to untreated (control) cells.
3.4. Validation of RNA-seq Results
To validate RNA-seq results using qRT-PCR, we selected three different sets of representative genes based on differential gene expression and pathway enrichment results in each treatment. We investigated 18 genes in 13-HPODE-treated cells, 12 genes in LA-treated cells, and 11 genes in H2O2-treated PDiff Caco-2 cells.
Genes of PPAR signaling that regulate lipid and glucose metabolism including PLIN2, HMGCS2, CPT1A, FABP1, and PCK1 were upregulated in 13-HPODE-treated cells (Figure S5a–e). We also tested key genes involved in stress response, detoxification, and peroxisome such as CREB3L3, DDIT4, RDH10, AKR1C3, PEX6, CAT, and CROT, which were upregulated in 13-HPODE-treated cells (Figure S5f–l). We observed upregulation of the brush border component TTN, and downregulation of RGCC and ODC1 indicating reduced proliferation and shift toward differentiation (Figure S5m–o). Among downregulated genes in 13-HPODE-treated cells were COX20, STX7, and CXCL1 involved in oxidative phosphorylation, phagosome and inflammation, respectively (Figure S5p–r).
In LA-treated PDiff cells, we tested genes of lipid metabolism FABP1 and HMGCS2, glycogen branching enzyme GBE1, peroxisome-related genes PEX6 and CROT, and stress response genes DDIT4 and CHAC1, which showed increased expression due to LA treatment (Figure S6a–g). Upregulation of CEBPB and downregulation of RGCC and DUSP4 was observed indicating altered proliferative phenotype and physiological processes (Figure S6h–j). Inflammatory genes IL18 and CXCL1 were also downregulated upon LA treatment suggesting anti-inflammatory effect of LA on PDiff cells (Figure S6k,l).
In H2O2-treated PDiff cells, we tested genes of detoxification including peroxisome genes CROT and CAT, glutathione transferase GSTA4, and aldo/keto reductase AKR1C3, which were upregulated upon H2O2 treatment (Figure S7a–d). In addition, DDIT4, involved in DNA damage stress response, was upregulated (Figure S7e). Genes of cholesterol synthesis and lipid metabolism were also upregulated due to H2O2 treatment including MVK, HMGCS2, and FABP1 (Figure S7f–h). Among downregulated genes were STX7, CXCL1, and DKK1 involved in phagosome, inflammation, and intestinal homeostasis, respectively (Figure S7i–k). The results of qRT-PCR were consistent with RNA-seq data.
4. Discussion
Linoleic acid, the most common dietary PUFA, can be ingested in the peroxidized form when vegetable oil is overheated such as in fried food. Studies have reported both harmful and possibly beneficial effects of LOOHs. DNA damage has been observed upon treating Caco-2 cells with subcytotoxic levels of LOOHs [60]. Peroxidized LA has also shown to exert pro-inflammatory effects and cause cell death in both Diff and PDiff Caco-2 cells [6]. Moreover, LOOHs have been strongly linked to atherogenesis [61,62], inflammatory disease [63], aging [64], and cancer [65]. On the other hand, a protective effect of LOOHs that promotes apolipoprotein A1 (ApoA1) production, in Diff and PDiff Caco-2 cells, which reduces atherosclerosis has been reported [34,66]. Of note, oxidized metabolites of LA are known to be ligands of PPARs, which are important for intestinal cell differentiation and play a role in lipid metabolism [67]. In this study, we demonstrate, using RNA-seq, that treating PDiff Caco-2 cells with peroxidized linoleic acid, 13-HPODE, can alter their transcriptomic profile significantly, which may reflect the effects of dietary LOOHs on in vivo intestinal crypt or tumor cells. We also compare the effects of 13-HPODE on PDiff cells with the effects of unoxidized LA as well as H2O2, which is another source of oxidative stress.
We demonstrate that 13-HPODE treatment of PDiff Caco-2 cells caused upregulation of mechanisms that help cells in detoxification as PDiff cells are more sensitive to xenobiotics than Diff cells [29,32,68]. Among the upregulated pathways were retinol metabolism, peroxisome, and cytochrome P450. Upregulation of these pathway suggests that there is a need for induction of antioxidant defense response against 13-HPODE-derived oxidative stress to prevent cellular damage. Upregulated retinol metabolism might be a protective mechanism as retinol is considered as a non-enzymatic antioxidant [69]. Products of peroxidized lipids have significantly shown to affect retinoid metabolism [70]. We observed upregulation of beta-carotene oxygenase, BCO1, and retinol dehydrogenase, RDH10, in 13-HPODE-treated cells. The expression levels of these two genes have shown to be low in cirrhotic rat intestine, which indicates retinol’s link to disrupted intestinal epithelial homeostasis [71]. In addition to its role in triglyceride and cholesterol metabolism [72], retinol is important for cellular growth, proliferation, and differentiation of intestinal epithelium and presents in higher amounts in intestinal crypts than villus cells [73]; therefore, enhancement of retinol could be a benefit of 13-HPODE.
PDiff cells showed upregulation of peroxisomal pathway in 13-HPODE-treated cells, which metabolizes lipids, detoxifies radicals, and maintains redox homeostasis. PEX6 gene involved in peroxisome biosynthesis and protein import to peroxisomes was upregulated as well as the detoxifying CAT in 13-HPODE-treated cells. Increased peroxisome proliferation and activity has been reported when PPAR ligands such as fatty acids were used [74]. CROT, IDH1, and ABCD1 genes were also upregulated enhancing peroxisomal lipid metabolism, a mechanism that some cancer cell types use to maintain tumorigenicity and survival through fatty acid oxidation [75]. We observed marginal induction of peroxisomal pathway as well as upregulation of PEX6, ACOX2, and CROT genes in LA-treated cells. This supports studies reported induction of PPAR by LA [76] and induction of peroxisomal acyl-CoA oxidase, which is associated with increase peroxisome number, in LA-treated cultured rat hepatocytes [77]. We also showed that 13-HPODE upregulated the metabolism by cytochrome P450 in PDiff cells, which allows the cells to oxidize and metabolize lipids, and clear xenobiotic compounds including dietary LOOHs and their metabolites such as 4-HNE [78]. Among upregulated genes were UGT2B4, CYP2B6, MAOB, and SULT2A1. These results suggest that exposure of intestinal epithelial cells to 13-HPODE may promote pre-systemic metabolism and reduced bioavailability of xenobiotics as they pass through the intestinal epithelium. Although peroxisomes and cytochrome P450 have a role in detoxifying substrates and protecting cells from oxidative stress, they can also produce ROS and lipid peroxidation, which might form adducts creating an environment for development of carcinogenesis and disease progression [74,79]. In addition, peroxisomal proliferation has been linked to tumor development, and induction of intestinal cytochrome P450 has been linked to gastrointestinal cancer [80,81,82].
Treatment of PDiff Caco-2 cells with 13-HPODE caused upregulation of STS, CYP17A1, and AKR1C2/C3 genes involved in steroid hormone biosynthesis, which has been shown to affect intestinal function and transport of nutrients [83]. Aldo/keto reductases, AKR1Cs, also play a role in bile acid binding and detoxification by reducing lipid peroxide product, 4-HNE, to battle oxidative stress-related injury [84]. Thus, upregulated expression of AKR1Cs might be an indicative of increase intracellular toxic aldehydes. In addition, AKR1C3 has been studied as a marker for progression of colorectal and prostate cancers in which aldehyde levels appear to be elevated [8,85,86]. Therefore, AKR1Cs may provide a potential therapeutic strategy to treat aldehyde-related pathologies. Of note, increased expression of AKR1Cs has been implicated in the resistance of anti-colon cancer therapies [87], which proposes a mechanism by which dietary LOOHs could promote drug resistance through upregulation of the detoxifying AKR1Cs.
