Abstract
Using natural language processing tools, we investigate the semantic differences in medical guidelines for three decision problems: breast cancer screening, lower back pain and hypertension management. The recommendation differences may cause undue variability in patient treatments and outcomes. Therefore, having a better understanding of their causes can contribute to a discussion on possible remedies. We show that these differences in recommendations are highly correlated with the knowledge brought to the problem by different medical societies, as reflected in the conceptual vocabularies used by the different groups of authors. While this article is a case study using three sets of guidelines, the proposed methodology is broadly applicable. Technically, our method combines word embeddings and a novel graph-based similarity model for comparing collections of documents. For our main case study, we use the CDC summaries of the recommendations (very short documents) and full (long) texts of guidelines represented as bags of concepts. For the other case studies, we compare the full text of guidelines with their abstracts and tables, summarizing the differences between recommendations. The proposed approach is evaluated using different language models and different distance measures. In all the experiments, the results are highly statistically significant. We discuss the significance of the results, their possible extensions, and connections to other domains of knowledge. We conclude that automated methods, although not perfect, can be applicable to conceptual comparisons of different medical guidelines and can enable their analysis at scale.
1. The Problem and the Method
This article investigates a natural question. We are asking whether differences in medical recommendations arise from differences in knowledge brought to the problem by different medical societies. To answer this question at scale we need an automated method to measure such differences. The purpose of this article is to present such a computational method and use a collection of case studies to evaluate its performance.
Our method uses the standard natural language processing approach to represent words and documents as embeddings, and combines it with a graph comparison algorithm. We evaluate our approach on three sets of medical guidelines: for breast cancer screening, lower back pain management guidelines and hypertension management guidelines.
The answer to this question matters because physicians with different specialties follow different guidelines. This results in the undue variability of treatment. Therefore, understanding what drives the differences in recommendation should contribute to its reduction, and to better patient outcomes [1,2,3].
1.1. Motivation
There are over twenty thousand clinical practice guidelines indexed by PubMed (https://pubmed.ncbi.nlm.nih.gov/ (accessed on 24 February 2020)), with over 1500 appearing every year [4]. Since clinical practice guidelines are developed by different medical associations, which count on experts with different specialties and sub-specialties, there is a high possibility that there may be disagreement in the guidelines. Indeed, as noted by [3], and discussed in [5,6], breast cancer screening guidelines contradict each other. Besides breast cancer screening disagreements, which we model in this article, controversies over PSA screening, hypertension and other treatment and prevention guidelines are also well-known.
Figure 1 illustrates our point. We see disagreements in seven breast cancer screening recommendations produced by seven different medical organizations. The hypothesis we investigate is that the contradictory recommendations reflect the specialized knowledge brought to bear on the problem by different societies.
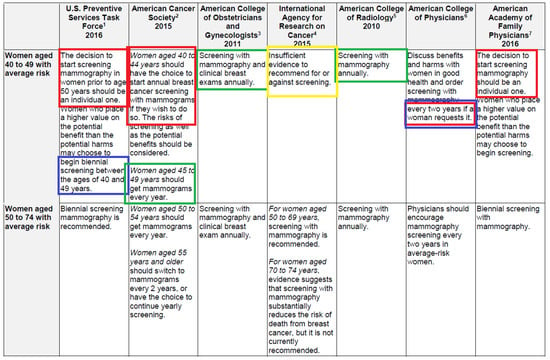
Figure 1.
Note the contradictory recommendations in green and blue boxes. The colors in the table come from [6], but the original table comes from the CDC [3]. Only a part of the table is reproduced here.
Notice that the dominant view is to see expertise as a shared a body of information, and experts as epistemic peers [7] with identical levels of competence. Under this paradigm of shared knowledge and inferential abilities, the medical bodies should not differ in their recommendations. That they do is interesting and worth investigating. Thus, this article is also motivated by the idea that epistemology of disagreement [7,8,9] can be modeled computationally. On the abstract level, we view medical disagreement as “near-peer” disagreement [10,11,12], where we see expert groups as having partly overlapping knowledge. This article shows that such more realistic and fine-grained models can also be studied computationally, quantitatively, and at scale.
1.2. Brief Description of the Proposed Method
In this article we investigate the question of whether differences in medical recommendations come from differences in specialized medical knowledge applied to specific classes of patients, and whether such differences in specialties can be modeled computationally.
Our idea is to model “specialized medical knowledge”, which we cannot easily observe, by the differences in vocabulary used in medical guidelines. We then show that these vocabularies, assembled in vector representations of these documents, produce the differences in recommendations. We evaluate our method using three case studies: breast cancer screening guidelines, lower back pain management guidelines and hypertension management guidelines. In the main track of this article, we use the breast cancer screening guidelines to present our approach and the evaluation, and the additional evaluations on the other two sets of guidelines are presented in the Appendix A.
More specifically, we computationally compare the full texts of guidelines with the their recommendation summaries. For breast cancer screening, the summaries come from the CDC [3]; for lower back pain management, they come from a summary article [13]; and, for hypertension management, where we lack a tabular comparison, we used the abstracts of the documents. That is, we see if the semantic similarities between the full documents follow the same pattern as semantic similarities between the summaries. Note that each computational comparison was made between two sets of documents and not individual documents.
This process involves several steps and is shown in Figure 2, for the breast cancer screening guidelines. Thus, the vector representations of full texts of the guidelines model the vocabularies as bags of concepts, and therefore cannot model specific recommendations: the concepts in the recommendations, such as “mammography” and “recommend”, appear in all full texts, but specific societies may be either for mammography or against it. The vector representations of recommendations model the differences in prescribed procedures, but not the vocabularies (see Table 1 and Table 2 below).
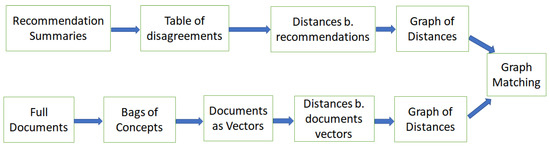
Figure 2.
The method of comparing concepts in full documents and recommendations contained in summaries. Note the difference in representations: the document are represented by a large number of high-dimensional (200) vectors with real valued features, whereas the disagreement representations can be low-dimensional vectors with discrete features (e.g., five-dimensional for the breast cancer screening guidelines). Our exposition will roughly follow the left-to-right order of this figure, using the breast cancer screening guidelines as the motivating example.

Table 1.
The table shows recommendations as follows: N—no recommendation; b—both patient and doctor, shared decision; r—recommending mammography.

Table 2.
This table shows the number of differing feature values for pair of guidelines, based on Table 1. The Jaccard distances between the documents are obtained by dividing the value in the table by five (the number of features).
How do we know if vocabularies determine recommendations? We compute pairwise distances (cosine or word mover’s distance) between the full text vectors. In parallel, we compute pairwise distances between the recommendation vectors. We thus get two graphs, and their shapes can be compared. We show that the resulting geometries are very similar and could not have been produced by chance.
This process is slightly modified for lower back pain management, where we start with the tables of disagreement from the summary article [13]. For the hypertension management guidelines, we use the graph of summaries that is generated from the abstracts of full documents, because we do not have any tabular sets of comparisons similar to [3,13]. However, even with this change the proposed method performs very well. Notice that to model full documents we use a large number of high dimensional (200), real valued, vectors. By contrast, the vectors representing the recommendations only have a smaller number of discrete-valued features (five for the breast cancer screening, and 12, 59 and 71 for lower back pain management).
1.3. Summary of Contributions
The main contribution of this article is in proposing an automated, and relatively straightforward, method of text analysis that (1) computes conceptual differences between documents addressing the same topic (for example, breast cancer screening), and (2) these automated judgments have a high correlation with recommendations extracted from these documents by a panel of experts. We test the approach on the already mentioned breast cancer screening recommendations, as well as in other sets of experiments on lower back pain management and hypertension management guidelines. As such, these results open the possibility of large-scale analysis of medical guidelines using automated tools.