ROS have been strongly linked to the initiation and progression of colorectal cancer (CRC) [88]. In addition, not only LOOHs have been implicated in the pathogenesis of CRC [89] but also levels of lipid peroxidation products due to oxidative stress have shown to be elevated in CRC tissue compared to normal colon mucosa [90]. Moreover, increased antioxidant enzyme activity in CRC has been reported which enhances adaptive response that enables cells to battle stress [90]. This is consistent with our results showing increased expression of genes involved in antioxidant defense mechanisms. Within this context, we observed upregulation of several Nrf2-regulated genes involved in antioxidant defense mechanisms in 13-HPODE-treated cells. Among those were genes involved in glutathione (GSH) antioxidant system such as SLC7A11, which facilitates cystine essential for GSH synthesis, GCLC, the rate-limiting step of GSH synthesis, GSR, which maintains high levels of reduced GSH, and GSTA2, that detoxifies compounds by conjugating them to GSH. We also observed upregulation of quinone reductase, NQO1, and CAT, which also have a role in detoxification of ROS and xenobiotics. This is consistent with previous studies demonstrated activation of Nrf2-mediated antioxidant response via oxidized lipids [91,92]. Moreover, cytochrome p450 oxidoreductase, POR, which catalyzes formation of tocopheryl hydroquinone, which is a powerful antioxidant, was upregulated in 13-HPODE-treated cells. This antioxidant response could be triggered by 13-HPODE exposure, disturbed redox status, oxidative stress, or a combination of them. In LA-treated cells, there was upregulation of some Nrf2-targeted antioxidant genes including SLC7A11, GCLC, GSR, and CAT; however, 13-HPODE triggered a more powerful antioxidant response than LA. It is worth to mention that none of the lipoxygenase genes were differentially expressed in either 13-HPODE- or LA-treated cells. However, PTGS2 (also called COX2), which has been shown to be overexpressed in some cancers including CRC [93], appeared to be downregulated in both 13-HPODE- and LA-treated cells. This is consistent with a previous study reported that 13-HPODE inhibited prostaglandin synthesis by inhibiting cyclooxygenase activity [94]. Additionally, several unsaturated FAs including LA have been previously reported as COX-2 inhibitors [95]. Our results implicate that dietary peroxidized LA may promote a strong antioxidant response in the proliferative intestinal tumor or crypt cells to battle the stress derived from the exposure to LOOHs. Of note, the ability of PDiff Caco-2 cells, which are considered cancerous, to induce antioxidant response upon 13-HPODE treatment might lead to progression of disease as it enables cancer cells to survive under stress, which has been implicated in anti-cancer drug resistance [96]. In normal crypt cells, antioxidant response might help in preventing cellular damage that can be caused by oxidative stress and lead to disease. However, Wistar rats treated with repeatedly heated cooking oil, which contains high levels of LOOHs, have shown abnormal colonic epithelium (polyp formation) with aberrant crypts despite the increased antioxidant enzyme activity [97]. Taken together, these observations warrant further studies to determine the role of LOOH-induced antioxidant response in protecting cells or induction of mutagenic changes.
We showed that 13-HPODE enhanced PPAR signaling in PDiff cells consisting with the studies reported oxidized LA as a PPAR ligand [34,67,98]. This might suggest the ability of PDiff cells to uptake 13-HPODE and induce PPAR signaling despite the lack of brush borders. Induction of PPAR signaling leads to activation of downstream genes involved in glucose and lipid metabolism including beta-oxidation. We observed upregulation of PPARA gene and some of its downstream genes including HMGCS2, FABP1, and CPT1A genes involved in ketogenesis, uptake of FA, and mitochondrial beta-oxidation, respectively, as well as PLIN2 that coats lipid droplets. We also observed upregulation of FGF21, which is required for activation of FA oxidation and ketogenic pathways and expression of PPARA target genes [99]. PCK1 and AQP7 were also upregulated suggesting that gluconeogenesis might be enhanced by 13-HPODE. Activation of PPAR could also be a protective anti-inflammatory response against 13-HPODE [100] as we also observed downregulation of proinflammatory genes including IL18, CCL20, and CXCL1/L3/L8. Previous studies have shown reduced expression of chemokines CXCL1/L8 when expression level of PPARα was increased indicating anti-inflammatory effect [101,102]. Another study has shown reduced expression of IL18 in mice treated with PPARα agonist, Wy14643 [103]. Moreover, it has been reported that fenofibrate, another PPARα agonist, also reduced expression of chemokines, including CCL20, in HT-29 cells [104]. On the other hand, previous reports suggested that PPAR activation in the intestinal stem cells may have pro-tumorigenic effect, which indicates that exposure of intestinal crypt cells or tumor cells to 13-HPODE might contribute to the development or progression of cancer [105]. It is worth to note that 13-HPODE caused upregulation of SRC proto-oncogene, which has shown to be upregulated in CRC [106]. Additionally, increased expression of PLIN2 might indicate increased lipid droplets, which have shown to play a role in colon cancer progression [107]. Other reports suggested that PPAR signaling induces cell differentiation or apoptosis [108] and reduces proliferation through downregulation of MYC proto-oncogene [109,110], which we observed, in our study, in 13-HPODE-treated cells. These conflicting studies and our results might indicate dual effects of 13-HPODE-derived PPAR signaling on PDiff cells although the role of PPARs in tumor development requires further investigations.
As seen in this study, starch and sucrose metabolic pathway was enhanced by 13-HPODE treatment of PDiff Caco-2 cells. GYS2 and GBE1 genes, involved in glycogen synthesis and solubility, AGL and PYGM, involved in glycogen degradation, appeared to be upregulated suggesting alteration of cellular glycogen contents. In addition, glycogen metabolism has shown to play an essential role in cancer cell survival [111]. We also observed upregulation of amino acid metabolism in 13-HPODE-treated cells, in particular glycine, serine, and threonine metabolism, which also plays a role in sustaining cancer cell survival. We also observed upregulation of PHGDH involved in serine biosynthesis, which has also shown to maintain survival [112]. The significant impact of 13-HPODE on glycogen and amino acid metabolism of PDiff cells proposes a mechanism by which dietary LOOH may alter the metabolic processes of intestinal crypt or tumor cells to create an environment that initiates or promotes gut disease. Of note, intestinal crypt cells have the ability to initiate tumors especially if exposed to oxidative stress [113].
Components involved in physiological processes such as cell growth, differentiation, and motility were enriched in all three treatment groups. Among those were focal adhesion, cell–substrate junction, cell–cell junction, and cell leading edge. Previous reports indicated that oxidative stress including oxidized lipids could affect cellular barrier function through alteration of focal adhesion, adherens junction, and cell cytoskeleton [114,115]. We also observed downregulation of CLDN1, which is a barrier-forming tight junction gene and upregulation of the pore-forming CLDN2 in 13-HPODE-treated cells, a characteristic of IBD [116]. In addition, it has been reported that LA and its oxidized metabolite could modulate cellular differentiation of Caco-2 cells [117,118,119].
PDiff Caco-2 cells showed downregulation of pathways involved in RNA processing and translation including ribosome and spliceosome as well as pyrimidine metabolism in the three treatment groups. This alteration might indicate reduced proliferation as these pathways have shown to be downregulated when Caco-2 cells lose their proliferative potential and shift towards differentiation [120]. In addition, induction of intestinal cell differentiation by 13-HPODE or LA has been previously reported [34,121]. Alternatively, downregulation of the translation machinery could be due to oxidative damage to the ribosomes to which the cells needed a feedback inhibition of ribosome biogenesis to prevent translation errors and further ribosomal damage [122]. Of note, oxidative stress-induced modification of RNA has been linked to age-related diseases including neurodegenerative disorders [122]. Moreover, we observed increased expression of pro-apoptotic genes such as DDIT4, BMF, BNIP3L, and BCL2L14 and decreased expression of anti-apoptotic BCL2L1 in both 13-HPODE- and LA-treated groups suggesting reduced cell viability. This is consistent with previous studies reported that 13-HPODE increases cell death [6] and that LA induces apoptosis and cell differentiation [121] in Caco-2 cells. In addition, our results at the gene expression level showed downregulation of several genes involved in proliferation such as CDK10, PIM1, CCN1/N2, and CYTOR in both 13-HPODE- and LA-treated cells, which suggests reduced proliferation. Although our results suggest, in part, that 13-HPODE promotes cell death, they may also indicate that 13-HPODE could trigger antioxidant response and modulate metabolic processes in a way that creates a carcinogenic environment and renders cells fight stress which could explain the resistance of some cancer types to anticancer therapies.
13-HPODE treatment of PDiff Caco-2 cells caused downregulation of oxidative phosphorylation. We observed downregulation of genes involved in electron transport chain complexes including complex I, III, IV, and V in 13-HPODE-treated cells. Our results support studies that demonstrated that oxidized LA products can damage the mitochondria and interfere with oxidative phosphorylation as these products can incorporate into mitochondrial membrane lipids and produce more radicals [123,124]. In addition, PUFAs have shown to increase mitochondrial -oxidation and ROS production, which might cause mitochondrial dysfunction [124]. Of note, lipid peroxidation-related mitochondrial dysfunction has been linked to several pathologies including non-alcoholic steatohepatitis, ischemic cardiac reperfusion, and IBD [125,126,127].