Another contribution is the articulation of a very natural graph clique-based algorithm/method for comparing the similarity of two collections of documents. Given two sets of documents, each of the same cardinality, and a mapping between nodes, we compute the percent of similarity (or, equivalently, distortion between the shapes of the two cliques), and the chances that the mapping arose from a random process.
We also document all steps of the process and provide the data and the code to facilitate both extensions of this work and its replication (the GitHub link is provided in Section 8).
1.4. Organization of the Article
In Section 2, we provide a brief overview of applications of natural language processing to texts of medical guidelines, word embedding, and some relevant work on disagreement. Afterwards, we follow the left-to-right order of Figure 2 using the breast cancer screening guidelines as the motivating example (other experiments are described in the Appendix A). Thus, Section 3 and Section 4 explain our example data sources: a CDC summary table of breast cancer screening guidelines and the corresponding full text documents. In these two sections, we also discuss the steps in the conceptual analysis of the table. First, the creation of a graph of conceptual distances between the columns of the table, and then the encoding of full documents as vectors, using two standard vectorization procedures. Our method of comparing summarized recommendations and full guideline documents is presented in three algorithms and discussed in Section 5.
After observing a roughly 70% similarity between the distances in the summaries and the distances in the full documents, we prove in Section 6 that this similarity is not accidental. We conclude in Section 6 and Section 8 that this case study shows that NLP methods are capable of approximate conceptual analysis in this space (using the Appendix A for additional support). This opens the possibility of deepening this exploration using more sophisticated tools such as relationship extraction, other graph models, and automated formal analysis (as discussed in Section 7 and Section 8).
In the Appendix A, we provide information about additional experiments we performed to validate the proposed method. (We decided to put this information in an appendix in order to simplify the main thread of the presentation). There, we first discuss a few variants of the main experiment, where we filtered out some sentences from the full guidelines’ texts. Then, we apply our method to two other collections of guidelines: namely, to hypertension and low back pain management guidelines. All of these experiments confirm the robustness of the proposed method and the system’s ability to computationally relate background knowledge to actual recommendations.
2. Discussion of Prior Art
We are not aware of any work directly addressing the issue we are tackling in this article; namely, the automated conceptual analysis of medical screening recommendations. However, there is a body of knowledge addressing similar issues individually, which we summarize in this section.
2.1. Text Analysis of Medical Guidelines
An overview article [14], from a few years ago, states that different types of analysis of medical guidelines are both a central theme in applications of artificial intelligence to medicine and a domain of research with many challenges. The latter include building formal, computational representations of guidelines and a wider application of natural language processing. From this perspective, our work is relevant to these central and general themes.
A more recent and more technical work [15] focuses on finding and resolving conflicting recommendations using a formal model and automated proof systems—it relies on a manual translation into a formal language, Labelled Event Structure. This is a very interesting work, somewhat in the spirit of our own attempts, using a combination of NLP and information retrieval tools [6]. Another article [16], dealing with contradictory recommendations, focuses on the semi-automatic detection of inconsistencies in guidelines; these tools are applied to antibiotherapy in primary care. Another recent application of natural language processing [17,18] shows that one can accurately measure adherence to best practice guidelines in a context of palliative care, as well as try to assess the quality of care from discharge summaries.
More broadly, modern NLP methods have been applied to clinical decision support, e.g., [19], with ontologies and semantic webs for concept representation; to clinical trials [20]; and to automatic extraction of adverse drug events and drug related entities, e.g., using a neural networks model [21]. For document processing, we have, e.g., a knowledge-based technique for inter-document similarity computation [22], and a successful application of conceptual representations to document retrieval [23].
All of these show that the state-of-the-art systems are capable of both performing statistical analysis of sets of documents and a semantic analysis fitting the need of a particular application. Our work extends both of these in a new direction, and connects statistics with semantics, for the purpose of analysis of medical guidelines.
2.2. Vector Representations of Documents Using Word Embeddings
Over the last 10 years, we have witnessed a new era in automated semantic analysis of textual documents [24]. While no system can claim to “really” understand natural language, in several domains, such as data extraction, classification and question answering, automated systems dramatically improved their performance, and in some cases performed better than humans, due to the unmatched pattern recognition and memorization capabilities of deep neural networks (see, e.g., [25] for an overview).
Some of the simplest, easiest to use and effective of these new methods are different types of word and concept embeddings [26,27,28,29]. Embeddings represent words and concepts as dense vectors (i.e., a few hundred dimensional real-valued vectors), and are a preferred tool to make similarity judgments on the level of words, phrases, sentences and whole documents. They have been applied to medical texts—see [30] for a survey.
Word embeddings have been widely used to compare documents, and in particular to compute their degree of similarity [31,32]. Other methods proposed to compute document similarity are based on using background knowledge [22].
This work uses both methods, namely human knowledge encoded in the CDC table (Figure 1), and embeddings. For the former, we use five-dimensional feature vectors representing differences in recommendations (Section 3). For the latter, we use (several versions of) 200-dimensional embeddings of full documents (Section 4).
2.3. Other Work on Disagreements and Contradictions
Disagreements among medical experts are clearly very relevant to work. A comprehensive review of medical disagreement with a focus on intervention risks and the standards of care can be found in [33]. Once medical experts express their disagreements, what happens next? Observations from disagreement adjudication are analyzed in [34,35], where the authors observe (among other things) that the differences in experts’ backgrounds increase the degree of disagreement.
If we broaden the context beyond medical disagreements, to artificial intelligence, there is a substantial amount of work on contradictory knowledge bases, as exemplified by [36,37,38]. Of particular interest may be proposals for real valued measures of contradictions in knowledge bases [38,39]. However, in that particular research avenue the starting points are collections of facts, and not recommendations; moreover, natural language texts are not mentioned. We believe this type of work will become more relevant as our capabilities to extract knowledge from text improve.
3. From Recommendations to Vectors of Differences and a Graph
We start with the simpler task of transforming the screening recommendations (referenced above in Figure 1) to vectors of differences, representing the disagreements in the recommendations, and then to a graph of their conceptual distances, where, intuitively, the larger the number of recommendation differences, the bigger the distance.
We will proceed in three steps: First, using a diagram (Figure 3) and a table (Table 1) we make explicit the difference in recommendations in Figure 1. Second, we transform the table into a count of differences (Table 2) and from that we derive distances between pairs of recommendations (Table 3). The graph representing the recommendations will have nodes named after each organization (e.g., AAFP, ACOG, etc.) and edges labeled and drawn with distances (Figure 4).
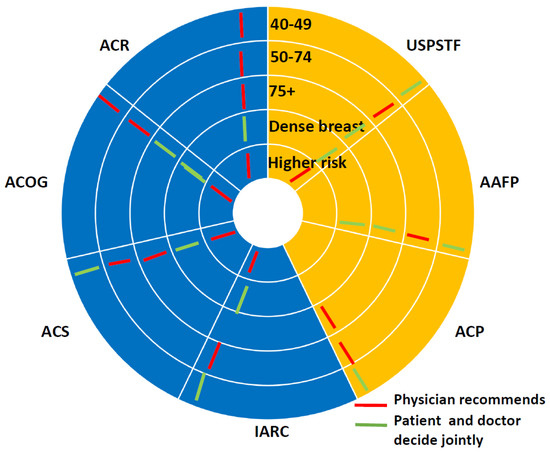
Figure 3.
Similarities and disagreements in summarized recommendations. The yellow coloring shows patient making decisions, the blue coloring shows explicit screening recommendations. The concentric circles show different age groups. Red marks—physician recommends, green marks—patient decides.