We observed downregulation of the cellular phagosomal pathway in PDiff Caco-2 cells for all treatment groups. Among downregulated genes were those that encode subunits of vacuolar-ATPase involved in organelle acidification, such as ATP6V1D/V0B. Intestinal epithelial cells are considered non-professional phagocytic cells and this effect of reduced phagosomal pathway could be attributed to the reduction of oxidative phosphorylation, particularly in 13-HPODE-treated cells, as phagocytosis is known to be an energy-dependent process [128,129]. It has been demonstrated that modification of photoreceptor outer segments by lipid peroxidation products causes impairment of lysosomal degradation [130], suggesting the negative regulation of lysosomal/phagosomal degradation by LOOH adducts. Of note, impaired intestinal epithelial autophagy has been implicated in the pathology of IBD [131], and impaired intestinal epithelial phagosomal function may affect the ability of epithelial cells, the first line of defense in the gut, to clear the pathogens they encounter [128]. Thus, disruption of both oxidative phosphorylation and phagosomal pathway could propose a mechanism by which dietary LOOHs contribute to IBD and/or IBD-related carcinogenesis in intestinal crypt or tumor cells.
On the other hand, PDiff Caco-2 cells showed an anti-inflammatory response to either treatment evidenced by downregulation of inflammatory chemokines, CXCL1/L3 and CCL20, and NOD-like receptor signaling. We demonstrated that upregulation of PPAR signaling by 13-HPODE might exert an anti-inflammatory response, which is consistent with a previous study reported the protective anti-inflammatory effect of PPAR against lipopolysaccharide [132]. Furthermore, PPAR ligands have shown to reduce expression of TNF as well as chemokine receptors and chemokines [133] including CXCL1 [134]. Previous studies also reported anti-inflammatory properties of PUFAs [135,136,137] consisting with our results.
H2O2 is a source of oxidative stress that can be found in food preservatives, dental care products, and in some beverages consumed daily such as coffee and tea, and might predispose the intestinal epithelium to oxidative stress [138]. To evaluate whether H2O2 has a similar impact of 13-HPODE on the transcriptomic profile of Caco-2 cells, we treated PDiff cells with 100 μM H2O2. A previous study reported that PDiff Caco-2 cells maintained 95% viability when treated with 100 μM of H2O2 and that cellular and DNA damage was observed at 150 μM H2O2 or higher [58]. H2O2 is normally detoxified by peroxisomal catalase and glutathione defense mechanism [115]. Similar to what we observed in 13-HPODE-treated cells, H2O2-treated cells showed upregulation of multiple pathways involved in detoxification such as peroxisome, cytochrome P450, and glutathione metabolism suggesting disrupted redox status in both treatment groups. We observed upregulation of CAT, PEX6, and PEX11A, as well as peroxisomal lipid metabolism genes including IDH1, ABCD1, and CROT in H2O2-treated cells. It has been reported that H2O2 may cause peroxisomal proliferation supporting the antioxidant role of peroxisome against ROS [74]. Upregulated genes also included alcohol dehydrogenases ADH5/6, and UGT2B4 involved in metabolism of toxic compounds and xenobiotics. Nrf2-mediated antioxidant response was also promoted by H2O2 evidenced by upregulation of MGST3 and GSTA4/A2/M4 involved in glutathione metabolism indicating disrupted redox status and promoted defense against oxidative stress. On the other hand, cellular cytoskeleton and integrity were disturbed by H2O2 as cell adhesion molecules, adherens/tight junction, and focal adhesion pathways were downregulated. This is consistent with previous studies reported Caco-2 cells’ injury and barrier dysfunction in response to oxidative stress, which contributes to IBD [139,140]. In addition, intestinal crypt cells have the ability to initiate tumors if exposed to oxidative stress [113].
We demonstrated that H2O2 treatment upregulated lipid uptake and metabolism as we observed upregulation of FABP1, MTTP, and DGAT2. We also observed that H2O2 induced steroid hormone and steroid biosynthesis, which has been observed in studies of liver cells treated with H2O2 [59,141]. LSS, DHCR24, and MSMO1 involved in steroid biosynthesis were upregulated in H2O2-treated cells. Upregulation of steroid biosynthesis might suggest sustained tumorigenicity as cholesterol synthesis is considered a hallmark feature of cancer [142]. Steroid biosynthesis was not affected by 13-HPODE treatment; in fact, it has been previously reported that oxidized lipids could inhibit cholesterol synthesis by binding to SREBPs and suppressing their nuclear localization [143]. We observed that H2O2 caused alteration of amino acids metabolism including upregulation of valine, leucine and isoleucine degradation pathway, which have also shown to be upregulated in MCF-7 cells treated with H2O2; this pathway has been linked to volatile organic compounds, which are biomarkers for cancer [144]. Our findings suggest that H2O2 might have a significant impact on intestinal tumor or crypt cells, which alters redox status and modifies lipid and amino acid metabolism as well as cell cytoskeleton affecting cellular physiology, integrity, and survival fates.
Table 4 demonstrates comparative differential expression of selected genes between the three treatment groups. Collectively, our results indicated that 13-HPODE, LA and H2O2 upregulated the. peroxisomal pathway and peroxisome-related genes including PEX6 and CROT. Although we observed upregulation of genes involved in PPAR signaling in both 13-HPODE- and LA-treated cells including PLIN2 and HMGCS2, the upregulation effect was more pronounced in 13-HPODE-treated cells. This may suggest that 13-HPODE is a more potent activator of PPAR signaling than LA although both oxidized and unoxidized LA have previously shown to activate PPARs [34,76]. Moreover, gene expression and pathway analysis indicated that 13-HPODE and H2O2 promoted a powerful antioxidant response in PDiff cells compared to LA treatment. This was evidenced by upregulation of genes involved in cytochrome P450, Nrf2-mediated response, and glutathione metabolism, including CAT and GSTA2, which might promote adaptive response. In addition, amino acid metabolism upregulation observed in 13-HPODE- and H2O2-treated cells, but not in LA-treated cells, might suggest that these two treatments could promote a tumorigenic environment as amino acid metabolism including serine biosynthesis has a role in cell survival [112]. 13-HPODE caused significant downregulation of oxidative phosphorylation, which was not seen in LA or H2O2 treatments indicating the damaging effect might be caused by 13-HPODE on mitochondrial function as reported by previous studies [123,124]. As we mentioned earlier, all three treatments caused downregulation of ribosome biogenesis, spliceosome, and pyrimidine synthesis. Downregulation of these pathways and some proliferation-related genes such as ODC1 and RGCC might indicate reduced proliferative potential as well as possible oxidative damage to ribosomes to which the cells responded by downregulating ribosome biogenesis in order to prevent translation errors. Despite the downregulation of some inflammatory genes including CXCL1 and IL18 in all three treatments, the phagosomal pathway also appeared to be downregulated, which has been implicated in the pathology of bowel disease [131]. Gene ontology analysis showed enrichment of focal adhesion and cellular junctions in all three treatments, however, pathway analysis showed significant downregulation of cell adhesion molecules, adherens junction, and focal adhesion in H2O2-treated cells indicating damaging effect of H2O2 on cellular cytoskeleton and junctions, which might affect barrier function [139,140].

Table 4.
Comparative gene expression. Comparison of expression of selected genes between cells treated with peroxidized linoleic acid (13-HPODE), non-peroxidized linoleic acid (LA), or hydrogen peroxide (H2O2) treatments (↑ upregulated; ↓ downregulated).
5. Conclusions
PDiff Caco-2 cells could simulate in vivo tumor cells and model intestinal epithelial crypt cells in their proliferative phenotype and lack of brush borders. In this study, we show how PDiff Caco-2 cells respond to peroxidized linoleic acid, 13-HPODE, in terms of differential gene expression, gene ontology, and pathway enrichment, which might reflect the response of in vivo intestinal tumor or crypt cells exposed to dietary LOOHs. Figure 7 summarizes the cellular events and processes altered by each treatment. The stress brought to PDiff cells by 13-HPODE caused the cells to upregulate genes and pathways involved in detoxification as rigorous activation of antioxidant response was required to maintain cellular function and prevent damage. Induction of peroxisomal pathway and cytochrome P450 may, on the one hand, metabolize lipids, detoxify radicals, and maintain redox homeostasis, but on the other hand, can increase ROS production and lipid peroxidation and contribute to the development or progression of disease including gastrointestinal cancer. Moreover, enhanced peroxisomal fatty acid oxidation due to either treatment might favor carcinogenesis. This could propose a mechanism by which 13-HPODE contributes to tumor progression or resistance to anticancer therapy. While induction of PPAR signaling by 13-HPODE promoted FA oxidation and lipid metabolism and might have anti-inflammatory and anti-proliferative effects on PDiff Caco-2 cells, it may maintain the tumorigenic phenotype of the cells. Moreover, upregulated glycogen metabolism by either 13-HPODE or LA and upregulated amino acid metabolism by 13-HPODE or H2O2 may promote cell survival. Although our results, in part, indicate reduced proliferation, the overall changes in cellular response to 13-HPODE and H2O2 may provide a carcinogenic environment. The negative impact we observed of 13-HPODE on electron transport chain and phago-lysosomal degradation pathways might link LOOHs to different intestinal pathologies including IBD and cancer, which could be initiated by intestinal crypt cells. Compared to 13-HPODE, H2O2 treatment not only enhanced antioxidant adaptive response, which protects against cytotoxicity, but also upregulated steroid biosynthesis, which is a hallmark of tumor development. This study expands the knowledge of how PDiff Caco-2 cells respond to 13-HPODE, which possibly resembles the response of in vivo intestinal tumor or epithelial crypt cells exposed to dietary LOOHs. In this context, our results suggest that dietary LOOHs, such as 13-HPODE, could trigger a strong antioxidant mechanism in intestinal tumor or epithelial crypt cells, which provides a defensive response to stress. Our results also indicate that dietary LOOHs may modify metabolic processes and alter energy production and degradation processes creating an environment that renders these proliferative cells to initiate or progress disease. Hence, this study provides possible mechanisms by which dietary LOOHs might contribute to disease development or progression in intestinal crypt or cancer cells. The hypotheses generated by our RNA-seq study can be further investigated using directed biomolecular experiments.
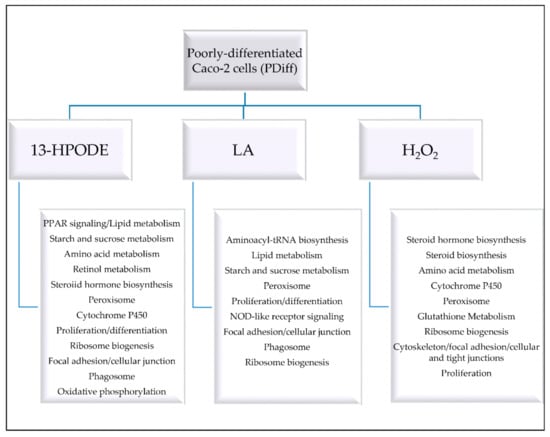
Figure 7.
A summary of the cellular events and processes altered in PDiff Caco-2 cells by 13-HPODE, LA, or H2O2.
6. Limitations and Future Work
In this study, we carried out our experiments on culture cells, and we used one concentration of treatment-100 μM, which, in our previous studies, has demonstrated significant differences between treated and untreated cells. Future transcriptomic studies might be dose-dependent and include in vivo experiments as well as testing other products of lipid peroxidation.
Supplementary Materials
Supplementary materials can be accessed at: https://www.mdpi.com/2076-3417/11/6/2678/s1. Figure S1. Leucomethylene blue (LMB) assay. Figure S2: Principal component analysis (PCA), Figure S3: Sample-to-sample distance heatmaps generated by DESeq2, Figure S4: MA plots of differentially expressed genes (DEGs) between untreated and treated PDiff Caco-2 cells, Figure S5: Quantitative RT-PCR validation of RNA-seq data in 13-HPODE-treated PDiff Caco-2 cells, Figure S6: Quantitative RT-PCR validation of RNA-seq data in LA-treated PDiff Caco-2 cells, Figure S7: Quantitative RT-PCR validation of RNA-seq data in H2O2-treated PDiff Caco-2 cells, Table S1: Primer sequences, Table S2: Number of sequence reads per sample after trimming, Table S3: Differential gene expression in 13-HPODE-treated cells, Table S4: Gene Ontology analysis (biological processes) in 13-HPODE-treated PDiff Caco-2 cells, Table S5: Gene Ontology analysis (molecular functions) in 13-HPODE-treated PDiff Caco-2 cells, Table S6: Gene Ontology analysis (cellular components) in 13-HPODE-treated PDiff Caco-2 cells, Table S7: Differential gene expression in LA-treated cells, Table S8: Gene Ontology analysis (biological processes) in LA-treated PDiff Caco-2 cells, Table S9: Gene Ontology analysis (molecular functions) in LA-treated PDiff Caco-2 cells, Table S10: Gene Ontology analysis (cellular components) in LA-treated PDiff Caco-2 cells, Table S11: Differential gene expression in H2O2-treated cells, Table S12: Gene Ontology analysis (biological processes) in H2O2-treated PDiff Caco-2 cells, Table S13: Gene Ontology analysis (molecular functions) in H2O2-treated PDiff Caco-2 cells, Table S14: Gene Ontology analysis (cellular components) in H2O2-treated PDiff Caco-2 cells.
Author Contributions
All authors have contributed substantially to the work reported. Conceptualization, N.F., S.Y., and S.P.; data curation, N.F. and S.Y.; formal analysis, N.F. and S.Y.; funding acquisition, S.Y. and S.P.; investigation, N.F., C.A.N., A.F., and S.Y.; methodology, N.F., C.A.N., A.F., S.Y., and S.P.; project administration, N.F., C.A.N., A.F., S.Y., and S.P.; resources, N.F., C.A.N., A.F., S.Y., and S.P.; software, N.F. and S.Y.; supervision, N.F., A.F., S.Y., and S.P.; validation, N.F., S.Y., and S.P.; visualization, N.F., S.Y., and S.P.; writing—original draft, N.F., S.Y., and S.P.; writing—review and editing, N.F., C.A.N., A.F., S.Y., and S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence files and metadata are available at NCBI under BioProject accession PRJNA694700. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA694700 (accessed on 25 January 2021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Penumetcha, M.; Khan, N.; Parthasarathy, S. Dietary oxidized fatty acids: An atherogenic risk? J. Lipid Res. 2000, 41, 1473–1480. [Google Scholar] [CrossRef]
- Penumetcha, M.; Khan-Merchant, N.; Parthasarathy, S. Enhanced solubilization and intestinal absorption of cholesterol by oxidized linoleic acid. J. Lipid Res. 2002, 43, 895–903. [Google Scholar] [CrossRef]
- Shichiri, M. The role of lipid peroxidation in neurological disorders. J. Clin. Biochem. Nutr. 2014, 54, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; Perluigi, M.; Butterfield, D.A. Lipid peroxidation triggers neurodegeneration: A redox proteomics view into the Alzheimer disease brain. Free Radic. Biol. Med. 2013, 62, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Hitchon, C.A.; El-Gabalawy, H.S. Oxidation in rheumatoid arthritis. Arthritis Res. Ther. 2004, 6, 265–278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Keewan, E.; Narasimhulu, C.A.; Rohr, M.; Hamid, S.; Parthasarathy, S. Are fried foods unhealthy? The dietary peroxidized fatty acid, 13-hpode, induces intestinal inflammation in vitro and in vivo. Antioxidants 2020, 9, 926. [Google Scholar] [CrossRef]
- Bhaskar, N.; Narasimhulu, C.A.; Keewan, E.; Rohr, M.; Parthasarathy, S. Proinflammatory Properties of Peroxidized Fat May Contribute to the Etiology of Crohn’s Disease. J. Med. Food. 2019, 22, 162–169. [Google Scholar] [CrossRef]
- Skrzydlewska, E.; Stankiewicz, A.; Sulkowska, M.; Sulkowski, S.; Kasacka, I. Antioxidant Status and Lipid Peroxidation in Colorectal Cancer. J. Toxicol. Environ. Health Part A. 2001, 64, 213–222. [Google Scholar] [CrossRef]
- Bergström, S. Autoxidation of Linoleic Acid. Nature 1945, 156, 717–718. [Google Scholar] [CrossRef]
- Raghavamenon, A.; Garelnabi, M.; Babu, S.; Aldrich, A.; Litvinov, D.; Parthasarathy, S. α-Tocopherol Is Ineffective in Preventing the Decomposition of Preformed Lipid Peroxides and May Promote the Accumulation of Toxic Aldehydes: A Potential Explanation for the Failure of Antioxidants to Affect Human Atherosclerosis. Antioxid. Redox Signal. 2009, 11, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Perše, M. Oxidative Stress in the Pathogenesis of Colorectal Cancer: Cause or Consequence? Biomed Res. Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, X.; Yang, R.; Ma, Q.; Qi, W.; Sanidad, K.Z.; Park, Y.; Kim, D.; Decker, E.A.; Zhang, G. Thermally Processed Oil Exaggerates Colonic Inflammation and Colitis-Associated Colon Tumorigenesis in Mice. Cancer Prev. Res. 2019, 12, 741–750. [Google Scholar] [CrossRef]
- Stierum, R.; Gaspari, M.; Dommels, Y.; Ouatas, T.; Pluk, H.; Jespersen, S.; Vogels, J.; Verhoeckx, K.; Groten, J.; van Ommen, B. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim. Biophys. Acta Proteins Proteom. 2003, 1650, 73–91. [Google Scholar] [CrossRef]
- Delie, F.; Rubas, W. A Human Colonic Cell Line Sharing Similarities With Enterocytes as a Model to Examine Oral Absorption: Advantages and Limitations of the Caco-2 Model. Crit. Rev. Ther. Drug Carr. Syst. 1997, 14, 66. [Google Scholar] [CrossRef]
- Sun, H.; Chow, E.C.Y.; Liu, S.; Du, Y.; Pang, K.S. The Caco-2 cell monolayer: Usefulness and limitations. Expert Opin. Drug Metab. Toxicol. 2008, 4, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Engle, M.J.; Goetz, G.S.; Alpers, D.H. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J. Cell. Physiol. 1998, 174, 362–369. [Google Scholar] [CrossRef]
- Peng, I.-W.; Kuo, S.-M. Research Communication: Flavonoid Structure Affects the Inhibition of Lipid Peroxidation in Caco-2 Intestinal Cells at Physiological Concentrations. J. Nutr. 2003, 133, 2184–2187. [Google Scholar] [CrossRef]