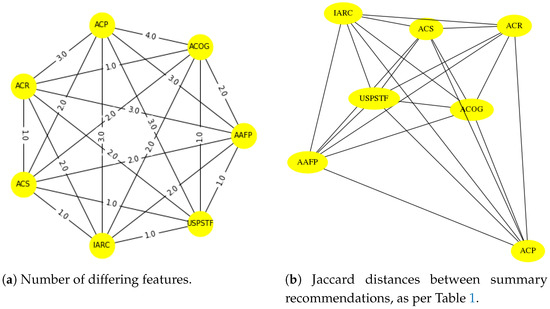
Figure 4.
In panel (a) we see a pictorial representation of the numbers of differing features, per Table 2 and Table 3. These differences between recommendations are converted into distances (using the Jaccard measure), resulting in panel (b). Can we replicate the geometric structure of panel (b) using automated tools? See Section 6 for an answer.
3.1. Computing the Differences in Recommendations
Figure 3 is another representation of the information in the CDC comparison of the recommendations [3], earlier presented in Figure 1. It clearly shows the differences between the guidelines (and it comes from [40]). As we can see, there are two sides to the circle. The yellow side indicates the scenario where patients will likely decide when breast cancer screening should be done, and the purple color side specifies the situation where breast cancer guideline providers most likely will demand screening interventions. White radial lines indicate boundaries between the different societies. The red color marks indicate that the physician decides. Green color marks indicate patients’ decisions.
3.2. From Differences to Distances and a Graph
Table 1 represent the content of this analysis as a collection of features. Table 2 encodes these differences in recommendations as numbers of differing features between pairs of recommendations. Then, Table 3 shows the distances between the guidelines derived from Table 1 and Table 2 using the Jaccard distance (the percentage of different elements in two sets):
Given two recommendation summaries A and B we compute the number of the differing feature values from Table 2 and divide it by five. For example, for the pair (AFP, ACR) we get . All these distances were normalized to sum to 1 and shown in Table 3 (we are not assuming that distances are always symmetric. In most cases they are, but later we will also report experiments with the search distances, which are not symmetric). The normalization does not change the relative distances, and in the comparisons with the geometry of full documents we only care about the relative distances.
Table 1, Table 2 and Table 3 represent the process of converting the information in Figure 3 into a set of distances. These distances are depicted graphically in Figure 4, where we display both Jaccard distances between the recommendations and the number of differing features as per Table 2.
In the following section we will create a graph representation for the full documents (Figure 5b). We will present our graph comparison method in Section 5. In Section 6, we will assign numerical values to the distance between the two graphs, and show that this similarity cannot be the result of chance.
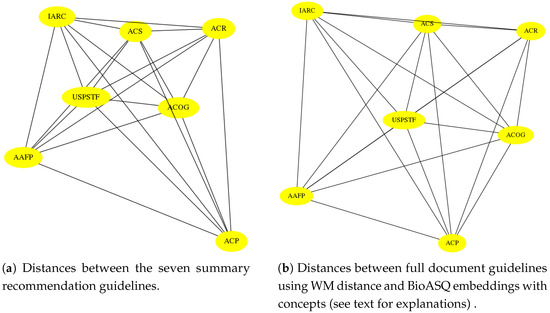
Figure 5.
Visual comparison of a the similarity/distance graphs based on human analysis is shown in panel (a), and computer generated comparison from Table 5 is shown in panel (b), which suggest a similar geometry. As we rigorously show in Section 6, this 69% similarity is not accidental; the distortion is about 31%. Notice that we are not pointing to the actual locations of similarities and difference in the guideline documents. Instead, we are pointing to global (latent) differences stemming from concepts appearing in them.
4. Transforming Full Guidelines Documents into Vectors and Graphs
In this article, we use both the CDC summaries ([3], reproduced and labeled in Figure 1 and Figure 3), and the full text of the guidelines used by the CDC to create the summaries. The focus of this section is on the full guideline documents. The detailed information about these guidelines is shown in Table 4.

Table 4.
Guidelines with references. All the sources were last retrieved in summer 2020.
Note that in this section we are using the same acronyms (of medical societies) to refer to full guideline documents. This will not lead to confusion, as in this section we are only discussing full documents.
4.1. Data Preparation for All Experiments
From the breast cancer screening guidelines listed in the CDC summary document [3], the texts of the USPSTF, ACS, ACP and ACR guidelines were extracted from their HTML format. We used Adobe Acrobat Reader to obtain the texts from the pdf format of the AAFP, ACOG, and IARC guidelines. Since the AAFP documents also included preventive service recommendations for other diseases (such as other types of cancers), we added a preprocess step to remove those recommendations, leaving the parts matching “breast cancer”.
4.2. Measuring Distances between Full Documents
When creating embedding representation of text, we replace each word or term with its embedding representation. Thus, the text full guideline documents are represented as a set of vectors. Our objective is to create a graph of conceptual distances between the documents.
The two most commonly used measures of distance, cosine distance and word mover’s distance, operate on different representations. The former operates on pairs of vectors, and the latter on sets of vectors. Thus, we need to create two types of representations.
Given a document, the first representation takes the average of all its word (term) embeddings. This creates a vector representing the guideline text. The second representation simply keeps the set of all its embedding vectors.
The cosine distance between two vectors v and w is defined as:
We will also use the following variant of cosine distance to argue that the geometries we obtain in our experiments are similar irrespective of distances measures (see Section 6):
The word mover’s distance (WMD, WM distance), introduced in [48], is a variant of the classic concept of “earth mover distance” from the transportation theory [49]. Sometimes, the term “Wasserstein distance” is also used. The intuition encoded in this metric is as follows. Given two documents represented by their set of vectors, each vector is viewed as a divisible object. We are allowed to “move” fractions of each vector in the first set to the other set. The WM distance is the minimal total distance accomplishing the transfer of all vector masses to the other set. More formally [48], WM distance minimizes:
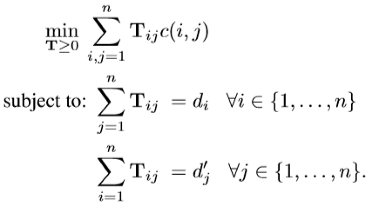 where is the fraction of word i in document d traveling to word j in document ; denotes the cost “traveling” from word i in document d to word j in document ; here the cost is the Euclidean distance between two words in the embedding space. Finally, is the normalized frequency of word i in document d (and same for ):
where is the fraction of word i in document d traveling to word j in document ; denotes the cost “traveling” from word i in document d to word j in document ; here the cost is the Euclidean distance between two words in the embedding space. Finally, is the normalized frequency of word i in document d (and same for ):
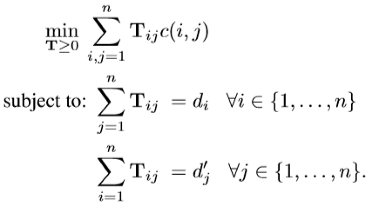
We used the n_similarity and wmdistance functions from Gensim [50] as a tool for generating vectors and calculating similarities/distances in our experiments.
4.3. Building Vector Representations of Full Documents
However, as there are multiple distance measures, there is more than one way to create word embeddings; we experimented with several methods. We used three language models of medical guidelines’ disagreement: “no concept”, conceptualized and BioASQ. (The details of these experiments appear later in Table 6). The first two were Word2Vec embedding models trained using the PubMed articles as the training data. The third one used pre-trained BioASQ word embeddings created for the BioASQ competitions [51] (http://BioASQ.org/news/BioASQ-releases-continuous-space-word-vectors-obtained-applying-word2vec-pubmed-abstracts (accessed on 24 February 2021)).