- Wingler, K.; Müller, C.; Schmehl, K.; Florian, S.; Brigelius-Flohé, R. Gastrointestinal glutathione peroxidase prevents transport of lipid hydroperoxides in CaCo-2 cells. Gastroenterology 2000, 119, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Taha, R.; Seidman, E.; Mailhot, G.; Boudreau, F.; Gendron, F.P.; Beaulieu, J.F.; Ménard, D.; Delvin, E.; Amre, D.; Levy, E. Oxidative stress and mitochondrial functions in the intestinal Caco-2/15 Cell Line. PLoS ONE 2010, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Deiana, M.; Corona, G.; Incani, A.; Loru, D.; Rosa, A.; Atzeri, A.; Paola Melis, M.; Assunta Dessì, M. Protective effect of simple phenols from extravirgin olive oil against lipid peroxidation in intestinal Caco-2 cells. Food Chem. Toxicol. 2010, 48, 3008–3016. [Google Scholar] [CrossRef]
- Bernotti, S.; Seidman, E.; Sinnett, D.; Brunet, S.; Dionne, S.; Delvin, E.; Levy, E. Inflammatory reaction without endogenous antioxidant response in Caco-2 cells exposed to iron/ascorbate-mediated lipid peroxidation. Am. J. Physiol. Liver Physiol. 2003, 285, G898–G906. [Google Scholar] [CrossRef][Green Version]
- Faizo, N.; Narasimhulu, C.A.; Forsman, A.; Yooseph, S.; Parthasarathy, S. Peroxidized Linoleic Acid, 13-HPODE, Alters Gene Expression Profile in Intestinal Epithelial Cells. Foods 2021, 10, 314. [Google Scholar] [CrossRef] [PubMed]
- Sääf, A.M.; Halbleib, J.M.; Chen, X.; Yuen, S.T.; Leung, S.Y.; Nelson, W.J.; Brown, P.O. Parallels between Global Transcriptional Programs of Polarizing Caco-2 Intestinal Epithelial Cells In Vitro and Gene Expression Programs in Normal Colon and Colon Cancer. HMol. Biol. Cell 2007, 18, 4245–4260. [Google Scholar] [CrossRef]
- Tadjali, M.; Seidelin, J.B.; Olsen, J.; Troelsen, J.T. Transcriptome changes during intestinal cell differentiation. Biochim. Biophys. Acta Mol. Cell Res. 2002, 1589, 160–167. [Google Scholar] [CrossRef]
- Dai, K.; Khatun, I.; Hussain, M.M. NR2F1 and IRE1β Suppress Microsomal Triglyceride Transfer Protein Expression and Lipoprotein Assembly in Undifferentiated Intestinal Epithelial Cells. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 568–574. [Google Scholar] [CrossRef]
- Iqbal, J.; Rudel, L.L.; Hussain, M.M. Microsomal Triglyceride Transfer Protein Enhances Cellular Cholesteryl Esterification by Relieving Product Inhibition. J. Biol. Chem. 2008, 283, 19967–19980. [Google Scholar] [CrossRef] [PubMed]
- Saraswathi, V.; Wu, G.; Toborek, M.; Hennig, B. Linoleic acid-induced endothelial activation. J. Lipid Res. 2004, 45, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Beeharry, N.; Lowe, J.E.; Hernandez, A.R.; Chambers, J.A.; Fucassi, F.; Cragg, P.J.; Green, M.H.; Green, I.C. Linoleic acid and antioxidants protect against DNA damage and apoptosis induced by palmitic acid. Mutat. Res. Mol. Mech. Mutagen. 2003, 530, 27–33. [Google Scholar] [CrossRef]
- Proquin, H.; Jonkhout, M.C.M.; Jetten, M.J.; van Loveren, H.; de Kok, T.M.; Briedé, J.J. Transcriptome changes in undifferentiated Caco-2 cells exposed to food-grade titanium dioxide (E171): Contribution of the nano- and micro- sized particles. Sci. Rep. 2019, 9, 18287. [Google Scholar] [CrossRef]
- Johnson, I. Anticarcinogenic effects of diet-related apoptosis in the colorectal mucosa. Food Chem. Toxicol. 2002, 40, 1171–1178. [Google Scholar] [CrossRef]
- Meilhac, O.; Zhou, M.; Santanara, N.; Parthasarathy, S. Lipid peroxides induce expression of catalase in cultured vascular cells. J. Lipid Res. 2000, 41, 1205–1213. [Google Scholar] [CrossRef]
- Rohr, M.; Narasimhulu, C.A.; Keewan, E.; Hamid, S.; Parthasarathy, S. The dietary peroxidized lipid, 13-HPODE, promotes intestinal inflammation by mediating granzyme B secretion from natural killer cells. Food Funct. 2020, 11, 9526–9534. [Google Scholar] [CrossRef] [PubMed]
- Khan-Merchant, N.; Penumetcha, M.; Meilhac, O.; Parthasarathy, S. Oxidized Fatty Acids Promote Atherosclerosis Only in the Presence of Dietary Cholesterol in Low-Density Lipoprotein Receptor Knockout Mice. J. Nutr. 2002, 132, 3256–3262. [Google Scholar] [CrossRef] [PubMed]
- Rong, R.; Ramachandran, S.; Penumetcha, M.; Khan, N.; Parthasarathy, S. Dietary oxidized fatty acids may enhance intestinal apolipoprotein A-I production. J. Lipid Res. 2002, 43, 557–564. [Google Scholar] [CrossRef]
- Auerbach, B.J.; Kiely, J.S.; Cornicelli, J.A. A spectrophotometric microtiter-based assay for the detection of hydroperoxy derivatives of linoleic acid. Anal. Biochem. 1992, 201, 375–380. [Google Scholar] [CrossRef]
- Andrews, S. FastQC—A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 March 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omi. A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 2009, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Vitale, S.G.; Laganà, A.S.; Nigro, A.; La Rosa, V.L.; Rossetti, P.; Rapisarda, A.M.C.; La Vignera, S.; Condorelli, R.A.; Corrado, F.; Buscema, M.; et al. Peroxisome Proliferator-Activated Receptor Modulation during Metabolic Diseases and Cancers: Master and Minions. PPAR Res. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Gheorghiu, I.; Deschênes, C.; Blais, M.; Boudreau, F.; Rivard, N.; Asselin, C. Role of Specific CCAAT/Enhancer-binding Protein Isoforms in Intestinal Epithelial Cells. J. Biol. Chem. 2001, 276, 44331–44337. [Google Scholar] [CrossRef]
- Ashizuka, S.; Inagaki-Ohara, K.; Kuwasako, K.; Kato, J.; Inatsu, H.; Kitamura, K. Adrenomedullin treatment reduces intestinal inflammation and maintains epithelial barrier function in mice administered dextran sulphate sodium. Microbiol. Immunol. 2009, 53, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Halbleib, J.M.; Sääf, A.M.; Brown, P.O.; Nelson, W.J. Transcriptional Modulation of Genes Encoding Structural Characteristics of Differentiating Enterocytes During Development of a Polarized Epithelium In Vitro. HMol. Biol. Cell 2007, 18, 4261–4278. [Google Scholar] [CrossRef][Green Version]
- Eilertsen, K.J.; Kazmierski, S.T.; Keller, T.C.S. Cellular titin localization in stress fibers and interaction with myosin II filaments in vitro. J. Cell Biol. 1994, 126, 1201–1210. [Google Scholar] [CrossRef]
- Vangaveti, V.N.; Jansen, H.; Kennedy, R.L.; Malabu, U.H. Hydroxyoctadecadienoic acids: Oxidised derivatives of linoleic acid and their role in inflammation associated with metabolic syndrome and cancer. Eur. J. Pharmacol. 2016, 785, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Boivin, M.A.; Ye, D.; Kennedy, J.C.; Al-Sadi, R.; Shepela, C.; Ma, T.Y. Mechanism of glucocorticoid regulation of the intestinal tight junction barrier. Am. J. Physiol. Liver Physiol. 2007, 292, G590–G598. [Google Scholar] [CrossRef]
- Kosinski, C.; Stange, D.E.; Xu, C.; Chan, A.S.; Ho, C.; Yuen, S.T.; Mifflin, R.C.; Powell, D.W.; Clevers, H.; Leung, S.Y.; et al. Indian hedgehog regulates intestinal stem cell fate through epithelial-mesenchymal interactions during development. Gastroenterology 2010, 139, 893–903. [Google Scholar] [CrossRef]
- Rosenberg, I.M.; Göke, M.; Kanai, M.; Reinecker, H.C.; Podolsky, D.K. Epithelial cell kinase-B61: An autocrine loop modulating intestinal epithelial migration and barrier function. Am. J. Physiol. 1997, 273, G824–G832. [Google Scholar] [CrossRef]
- Simile, M.M.; Latte, G.; Pascale, R.M. MAF proteins: A family of regulating and regulated molecules. Dig. Med. Res. 2018, 1, 22. [Google Scholar] [CrossRef]
- Lund, I.; El Kertaoui, N.; Izquierdo, M.S.; Dominguez, D.; Hansen, B.W.; Kestemont, P. The importance of phospholipids combined with long-chain PUFA in formulated diets for pikeperch (Sander lucioperca) larvae. Br. J. Nutr. 2018, 120, 628–644. [Google Scholar] [CrossRef]
- Hudson, B.H.; York, J.D. Tissue-specific regulation of 3′-nucleotide hydrolysis and nucleolar architecture. Adv. Biol. Regul. 2014, 54, 208–213. [Google Scholar] [CrossRef]
- Al-Khallaf, H. Isocitrate dehydrogenases in physiology and cancer: Biochemical and molecular insight. Cell Biosci. 2017, 7, 37. [Google Scholar] [CrossRef]
- Valverde, M.; Lozano-Salgado, J.; Fortini, P.; Rodriguez-Sastre, M.A.; Rojas, E.; Dogliotti, E. Hydrogen Peroxide-Induced DNA Damage and Repair through the Differentiation of Human Adipose-Derived Mesenchymal Stem Cells. Stem Cells Int. 2018, 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Driessens, N.; Versteyhe, S.; Ghaddhab, C.; Burniat, A.; De Deken, X.; Van Sande, J.; Dumont, J.-E.; Miot, F.; Corvilain, B. Hydrogen peroxide induces DNA single- and double-strand breaks in thyroid cells and is therefore a potential mutagen for this organ. Endocr. Relat. Cancer 2009, 16, 845–856. [Google Scholar] [CrossRef]
- Wijeratne, S.S.K.; Cuppett, S.L.; Schlegel, V. Hydrogen Peroxide Induced Oxidative Stress Damage and Antioxidant Enzyme Response in Caco-2 Human Colon Cells. J. Agric. Food Chem. 2005, 53, 8768–8774. [Google Scholar] [CrossRef] [PubMed]
- Seo, E.; Kang, H.; Choi, H.; Choi, W.; Jun, H.-S. Reactive oxygen species-induced changes in glucose and lipid metabolism contribute to the accumulation of cholesterol in the liver during aging. Aging Cell 2019, 18, e12895. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, Y.; Noda, T.; Iwakiri, R.; Fujimoto, K.; Rhoads, C.A.; Aw, T.Y. Lipid peroxide-induced redox imbalance differentially mediates CaCo-2 cell proliferation and growth arrest. Cell Prolif. 2002, 35, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Epstein, F.H.; Steinberg, D.; Parthasarathy, S.; Carew, T.E.; Khoo, J.C.; Witztum, J.L. Beyond Cholesterol. N. Engl. J. Med. 1989, 320, 915–924. [Google Scholar] [CrossRef]
- Esterbauer, H.; Gebicki, J.; Puhl, H.; Jürgens, G. The role of lipid peroxidation and antioxidants in oxidative modification of LDL. Free Radic. Biol. Med. 1992, 13, 341–390. [Google Scholar] [CrossRef]
- Sottero, B.; Rossin, D.; Poli, G.; Biasi, F. Lipid Oxidation Products in the Pathogenesis of Inflammation-related Gut Diseases. Curr. Med. Chem. 2018, 25, 1311–1326. [Google Scholar] [CrossRef] [PubMed]
- Spiteller, G. Lipid peroxidation in aging and age-dependent diseases. Exp. Gerontol. 2001, 36, 1425–1457. [Google Scholar] [CrossRef]
- Barrera, G. Oxidative stress and lipid peroxidation products in cancer progression and therapy. ISRN Oncol. 2012, 2012, 137289. [Google Scholar] [CrossRef]
- Plump, A.S.; Scott, C.J.; Breslow, J.L. Human apolipoprotein A-I gene expression increases high density lipoprotein and suppresses atherosclerosis in the apolipoprotein E-deficient mouse. Proc. Natl. Acad. Sci. USA 1994, 91, 9607–9611. [Google Scholar] [CrossRef]
- Bull, A.W.; Steffensen, K.R.; Leers, J.; Rafter, J.J. Activation of PPAR γ in colon tumor cell lines by oxidized metabolites of linoleic acid, endogenous ligands for PPAR γ. Carcinogenesis 2003, 24, 1717–1722. [Google Scholar] [CrossRef]
- Gerloff, K.; Pereira, D.I.A.; Faria, N.; Boots, A.W.; Kolling, J.; Förster, I.; Albrecht, C.; Powell, J.J.; Schins, R.P.F. Influence of simulated gastrointestinal conditions on particle-induced cytotoxicity and interleukin-8 regulation in differentiated and undifferentiated Caco-2 cells. Nanotoxicology 2013, 7, 353–366. [Google Scholar] [CrossRef]
- Jakubczyk, K.; Kałduńska, J.; Dec, K.; Kawczuga, D.; Janda, K. Antioxidant properties of small-molecule non-enzymatic compounds. Pol. Merkur. Lekarski. 2020, 48, 128–132. [Google Scholar]
- Lee, S.-A.; Belyaeva, O.V.; Kedishvili, N.Y. Effect of lipid peroxidation products on the activity of human retinol dehydrogenase 12 (RDH12) and retinoid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2008, 1782, 421–425. [Google Scholar] [CrossRef]
- Natarajan, S.K.; Amirtharaj, G.J.; Ramachandran, A.; Pulimood, A.B.; Balasubramanian, K.A. Retinoid metabolism in the small intestine during development of liver cirrhosis. J. Gastroenterol. Hepatol. 2009, 24, 821–829. [Google Scholar] [CrossRef]
- D’Ambrosio, D.N.; Clugston, R.D.; Blaner, W.S. Vitamin A Metabolism: An Update. Nutrients 2011, 3, 63–103. [Google Scholar] [CrossRef]
- Thomas, S.; Prabhu, R.; Balasubramanian, K.A. Retinoid metabolism in the rat small intestine. Br. J. Nutr. 2005, 93, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Fahimi, H.D. Peroxisomes and oxidative stress. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-A. Peroxisome Metabolism in Cancer. Cells 2020, 9, 1692. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.N.; Huong, H.T.; Song, C.H.; Lee, J.H.; Han, H.J. Linoleic acid stimulates gluconeogenesis via Ca 2+/PLC, cPLA 2, and PPAR pathways through GPR40 in primary cultured chicken hepatocytes. Am. J. Physiol. Physiol. 2008, 295, C1518–C1527. [Google Scholar] [CrossRef]
- Intrasuksri, U.; Rangwala, S.M.; O’Brien, M.; Noonan, D.J.; Feller, D.R. Mechanisms of peroxisome proliferation by perfluorooctanoic acid and endogenous fatty acids. Gen. Pharmacol. 1998, 31, 187–197. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 1–31. [Google Scholar] [CrossRef]
- Veith, A.; Moorthy, B. Role of cytochrome P450s in the generation and metabolism of reactive oxygen species. Curr. Opin. Toxicol. 2018, 7, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Kaminsky, L.S.; Fasco, M.J. Small Intestinal Cytochromes P450. Crit. Rev. Toxicol. 1992, 21, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.K.; Lalvvai, N.D.; Farber, E. Carcinogenesis by Hepatic Peroxisome Proliferators: Evaluation of the Risk of Hypolipidemic Drugs and Industrial Plasticizers to Humans. CRC Crit. Rev. Toxicol. 1983, 12, 1–58. [Google Scholar] [CrossRef]
- Reddy, J.K.; Qureshi, S.A. Tumorigenicity of the hypolipidaemic peroxisome proliferator ethyl-α-p-chlorophenoxyisobutyrate (clofibrate) in rats. Br. J. Cancer 1979, 40, 476–482. [Google Scholar] [CrossRef]