Our first model, trained on PubMed, included only words, and no additional conceptual analysis with MeSH (https://www.nlm.nih.gov/mesh/meshhome.html (accessed on 24 February 2021)) was done. In the second, which was a more complex model, MeSH terms were replaced with n-grams. For example, if breast and cancer appeared next to each other in the text, they were replaced with breast-neoplasms and treated as a concept.
4.4. Our Best Model: Using BioASQ Embeddings and Word Mover’s Distance
Table 5 shows (unnormalized) WM distances between the seven guidelines using BioASQ embeddings. Figure 5 shows side by side the geometries of the two graphs: one generated from the summary of full documents, using features derived from the CDC summaries, and the second one based on the machine-generated representations of the full guideline documents. To create Figure 5, for each metric, a diagram representing the distance between the nodes (guidelines) and a diagram with the labeled edges were drawn using Python the networkx library (https://networkx.github.io/ (accessed on 24 February 2021)). All values were normalized to the same scale to allow visual comparison.

Table 5.
This table shows the word mover’s distances between the guidelines using BioASQ embeddings. This model also performed very well on the datasets in the Appendix A.
The similarity is visible in a visual inspection, and will be quantified in Section 6 to be about 70%. However, before we provide the details of the experiments, we will also answer two questions:
- —
- How do we measure the distortion/similarity between the two graphs?
- —
- Could this similarity of shapes be accidental? How do we measure such probability?
5. Graph-Based Method for Comparing Collections of Documents
At this point we have created two graphs, one showing the distances between summary recommendations, and the other representing conceptual distances between documents. The procedure we used so far can be concisely expressed as Algorithm 1, where given a set of documents, after specifying the Model (type of embeddings) and a distance metric, we get an adjacency matrix containing the distances between the nodes representing the documents. An example output of Algorithm 1 is shown in Figure 4 above.
What remains to be done is to quantify the difference in shapes of these two graphs, and then to show that the similarity we observe is not accidental. The methods used in these two steps are described in Algorithms 2 and 3. The experiments and the details of the performed computations will be presented in Section 6.
| Algorithm 1 Computing Graph of Distances Between Documents. |
| Input:Guidelines: a set of guideline documents in textual format. |
| Model: a model to compute distances between two documents. |
Output:—Adjacency matrix of distances between document guidelines.
|
We use a very natural, graph clique-based method for comparing similarity of two collections of documents. Given two sets of documents, represented by graphs, and a one-to-one mapping between nodes, in Algorithm 2, we compute the percent distortion between the shapes of the two cliques—this is perhaps the most natural similarity measure (similarity = 1 − distortion) for comparing the shapes of two cliques of identical cardinality.
| Algorithm 2 Distance or Percentage Distortion between Two Complete Graphs (cliques of the same size). |
| Input: Adjacency Matrices , of equal dimensions |
Output: Graph distance/distortion , as a value between 0 and 1.
|
Next, we need to compute the chance that the mapping arose from a random process. This is because if the chances of the similarity arising from a random process are small, we can conclude that the conceptual vocabulary of a full document determines the type of recommendation given by a particular organization. In our case the nodes of both graphs have the same names (the names of the medical societies), but the shapes of the graphs are different, one coming from human summaries and comparison (Figure 1, Table 1) and the other from machine produced conceptual distances. Thus, the randomization can be viewed as a permutation on the nodes. When such permutations do not produce similar structures, we can conclude the similarity of the two graphs in Figure 5 is not accidental.
Next, in Algorithm 3, we compute the average distortion, and the standard deviation of distortions, under permutation of nodes. The input consists of two cliques of the same cardinality. The distance measure comes from Algorithm 2.
| Algorithm 3: Computing Graph Distortion Statistics. |
| Input: Normalized Adjacency Matrices , of equal dimensions |
Output: Baseline for the graph distance, standard deviation of graph distances under permutations of computed distances.
|
6. Details of Experiments and Their Results
In Section 4 we described the procedure of creating the graph of full documents and in Section 4.4 we referenced the best model, although the details of the methods were presented in Section 5. This was not the only model we tried, and we will now discuss other experiments; they all support the conclusion of the non-accidental similarity of the graph of recommendations and the graphs of concepts. (As shown later in Appendix A, this model also performs very well on other sets of guidelines).
6.1. Steps Used in All Our Experiments and Evaluation
In all our experiments we used the procedure in Algorithm 2 to compute the distance/distortion between the two labeled graphs, using the matrix of conceptual distanced between full documents, and the matrix in Table 3. As mentioned earlier, for our best model the distortion was 0.31 and therefore the similarity was 0.69 (or 69%). We then asked the question: Could this distortion be accidental? In other words, could it be the case that we were lucky? If so, how lucky would we have to be? Since the distance between nodes of both graphs are fixed (in a given experiment), the only variable we can manipulate is the mapping from the nodes of one graph to another. In other words, if we did not have the labels, what are the chances of finding the right match from all possible labelings. We thus asked: Can other mappings produce similar results? To answer this question, we computed the average distortion and the standard deviation, based on all possible permutation of nodes ( permutations). The pseudo-code for this computation is shown in Algorithm 3.
In all experiments, the difference between our results and average distortion was seven (or more) standard deviations. Therefore, we can conclude the that the matching of the two geometries is not accidental and is highly significant.
6.2. Results of the Experiments
In this section we first discuss the statistical properties of the experiments to show that our models capture statistically significant geometric correspondences between the graph of recommendation summaries and the graph of conceptual distances between the full document guidelines. Table 6 shows results of the main series of experiments we performed. Additional experiments are reported in Appendix A.

Table 6.
This table shows the values obtained in multiple experiments. Column 2, Distortion, shows the distortions of graphs produced using corresponding models from Column 1. Average distortions per permutation are shown in Column 3. STD is the standard deviation of the distortion per permutation of vertices. Note that the distortion is somewhat depended on how we measure distances; however, the shapes of the distributions are very similar. (The cosine measures are capitalized for readability).
Table 6 shows the results of the experiments with full text of the guidelines. For our best model, BioASQ_WMD, we found a 69% similarity (top line), or 0.31 distance (distortion). As can be seen, the average distortion of permutations (using the distances produced by BioASQ_WMD) is 38%; however the standard deviation of the distortions is less than 1%. Thus, the distance between our model and the mean is about seven standard deviations. Therefore, we conclude that the similarity between the shapes of the two graphs is extremely unlikely to be coincidental. Hence the model represents a non-trivial similarity. Moreover, we performed the same kind of analysis using different models, i.e., different embeddings and different distance measures. The table also shows experiments using the normalized search distance implemented in Solr (https://lucene.apache.org/solr/ (accessed on 24 February 2021)) and the distortion using a transformed cosd distance. (Note that there is no natural transformation of WM distance applicable here). Additionally, while the distances and distortions change, the chances of similarities arising by accident are always smaller than 1/1000 (four standard deviations from the mean of distortions). By this standard statistical criterion, no matter what measures of distance we use, the similarity between two graphs, one from human analysis [3] and the other from automated concept modeling, is non-trivial and not accidental. This observation is amplified by the additional experiments reported in Appendix A. We conclude that vector-based representation are capable of detecting conceptual differences, i.e., the types and densities of concepts brought to the writing of medical recommendations.
7. Discussion and Possible Extensions of This Work
Our broad research objective is to create a computational model accurately representing medical guidelines’ disagreements. Since the creation of such accurate models is beyond the current state of the art, in this article, we focused on an approximation, i.e., a model that is simple and general enough to be potentially applicable in other situations, and which was useful for the question at hand, namely, whether conceptual vocabulary determines recommendations.
As mentioned earlier, this article was partly motivated by epistemology of disagreement, and more specifically medical disagreement, viewed as “near-peer” disagreement. Our results show that it is possible to build computational models of “near-peer” disagreement. Additionally, they provide support for the empirical observations of disagreement adjudication among medical experts [34,35], where the authors observe that the differences in experts’ backgrounds increase the degree of disagreement.
A limitation of the article lies in testing the proposed method on a small number of case studies. In the main track, we focused on the CDC summaries of the breast cancer screening guidelines, and, in Appendix A, we discuss our experiments on the lower back pain management and hypertension guidelines. We showed that the method is robust in the case of these sample guidelines, because even with the change of metrics, the similarities remain statistically significant. However, this article only describes a few case studies, and leaves it as an open question whether it will work equally well in other cases. Thus, an obvious extension of this work would be to compare other groups of guidelines, e.g., European medical societies vs. US medical societies. We know that for years their recommendations, e.g., on managing blood cholesterol, differed.
Another potential extension would be to experiment with other representations, such as more complex word and document embeddings, or with more subtle semantic representations based on entity and relationship extraction or formal models, cf. [52], and on formal modeling of contradictions, like the ones discussed in [5,6]. This, however, would introduce several complications. Attention-based models, such as BERT, GPT or Universal Encoder [53,54,55], would have to be used very carefully, since they encode the order of terms in documents, and indirectly relations between words. Therefore, they would not be appropriate for the experiments described in this article. More subtle formal models, on the other hand, are very brittle, and incapable of operating on real documents, with all the complications arising from interaction between sentences and the use of discourse constructions such as lists and tables. Perhaps one solution to this problem could be to represent the full text of each guideline document as a graph, and not a bag of word embeddings. There is a vast amount of applicable empirical work on graph representations, including representations of recommendations (e.g., [56,57]) and various types of knowledge (e.g., [58,59]). The algorithms proposed in Section 5 would still be directly applicable, and only the distances between pairs of documents would have to be modified and computed on graph representations. These representations could vary, but in all cases we could use applicable algorithms for computing distances in graphs (e.g., [60]), similar to the word mover’s distance (WMD) used in this article. In addition, by experimenting with matching corresponding subgraphs, we could develop new distance measures.
Unlike our earlier work [6], in this article we have not performed any logical analysis of the guidelines. We also did not use text mining to extract relations from the content of the guidelines, and although our focus was on concepts appearing in guidelines, we did not point to specific vocabulary differences. Instead, we measured semantic differences between guidelines using the distances between their vectorial representations. This has to do with the fact that, even though NLP methods have progressed enormously over the last decade [24], they are far from perfect. In our experiments, we used some of the simplest semantic types of words and simple collocations represented as vectors in high-dimensional spaces. This simplicity is helpful, as we can run several experiments, and compare the effects of using different representations and metrics. This gives us the confidence that the similarities we are discovering tell us something interesting about guideline documents.
8. Conclusions
This article investigates the question whether the disagreements in medical recommendations, for example in breast cancer screening or back pain management guidelines, can be attributed to the differences in concepts brought to the problem by specific medical societies (and not, e.g., the style or formalization of recommendations). Our experiments answered this question in the affirmative, and showed that a simple model using word embeddings to represent concepts can account for about 70% to 85% of disagreements in the recommendations. Another contribution is the articulation of a very natural graph clique-based algorithm/method for comparing the similarity of two collections of documents. Given two sets of documents, each of the same cardinality, and a mapping between nodes, we computed the percent of distortion between the shapes of the two cliques, and the chances that the mapping arose from a random process. We also documented all of the steps of the process and provided the data and the code (https://github.com/hematialam/Conceptual_Distances_Medical_Recommendations (accessed on 24 February 2021)) to facilitate both extensions of this work and its replication.
Our work extends the state-of-the-art computational analysis of medical guidelines. Namely, instead of semi-automated conceptual analysis, we demonstrated the feasibility of automated conceptual analysis. That is, in our study, we used a representation derived from a (relatively shallow) neural network (BioASQ embeddings [51]), and knowledge-based annotations derived from MetaMap (https://metamap.nlm.nih.gov/ (accessed on 24 February 2021)). Our results, detailed in Section 6 and in Appendix A, show that both can be useful as representations of our set of guidelines. Overall, they show similar performance in modeling conceptual similarities. However, the BioAsq_WMD model, using the BioASQ embeddings and the Word Mover’s Distance, seems to be most stable, as it performed very well in all our experiments.
Although this article is a collection of three case studies, bound by a common method, it could be a good starting point for an analysis of other medical guidelines and perhaps other areas of expert disagreement. The methods described in this article are easy to use and rely on well-known tools such as word embeddings and MetaMap. They can also be extended and improved to produce more accurate and deeper analyses, due to the fast progress in text mining and deep learning techniques. From the point of view of methodology of analyzing medical guidelines, this article contains the first computational implementation of the “near-peer” model mentioned earlier. To our knowledge, ours is the first proposal to use automated methods of text analysis to investigate differences in recommendations.
Author Contributions
H.H. performed the majority of experiments discussed in this article, and substantially contributed to writing. The idea to computationally investigate the conceptual difference between the guidelines comes from L.G.; she also contributed with graphing and overall analysis and writing. S.G. performed several experiments allowing us to progress with the evaluation. W.W.Z. provided the project supervision, suggested the clique-based method for computing distortions, and did the majority of the writing. The overall progress was achieved during multiple team discussions, with equal contributions. This research has not been supported by a grant. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Our code and data are available at https://github.com/hematialam/Conceptual_Distances_Medical_Recommendations (accessed on 24 February 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| NLP | Natural Language Processing |
| CDC | Centers for Disease Control and Prevention |
Appendix A. Additional Experiments
We performed several additional experiments, and we report on three of them in this appendix. The first experiment is a variant of the one reported above using the CDC table in Figure 1. The second experiment is on lower back pain management guidelines, for which we could find an online summary table similar to the one in Figure 1. The third one is a different one, applying the graph comparison not to a table, but to the guideline abstracts. We could not find a tabular comparison of the hypertension management guidelines, so instead we compared the concepts in full and abstracted texts. This shows the potential applicability of the proposed approach to other situations, where we might be interested in conceptual comparisons of related collections of documents.
Appendix A.1. Experimenting with the Full Texts of the Guidelines
We performed additional experiments with modified views of the full guideline documents, as enumerated below. This was driven by the fact that the levels of distances between full documents may change if we compute the similarities/distances between selected sentences, which are explicitly related to the statements from the CDC table in Figure 1. For these additional experiments we split each full text guideline document into two different subsets:
- Related: containing sentences that are related to the CDC table, by having common concepts, as represented by UMLS concepts. This was done in multiple ways, giving us six possible experiments:
- (a)
- The CDC recommendations table was considered as a single bag of concepts. If a sentence in the full text had a minimum number of mutual concepts with this bag, that sentence was considered a related sentence.
- (b)
- If a sentence in the full text had a minimum number of mutual concepts with at least one statement from the CDC table (again, viewed as a bag of concepts), that sentence was considered a related sentence.
Different minimum numbers of mutual concept(s) were examined in our experiment, that is the minimum was set to 1, 2 and 3. - Unrelated: the other sentences.Unrelated sentences were not used for these additional experiments.
Concept extraction: For all experiments, we used MetaMap (https://metamap.nlm.nih.gov/ (accessed on 24 February 2021)) to extract UMLS concepts (https://www.nlm.nih.gov/research/umls/index.html (accessed on 24 February 2021)) and semantic types (https://www.nlm.nih.gov/research/umls/META3_current_semantic_types.html (accessed on 24 February 2021)) in sentences. We only considered concepts with informative (in our opinion) semantic types. This meant using concepts related to diagnosis and prevention, for example “findings”, and not using ones related, e.g., to genomics. Our final list had the following: [[diap], [hlca], [dsyn], [neop], [qnco], [qlco], [tmco], [fndg], [geoa], [topp], [lbpr]].