- Pfaffl, M.; Lange, I.; Meyer, H.H. The gastrointestinal tract as target of steroid hormone action: Quantification of steroid receptor mRNA expression (AR, ERα, ERβ and PR) in 10 bovine gastrointestinal tract compartments by kinetic RT-PCR. J. Steroid Biochem. Mol. Biol. 2003, 84, 159–166. [Google Scholar] [CrossRef]
- Singh, M.; Kapoor, A.; Bhatnagar, A. Oxidative and reductive metabolism of lipid-peroxidation derived carbonyls. Chem. Biol. Interact. 2015, 234, 261–273. [Google Scholar] [CrossRef]
- Nakarai, C.; Osawa, K.; Akiyama, M.; Matsubara, N.; Ikeuchi, H.; Yamano, T.; Hirota, S.; Tomita, N.; Usami, M.; Kido, Y. Expression of AKR1C3 and CNN3 as markers for detection of lymph node metastases in colorectal cancer. Clin. Exp. Med. 2015, 15, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhao, L.; Zhang, H.; Liu, X.; Zhao, L.; Zhao, X.; Li, Y.; Li, J. AKR1C3 overexpression may serve as a promising biomarker for prostate cancer progression. Diagn. Pathol. 2014, 9, 42. [Google Scholar] [CrossRef]
- Matsunaga, T.; Hojo, A.; Yamane, Y.; Endo, S.; El-Kabbani, O.; Hara, A. Pathophysiological roles of aldo–keto reductases (AKR1C1 and AKR1C3) in development of cisplatin resistance in human colon cancers. Chem. Biol. Interact. 2013, 202, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Carini, F.; Mazzola, M.; Rappa, F.; Jurjus, A.; Geagea, A.G.; Al Kattar, S.; Bou-Assi, T.; Jurjus, R.; Damiani, P.; Leone, A.; et al. Colorectal Carcinogenesis: Role of Oxidative Stress and Antioxidants. Anticancer Res. 2017, 37, 4759–4766. [Google Scholar]
- Demeyer, D.; Mertens, B.; De Smet, S.; Ulens, M. Mechanisms Linking Colorectal Cancer to the Consumption of (Processed) Red Meat: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2747–2766. [Google Scholar] [CrossRef]
- Skrzydlewska, E.; Sulkowski, S.; Koda, M.; Zalewski, B.; Kanczuga-Koda, L.; Sulkowska, M. Lipid peroxidation and antioxidant status in colorectal cancer. World J. Gastroenterol. 2005, 11, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Itoh, K.; Ruiz, E.; Leake, D.S.; Unoki, H.; Yamamoto, M.; Mann, G.E. Role of Nrf2 in the Regulation of CD36 and Stress Protein Expression in Murine Macrophages. Circ. Res. 2004, 94, 609–616. [Google Scholar] [CrossRef]
- Akazawa-Ogawa, Y.; Shichiri, M.; Nishio, K.; Yoshida, Y.; Niki, E.; Hagihara, Y. Singlet-oxygen-derived products from linoleate activate Nrf2 signaling in skin cells. Free Radic. Biol. Med. 2015, 79, 164–175. [Google Scholar] [CrossRef]
- Koehne, C.-H.; Dubois, R.N. COX-2 inhibition and colorectal cancer. Semin. Oncol. 2004, 31, 12–21. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Yonemura, T.; Sakuma, S. Role of Linoleic Acid Hydroperoxide Preformed by Cyclooxygenase-1 or -2 on the Regulation of Prostaglandin Formation from Arachidonic Acid by the Respective Enzyme. J. Clin. Biochem. Nutr. 2008, 43, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Ringbom, T.; Huss, U.; Stenholm, Å.; Flock, S.; Skattebøl, L.; Perera, P.; Bohlin, L. COX-2 inhibitory effects of naturally occurring and modified fatty acids. J. Nat. Prod. 2001, 64, 745–749. [Google Scholar] [CrossRef]
- Ozdemirler, G.; Pabuççuoglu, H.; Bulut, T.; Bugra, D.; Uysal, M.; Toker, G. Increased lipoperoxide levels and antioxidant system in colorectal cancer. J. Cancer Res. Clin. Oncol. 1998, 124, 555–559. [Google Scholar] [CrossRef]
- Perumalla Venkata, R.; Subramanyam, R. Evaluation of the deleterious health effects of consumption of repeatedly heated vegetable oil. Toxicol. Rep. 2016, 3, 636–643. [Google Scholar] [CrossRef]
- Delerive, P.; Furman, C.; Teissier, E.; Fruchart, J.-C.; Duriez, P.; Staels, B. Oxidized phospholipids activate PPARα in a phospholipase A2-dependent manner. FEBS Lett. 2000, 471, 34–38. [Google Scholar] [CrossRef]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef]
- Necela, B.M.; Thompson, E.A. Pathophysiological Roles of PPAR γ in Gastrointestinal Epithelial Cells. PPAR Res. 2008, 2008, 1–8. [Google Scholar] [CrossRef]
- Escudero, P.; Martinez de Marañón, A.; Collado, A.; Gonzalez-Navarro, H.; Hermenegildo, C.; Peiró, C.; Piqueras, L.; Sanz, M.-J. Combined Sub-Optimal Doses of Rosuvastatin and Bexarotene Impair Angiotensin II-Induced Arterial Mononuclear Cell Adhesion Through Inhibition of Nox5 Signaling Pathways and Increased RXR/PPARα and RXR/PPARγ Interactions. Antioxid. Redox Signal. 2015, 22, 901–920. [Google Scholar] [CrossRef] [PubMed]
- Jiao, M.; Ren, F.; Zhou, L.; Zhang, X.; Zhang, L.; Wen, T.; Wei, L.; Wang, X.; Shi, H.; Bai, L.; et al. Peroxisome proliferator-activated receptor α activation attenuates the inflammatory response to protect the liver from acute failure by promoting the autophagy pathway. Cell Death Dis. 2014, 5, e1397. [Google Scholar] [CrossRef] [PubMed]
- Stienstra, R.; Mandard, S.; Tan, N.S.; Wahli, W.; Trautwein, C.; Richardson, T.A.; Lichtenauer-Kaligis, E.; Kersten, S.; Müller, M. The Interleukin-1 receptor antagonist is a direct target gene of PPARα in liver. J. Hepatol. 2007, 46, 869–877. [Google Scholar] [CrossRef]
- Lee, J.W.; Bajwa, P.J.; Carson, M.J.; Jeske, D.R.; Cong, Y.; Elson, C.O.; Lytle, C.; Straus, D.S. Fenofibrate Represses Interleukin-17 and Interferon-γ Expression and Improves Colitis in Interleukin-10–Deficient Mice. Gastroenterology 2007, 133, 108–123. [Google Scholar] [CrossRef]
- Beyaz, S.; Yilmaz, Ö.H. Molecular Pathways: Dietary Regulation of Stemness and Tumor Initiation by the PPAR-δ Pathway. Clin. Cancer Res. 2016, 22, 5636–5641. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Regulation of Src Family Kinases during Colorectal Cancer Development and Its Clinical Implications. Cancers 2020, 12, 1339. [Google Scholar] [CrossRef]
- Koizume, S.; Miyagi, Y. Lipid Droplets: A Key Cellular Organelle Associated with Cancer Cell Survival under Normoxia and Hypoxia. Int. J. Mol. Sci. 2016, 17, 1430. [Google Scholar] [CrossRef]
- Tachibana, K.; Yamasaki, D.; Ishimoto, K.; Doi, T. The Role of PPARs in Cancer. PPAR Res. 2008, 2008, 1–15. [Google Scholar] [CrossRef]
- Grabacka, M.; Reiss, K. Anticancer Properties of PPAR α -Effects on Cellular Metabolism and Inflammation. PPAR Res. 2008, 2008, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cerbone, A.; Toaldo, C.; Laurora, S.; Briatore, F.; Pizzimenti, S.; Dianzani, M.U.; Ferretti, C.; Barrera, G. 4-Hydroxynonenal and PPARgamma ligands affect proliferation, differentiation, and apoptosis in colon cancer cells. Free Radic. Biol. Med. 2007, 42, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Zois, C.E.; Harris, A.L. Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy. J. Mol. Med. 2016, 94, 137–154. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Oberreuther-Moschner, D.L.; Rechkemmer, G.; Pool-Zobel, B.L. Basal colon crypt cells are more sensitive than surface cells toward hydrogen peroxide, a factor of oxidative stress. Toxicol. Lett. 2005, 159, 212–218. [Google Scholar] [CrossRef]