For full text guidelines (as per Table 4), the results of the experiments are shown in Table 6, and discussed in Section 6 and Section 8. Table A1 and Table A2 are based on the same type of comparisons as discussed in Section 6, except that we subtract the Unrelated sentences from the full guidelines. Again, we observed that the similarity is not accidental, and that BioASQ embeddings with WM distance seem on average to give the best performance.

Table A1.
Using sentences in recommendations and minimum mutual concepts. This table shows the values obtained in additional experiments, where full document guidelines were modified by attending to concepts in sentences (see above). Column 1 refers to the number of concepts overlapping with summaries. Distortion shows the distortions of graphs produced using corresponding models from Column 1. As before, in Table 6, the distortion depends on how we measure the distances; however, the shapes of the distributions are very similar.
Table A1.
Using sentences in recommendations and minimum mutual concepts. This table shows the values obtained in additional experiments, where full document guidelines were modified by attending to concepts in sentences (see above). Column 1 refers to the number of concepts overlapping with summaries. Distortion shows the distortions of graphs produced using corresponding models from Column 1. As before, in Table 6, the distortion depends on how we measure the distances; however, the shapes of the distributions are very similar.
| Min. Mutual Concepts | Model | Distortion | Distortion of Permutations | STD |
|---|---|---|---|---|
| 1 | BioASQ CosD | 0.526380991 | 0.602890558 | 0.011735664 |
| Conceptualized_CosD | 0.635564038 | 0.646721788 | 0.011417208 | |
| NoConcept_CosD | 0.626087519 | 0.646906954 | 0.011131221 | |
| NoConcept_WMD | 0.352402031 | 0.383852647 | 0.006550777 | |
| Conceptualized_WMD | 0.359296888 | 0.390059373 | 0.006626223 | |
| BioASQ_WMD | 0.336903254 | 0.384735148 | 0.006498348 | |
| 2 | BioASQ_CosD | 0.449264689 | 0.572620976 | 0.010916054 |
| Conceptualized_CosD | 0.384945443 | 0.488740293 | 0.008608367 | |
| NoConcept_CosD | 0.433167046 | 0.501788823 | 0.008699466 | |
| NoConcept_WMD | 0.34284288 | 0.376371094 | 0.006467164 | |
| Conceptualized_WMD | 0.330059701 | 0.373155641 | 0.006466969 | |
| BioASQ_WMD | 0.32446554 | 0.38365857 | 0.006428759 | |
| 3 | BioASQ_CosD | 0.468163076 | 0.537093759 | 0.010040669 |
| Conceptualized_CosD | 0.564019791 | 0.57488789 | 0.010091071 | |
| NoConcept_CosD | 0.594326474 | 0.596293202 | 0.010300973 | |
| NoConcept_WMD | 0.360513492 | 0.375067469 | 0.006461442 | |
| Conceptualized_WMD | 0.37193217 | 0.383126986 | 0.006477258 | |
| BioASQ_WMD | 0.34276229 | 0.375886963 | 0.006455091 |
Note the potentially important observation about Table 6, Table A1 and Table A2: They jointly show that the property we investigate, i.e., the conceptual distances between guidelines, is indeed geometric, and therefore the word “distances” is not merely a metaphor. The correspondence between the two graphs is preserved no matter how we set up the experiments. That is, as with geometric properties such as being collinear or parallel, the structure remains the same when a transformation (such as a projection) is applied to the points, even though some of the measurements might change (e.g., measured distances, or the area of a parallelogram). The same happens when we transform the documents by removing Unrelated sentences: the values of distortions change, but the non-accidental correspondence with the summary graph (Figure 5) remains invariant.

Table A2.
Using the whole summary recommendations and minimum mutual concepts. This table shows the values obtained in additional experiments, where the whole CDC summary was used to obtain sets of mutual concepts (see above). Column 1 refers to the number of concepts overlapping with the summary. Distortion shows the distortions of graphs produced using corresponding models from Column 1. As before, in Table 6 and Table A1 the distortion is somewhat depended on how we measure distances; however, the shapes of the distributions are very similar.
Table A2.
Using the whole summary recommendations and minimum mutual concepts. This table shows the values obtained in additional experiments, where the whole CDC summary was used to obtain sets of mutual concepts (see above). Column 1 refers to the number of concepts overlapping with the summary. Distortion shows the distortions of graphs produced using corresponding models from Column 1. As before, in Table 6 and Table A1 the distortion is somewhat depended on how we measure distances; however, the shapes of the distributions are very similar.
| Min. Mutual Concepts | Model | Distortion | Distortion of Permutations | STD |
|---|---|---|---|---|
| 1 | BioASQ_CosD | 0.52638099 | 0.60289056 | 0.01173566 |
| Conceptualized_CosD | 0.63556404 | 0.64672179 | 0.01141721 | |
| NoConcept_CosD | 0.62608752 | 0.64690695 | 0.01113122 | |
| NoConcept_WMD | 0.35240203 | 0.38385265 | 0.00655078 | |
| Conceptualized_WMD | 0.35929689 | 0.39005937 | 0.00662622 | |
| BioASQ_WMD | 0.33690325 | 0.38473515 | 0.00649835 | |
| 2 | BioASQ_CosD | 0.44926469 | 0.57262098 | 0.01091605 |
| Conceptualized_CosD | 0.38494544 | 0.48874029 | 0.00860837 | |
| NoConcept_CosD | 0.43316705 | 0.50178882 | 0.00869947 | |
| NoConcept_WMD | 0.34284288 | 0.37637109 | 0.00646716 | |
| Conceptualized_WMD | 0.3300597 | 0.37315564 | 0.00646697 | |
| BioASQ_WMD | 0.32446554 | 0.38365857 | 0.00642876 | |
| 3 | BioASQ_CosD | 0.46816308 | 0.53709376 | 0.01004067 |
| Conceptualized_CosD | 0.56401979 | 0.57488789 | 0.01009107 | |
| NoConcept_CosD | 0.59432647 | 0.5962932 | 0.01030097 | |
| NoConcept_WMD | 0.36051349 | 0.37506747 | 0.00646144 | |
| Conceptualized_WMD | 0.37193217 | 0.38312699 | 0.00647726 | |
| BioASQ_WMD | 0.34276229 | 0.37588696 | 0.00645509 |
Appendix A.2. Lower Back Pain Management Guidelines
In this experiment we used the summary tables on “clinical practice guidelines for the management of non-specific low back pain in primary care” from [13]. In the cited paper, several comparisons are made between 15 clinical practice guidelines from multiple continents and countries (Africa (multinational), Australia, Brazil, Belgium, Canada, Denmark, Finland, Germany, Malaysia, Mexico, the Netherlands, Philippine, Spain, the USA and the UK). In our experiments we used all of those for which an English text was available: (GER) [61], (MAL) [62], (SPA) [63], (UK) [64], (AUS) [65], (USA) [66], (CAN) [67], (DEN) [68], and (BEL) [69]. For this total of nine guideline texts, we experimented with Table 1 (describing methodologies for diagnosis) and Table 2 (treatment recommendations) from the article [13] containing, respectively, 12 and 60 features; in addition, we created a super-table combining the two tables, and applied our method to it as well.
With the same process as described in Section 3 we converted the Tables 1 and 2 of [13] into Jaccard distances. Then, as before, we computed the distortion between the graphs of the full text and the graphs of distances between the extracted features; and, as before, we established that the probability of obtaining high similarity by chance is extremely small. For Table 1 of [13] our best model BioAsq_WMD produced about 28% distortion (or 72% similarity). Similar results hold for Table 2 of and for the combined table, although the actual distortion numbers differ. In all cases, for the model BioAsq_WMD we found about 10-fold standard deviation, with distortion of about 16% for Table 2 and about 14% for the aggregated tables combining Tables 1 and 2 of [13].