- Usatyuk, P.V.; Natarajan, V. Hydroxyalkenals and oxidized phospholipids modulation of endothelial cytoskeleton, focal adhesion and adherens junction proteins in regulating endothelial barrier function. Microvasc. Res. 2012, 83, 45–55. [Google Scholar] [CrossRef]
- Rao, R. Oxidative stress-induced disruption of epithelial and endothelial tight junctions. Front. Biosci. 2008, 13, 7210–7226. [Google Scholar] [CrossRef]
- Ahmad, R.; Rah, B.; Bastola, D.; Dhawan, P.; Singh, A.B. Obesity-Induces Organ and Tissue Specific Tight Junction Restructuring and Barrier Deregulation by Claudin Switching. Sci. Rep. 2017, 7, 5125. [Google Scholar] [CrossRef] [PubMed]
- Levy, P.; Robin, H.; Kornprobst, M.; Capeau, J.; Cherqui, G. Enterocytic differentiation of the human Caco-2 cell line correlates with alterations in integrin signaling. J. Cell. Physiol. 1998, 177, 618–627. [Google Scholar] [CrossRef]
- Laprise, P.; Chailler, P.; Houde, M.; Beaulieu, J.-F.; Boucher, M.-J.; Rivard, N. Phosphatidylinositol 3-Kinase Controls Human Intestinal Epithelial Cell Differentiation by Promoting Adherens Junction Assembly and p38 MAPK Activation. J. Biol. Chem. 2002, 277, 8226–8234. [Google Scholar] [CrossRef]
- Barron, L.; Sun, R.C.; Aladegbami, B.; Erwin, C.R.; Warner, B.W.; Guo, J. Intestinal Epithelial-Specific mTORC1 Activation Enhances Intestinal Adaptation After Small Bowel Resection. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 231–244. [Google Scholar] [CrossRef]
- Mariadason, J.M.; Arango, D.; Corner, G.A.; Arañes, M.J.; Hotchkiss, K.A.; Yang, W.; Augenlicht, L.H. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002, 62, 4791–4804. [Google Scholar] [PubMed]
- LLOR, X. The effects of fish oil, olive oil, oleic acid and linoleic acid on colorectal neoplastic processes. Clin. Nutr. 2003, 22, 71–79. [Google Scholar] [CrossRef]
- Shcherbik, N.; Pestov, D.G. The Impact of Oxidative Stress on Ribosomes: From Injury to Regulation. Cells 2019, 8, 1379. [Google Scholar] [CrossRef]
- Schuster, S.; Johnson, C.D.; Hennebelle, M.; Holtmann, T.; Taha, A.Y.; Kirpich, I.A.; Eguchi, A.; Ramsden, C.E.; Papouchado, B.G.; McClain, C.J.; et al. Oxidized linoleic acid metabolites induce liver mitochondrial dysfunction, apoptosis, and NLRP3 activation in mice. J. Lipid Res. 2018, 59, 1597–1609. [Google Scholar] [CrossRef]
- Sullivan, E.M.; Pennington, E.R.; Green, W.D.; Beck, M.A.; Brown, D.A.; Shaikh, S.R. Mechanisms by Which Dietary Fatty Acids Regulate Mitochondrial Structure-Function in Health and Disease. Adv. Nutr. 2018, 9, 247–262. [Google Scholar] [CrossRef]
- Begriche, K.; Igoudjil, A.; Pessayre, D.; Fromenty, B. Mitochondrial dysfunction in NASH: Causes, consequences and possible means to prevent it. Mitochondrion 2006, 6, 1–28. [Google Scholar] [CrossRef]
- Lucas, D.T.; Szweda, L.I. Cardiac reperfusion injury: Aging, lipid peroxidation, and mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 1998, 95, 510–514. [Google Scholar] [CrossRef]
- Novak, E.A.; Mollen, K.P. Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell Dev. Biol. 2015, 3, 62. [Google Scholar] [CrossRef]
- Günther, J.; Seyfert, H.-M. The first line of defence: Insights into mechanisms and relevance of phagocytosis in epithelial cells. Semin. Immunopathol. 2018, 40, 555–565. [Google Scholar] [CrossRef]
- Mapother, M.E.; Songer, J.G. In vitro interaction of Mycobacterium avium with intestinal epithelial cells. Infect. Immun. 1984, 45, 67–73. [Google Scholar] [CrossRef]
- Kaemmerer, E.; Schutt, F.; Krohne, T.U.; Holz, F.G.; Kopitz, J. Effects of Lipid Peroxidation-Related Protein Modifications on RPE Lysosomal Functions and POS Phagocytosis. Investig. Opthalmol. Vis. Sci. 2007, 48, 1342. [Google Scholar] [CrossRef]
- Wu, Y.; Yao, J.; Xie, J.; Liu, Z.; Zhou, Y.; Pan, H.; Han, W. The role of autophagy in colitis-associated colorectal cancer. Signal Transduct. Target. Ther. 2018, 3, 31. [Google Scholar] [CrossRef]
- Delerive, P.; De Bosscher, K.; Besnard, S.; Vanden Berghe, W.; Peters, J.M.; Gonzalez, F.J.; Fruchart, J.-C.; Tedgui, A.; Haegeman, G.; Staels, B. Peroxisome Proliferator-activated Receptor α Negatively Regulates the Vascular Inflammatory Gene Response by Negative Cross-talk with Transcription Factors NF-κB and AP-1. J. Biol. Chem. 1999, 274, 32048–32054. [Google Scholar] [CrossRef] [PubMed]
- Bochkov, V.N.; Leitinger, N. Anti-inflammatory properties of lipid oxidation products. J. Mol. Med. 2003, 81, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Keshamouni, V.G.; Arenberg, D.A.; Reddy, R.C.; Newstead, M.J.; Anthwal, S.; Standiford, T.J. PPAR-γ activation inhibits angiogenesis by blocking ELR+CXC chemokine production in non-small cell lung cancer. Neoplasia 2005, 7, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Radzikowska, U.; Rinaldi, A.O.; Çelebi Sözener, Z.; Karaguzel, D.; Wojcik, M.; Cypryk, K.; Akdis, M.; Akdis, C.A.; Sokolowska, M. The Influence of Dietary Fatty Acids on Immune Responses. Nutrients 2019, 11, 2990. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, M.; Liu, M.; Ji, Y.; Li, Z. TNF-alpha and IL-6 inhibit apolipoprotein A-IV production induced by linoleic acid in human intestinal Caco2 cells. J. Inflamm. 2015, 12, 22. [Google Scholar] [CrossRef]
- Virtanen, J.K.; Mursu, J.; Voutilainen, S.; Tuomainen, T.-P. The associations of serum n-6 polyunsaturated fatty acids with serum C-reactive protein in men: The Kuopio Ischaemic Heart Disease Risk Factor Study. Eur. J. Clin. Nutr. 2018, 72, 342–348. [Google Scholar] [CrossRef]
- Halliwell, B.; Clement, M.V.; Long, L.H. Hydrogen peroxide in the human body. FEBS Lett. 2000, 486, 10–13. [Google Scholar] [CrossRef]
- Katsube, T.; Tsuji, H.; Onoda, M. Nitric oxide attenuates hydrogen peroxide-induced barrier disruption and protein tyrosine phosphorylation in monolayers of intestinal epithelial cell. Biochim. Biophys. Acta 2007, 1773, 794–803. [Google Scholar] [CrossRef]
- Zhao, M.; Tang, S.; Xin, J.; Wei, Y.; Liu, D. Reactive oxygen species induce injury of the intestinal epithelium during hyperoxia. Int. J. Mol. Med. 2018, 41, 322–330. [Google Scholar] [CrossRef]
- Giudetti, A.M.; Damiano, F.; Gnoni, G.V.; Siculella, L. Low level of hydrogen peroxide induces lipid synthesis in BRL-3A cells through a CAP-independent SREBP-1a activation. Int. J. Biochem. Cell Biol. 2013, 45, 1419–1426. [Google Scholar] [CrossRef]
- Huang, B.; Song, B.; Xu, C. Cholesterol metabolism in cancer: Mechanisms and therapeutic opportunities. Nat. Metab. 2020, 2, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Leonarduzzi, G.; Sottero, B.; Poli, G. Oxidized products of cholesterol: Dietary and metabolic origin, and proatherosclerotic effects (review). J. Nutr. Biochem. 2002, 13, 700–710. [Google Scholar] [CrossRef]
- Liu, Y.; Li, W.; Duan, Y. Effect of H2O2 induced oxidative stress (OS) on volatile organic compounds (VOCs) and intracellular metabolism in MCF-7 breast cancer cells. J. Breath Res. 2019, 13, 036005. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).