All other models used in Table 6 performed in line with the previous results, with the only exception being the conceptualized models for Table 1 of [13], where for Conceptualized_CosD and Conceptualized_CosD’ the distortion was slightly worse than random. We do not have an explanation for this sub-par performance, but we have seen a relatively weak performance of this model in Table A1. Table A3 shows the Jaccard distances and Table A4 shows the performance of all models on the combined table. Thus the performance of the model does not seem to degrade with a large number of comparisons.

Table A3.
Jaccard distances based on the combined Tables 1 and 2 from [13]. The guidelines are about the management of non-specific lower back pain.
Table A3.
Jaccard distances based on the combined Tables 1 and 2 from [13]. The guidelines are about the management of non-specific lower back pain.
| US | DEN | MAL | CAN | BEL | GER | UK | SPA | AUS | |
|---|---|---|---|---|---|---|---|---|---|
| US | 0 | 46 | 43 | 35 | 39 | 32 | 43 | 41 | 46 |
| DEN | 46 | 0 | 50 | 45 | 47 | 44 | 47 | 49 | 39 |
| MAL | 43 | 50 | 0 | 33 | 36 | 39 | 40 | 36 | 42 |
| CAN | 35 | 45 | 33 | 0 | 33 | 23 | 33 | 34 | 32 |
| BEL | 39 | 47 | 36 | 33 | 0 | 26 | 15 | 36 | 38 |
| GER | 32 | 44 | 39 | 23 | 26 | 0 | 30 | 33 | 35 |
| UK | 43 | 47 | 40 | 33 | 15 | 30 | 0 | 41 | 36 |
| SPA | 41 | 49 | 36 | 34 | 36 | 33 | 41 | 0 | 44 |
| AUS | 46 | 39 | 42 | 32 | 38 | 35 | 36 | 44 | 0 |

Table A4.
The performance of the algorithms on the combined Tables 1 and 2 from [13] is in line with the results in Section 6.2, except for the weaker showing of the Conceptualized_WMD model.
Table A4.
The performance of the algorithms on the combined Tables 1 and 2 from [13] is in line with the results in Section 6.2, except for the weaker showing of the Conceptualized_WMD model.
| Model | Distortion | Distortion of Permutations | STD |
|---|---|---|---|
| BioASQ_WMD | 0.14157219 | 0.16717125 | 0.00186797 |
| Conceptualized_WMD | 0.15689518 | 0.16098291 | 0.00179856 |
| NoConcept_WMD | 0.13946498 | 0.16067458 | 0.00180108 |
| BioASQ_CosD’ | 0.44899108 | 0.49891074 | 0.00595033 |
| Conceptualized_CosD’ | 0.31577959 | 0.35477261 | 0.00389520 |
| NoConcept_CosD’ | 0.27595783 | 0.33897971 | 0.00418504 |
| BioASQ_CosD | 0.40412785 | 0.45347124 | 0.00511851 |
| Conceptualized_CosD | 0.28283583 | 0.32039879 | 0.00342097 |
| NoConcept_CosD | 0.25530361 | 0.31717921 | 0.00378427 |
Appendix A.3. Comparing Hypertension Management Guidelines
In an additional experiment, we used a collection of hypertension management guidelines from different countries, including the USA, Canada, Brasil, the UK and Ireland [70,71,72,73,74,75,76,77]. The corpus was created by searching PubMed for “practice guideline’’ as “publication type’’ and “hypertension’’ and as the “major MeSh’’ index. We selected eight of them from different medical bodies, where the full text of the guidelines was available. This corpus consists of the following eight documents: CHEP2007 [70], the 2007 Canadian Hypertension Education Program; AHA & ASH & PCNA [71], joint statement of the American Heart Association, American Society Of Hypertension, and Preventive Cardiovascular Nurses Association; BGAH [72], the Brazilian Guideline of Arterial Hypertension; CFP [73], the 2013 Canadian screening recommendations; AAGBI & BHS [74], the 2016 joint British and Irish guidelines; CHEP2009 [77], the 2009 Canadian Hypertension Education Program; AAP [75], 2017 guidelines focusing on children and adolescents; and JNC [76], the 2014 evidence-based guidelines focusing on adults.
Because we are not aware of any tabular summary of differences between hypertension guidelines, similar to the one shown earlier in Figure 1, we made the comparisons between full texts of the guidelines and their abstracts. That is, we created two graphs of embeddings, as shown in Figure A1, and measured their similarity, as well as the probability of the similarity arising by chance, as shown in Table A5. The experiment shows that the concepts appearing in the abstracts of the guidelines strongly correlate with the concepts used the full texts of the guidelines. Moreover, the method, described earlier in Section 5, which we used to find this correspondence, was very good at picking up this similarity; and, as before, a very good model was obtained by using BioASQ embeddings with the Word Mover Distance (WMD).
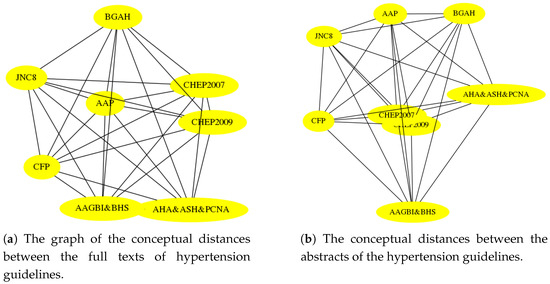

Table A5.
We see the robustness of the proposed method when comparing the conceptual distances of the abstracts and the full documents of the hypertension guidelines. The results are in line with those of Section 6.2.
Table A5.
We see the robustness of the proposed method when comparing the conceptual distances of the abstracts and the full documents of the hypertension guidelines. The results are in line with those of Section 6.2.
| Model | Distortion | Distortion of Permutations | STD |
|---|---|---|---|
| BioASQ_WMD | 0.10722113 | 0.15779628 | 0.00339429 |
| Conceptualized_WMD | 0.22486228 | 0.30471905 | 0.00416548 |
| NoConcept_WMD | 0.10659179 | 0.15903103 | 0.00339359 |
| BioASQ_CosD’ | 0.63552553 | 0.67228101 | 0.00891151 |
| Conceptualized_CosD’ | 0.34923088 | 0.48818741 | 0.006308838 |
| NoConcept_CosD’ | 0.51297572 | 0.54567338 | 0.00727428 |
| BioASQ_CosD | 0.53894790 | 0.58653238 | 0.00754606 |
| Conceptualized_CosD | 0.43294195 | 0.48253914 | 0.00626540 |
| Conceptualized_CosD | 0.34443154 | 0.47391939 | 0.00608174 |
| NoConcept_CosD | 0.42284040 | 0.45815692 | 0.00609919 |
References
- McClintock, A.H.; Golob, A.L.; Laya, M.B. Breast Cancer Risk Assessment: A Step-Wise Approach for Primary Care Providers on the Front Lines of Shared Decision Making. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2020; Volume 95, pp. 1268–1275. [Google Scholar]
- Pace, L.E.; Keating, N.L. A systematic assessment of benefits and risks to guide breast cancer screening decisions. JAMA 2014, 311, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- CDC. Breast Cancer Screening Guidelines for Women; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2017. [Google Scholar]
- Catillon, M. Medical Knowledge Synthesis: A Brief Overview; 2017; Available online: https://www.hbs.edu/ris/Publication%20Files/WhitePaper-Catillon10.2017_40a6683d-411b-4621-a121-8f5e93b13605.pdf (accessed on 24 February 2021).
- Zadrozny, W.; Garbayo, L. A Sheaf Model of Contradictions and Disagreements. Preliminary Report and Discussion. arXiv 2018, arXiv:1801.09036. [Google Scholar]
- Zadrozny, W.; Hematialam, H.; Garbayo, L. Towards Semantic Modeling of Contradictions and Disagreements: A Case Study of Medical Guidelines. arXiv 2017, arXiv:1708.00850. [Google Scholar]
- Christensen, D.; Lackey, J.; Kelly, T. The Epistemology of Disagreement: New Essays; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Lackey, J. Taking Religious Disagreement Seriously. In Religious Faith and Intellectual Virtue; Callahan, L., O’Connor, T., Eds.; Oxford University Press: Oxford, UK, 2014; pp. 299–316. [Google Scholar]
- Grim, P.; Modell, A.; Breslin, N.; Mcnenny, J.; Mondescu, I.; Finnegan, K.; Olsen, R.; An, C.; Fedder, A. Coherence and correspondence in the network dynamics of belief suites. Episteme 2017, 14, 233–253. [Google Scholar] [CrossRef]
- Garbayo, L. Epistemic Considerations on Expert Disagreement, Normative Justification, and Inconsistency Regarding Multi-criteria Decision Making. In Constraint Programming and Decision Making; Ceberio, M., Kreinovich, V., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 35–45. [Google Scholar]
- Garbayo, L.; Ceberio, M.; Bistarelli, S.; Henderson, J. On Modeling Multi-experts Multi-criteria Decision-Making Argumentation and Disagreement: Philosophical and Computational Approaches Reconsidered. In Constraint Programming and Decision Making: Theory and Applications; Ceberio, M., Kreinovich, V., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 67–75. [Google Scholar]
- Garbayo, L. Dependence logic & medical guidelines disagreement: An informational (in) dependence analysis. In Logic Colloquium 2019; AMCA: Praha, Czech Republic, 2019; p. 112. [Google Scholar]
- Oliveira, C.B.; Maher, C.G.; Pinto, R.Z.; Traeger, A.C.; Lin, C.W.C.; Chenot, J.F.; van Tulder, M.; Koes, B.W. Clinical practice guidelines for the management of non-specific low back pain in primary care: An updated overview. Eur. Spine J. 2018, 27, 2791–2803. [Google Scholar] [CrossRef]
- Peek, N.; Combi, C.; Marin, R.; Bellazzi, R. Thirty years of artificial intelligence in medicine (AIME) conferences: A review of research themes. Artif. Intell. Med. 2015, 65, 61–73. [Google Scholar] [CrossRef]
- Bowles, J.; Caminati, M.; Cha, S.; Mendoza, J. A framework for automated conflict detection and resolution in medical guidelines. Sci. Comput. Program. 2019, 182, 42–63. [Google Scholar] [CrossRef]
- Tsopra, R.; Lamy, J.B.; Sedki, K. Using preference learning for detecting inconsistencies in clinical practice guidelines: Methods and application to antibiotherapy. Artif. Intell. Med. 2018, 89, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Udelsman, B.V.; Streid, J.; Chang, D.C.; Salim, A.; Livingston, D.H.; Lindvall, C.; Cooper, Z. Natural Language Processing Accurately Measures Adherence to Best Practice Guidelines for Palliative Care in Trauma. J. Pain Symptom Manag. 2020, 59, 225–232. [Google Scholar] [CrossRef]
- Waheeb, S.A.; Ahmed Khan, N.; Chen, B.; Shang, X. Machine Learning Based Sentiment Text Classification for Evaluating Treatment Quality of Discharge Summary. Information 2020, 11, 281. [Google Scholar] [CrossRef]
- Seneviratne, O.; Das, A.K.; Chari, S.; Agu, N.N.; Rashid, S.M.; Chen, C.H.; McCusker, J.P.; Hendler, J.A.; McGuinness, D.L. Enabling Trust in Clinical Decision Support Recommendations through Semantics; 2019; Available online: http://ceur-ws.org/Vol-2477/paper_5.pdf (accessed on 24 February 2021).
- Chen, X.; Xie, H.; Cheng, G.; Poon, L.K.; Leng, M.; Wang, F.L. Trends and Features of the Applications of Natural Language Processing Techniques for Clinical Trials Text Analysis. Appl. Sci. 2020, 10, 2157. [Google Scholar] [CrossRef]
- Ju, M.; Nguyen, N.T.; Miwa, M.; Ananiadou, S. An ensemble of neural models for nested adverse drug events and medication extraction with subwords. J. Am. Med. Inform. Assoc. 2020, 27, 22–30. [Google Scholar] [CrossRef]
- Benedetti, F.; Beneventano, D.; Bergamaschi, S.; Simonini, G. Computing inter-document similarity with context semantic analysis. Inf. Syst. 2019, 80, 136–147. [Google Scholar] [CrossRef]
- Rospocher, M.; Corcoglioniti, F.; Dragoni, M. Boosting Document Retrieval with Knowledge Extraction and Linked Data. Semant. Web 2019, 10, 753–778. [Google Scholar] [CrossRef]
- Zhou, M.; Duan, N.; Liu, S.; Shum, H.Y. Progress in Neural NLP: Modeling, Learning, and Reasoning. Engineering 2020, 6, 275–290. [Google Scholar] [CrossRef]
- Smith, N.A. Contextual word representations: Putting words into computers. Commun. ACM 2020, 63, 66–74. [Google Scholar] [CrossRef]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems; 2013; pp. 3111–3119. Available online: https://arxiv.org/abs/1310.4546 (accessed on 24 February 2021).
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Shalaby, W.; Zadrozny, W.; Jin, H. Beyond word embeddings: Learning entity and concept representations from large scale knowledge bases. Inf. Retr. J. 2019, 22, 525–542. [Google Scholar] [CrossRef]
- Kalyan, K.S.; Sangeetha, S. SECNLP: A survey of embeddings in clinical natural language processing. J. Biomed. Inform. 2020, 101, 103323. [Google Scholar] [CrossRef] [PubMed]
- Khattak, F.K.; Jeblee, S.; Pou-Prom, C.; Abdalla, M.; Meaney, C.; Rudzicz, F. A survey of word embeddings for clinical text. J. Biomed. Inform. X 2019, 4, 100057. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Duong, P.H.; Cambria, E. Learning short-text semantic similarity with word embeddings and external knowledge sources. Knowl. Based Syst. 2019, 182, 104842. [Google Scholar] [CrossRef]
- Tien, N.H.; Le, N.M.; Tomohiro, Y.; Tatsuya, I. Sentence modeling via multiple word embeddings and multi-level comparison for semantic textual similarity. Inf. Process. Manag. 2019, 56, 102090. [Google Scholar] [CrossRef]
- Lie, R.K.; Chan, F.K.; Grady, C.; Ng, V.H.; Wendler, D. Comparative effectiveness research: What to do when experts disagree about risks. BMC Med Ethics 2017, 18, 1–9. [Google Scholar] [CrossRef]
- Schaekermann, M.; Beaton, G.; Habib, M.; Lim, A.; Larson, K.; Law, E. Capturing Expert Arguments from Medical Adjudication Discussions in a Machine-readable Format. In Proceedings of the Companion Proceedings of The 2019 World Wide Web Conference, San Francisco, CA, USA, 13–17 May 2019; pp. 1131–1137. [Google Scholar]
- Schaekermann, M.; Beaton, G.; Habib, M.; Lim, A.; Larson, K.; Law, E. Understanding Expert Disagreement in Medical Data Analysis through Structured Adjudication. Proc. ACM Hum. Comput. Interact. 2019, 3, 1–23. [Google Scholar] [CrossRef]
- Grant, J.; Hunter, A. Analysing inconsistent first-order knowledgebases. Artif. Intell. 2008, 172, 1064–1093. [Google Scholar] [CrossRef]
- Subrahmanian, V.S.; Amgoud, L. A General Framework for Reasoning about Inconsistency. In Proceedings of the 20th International Joint Conference on Artificial Intelligence, Hyderabad, India, 6–12 January 2007; pp. 599–604. [Google Scholar]
- Grant, J.; Hunter, A. Analysing inconsistent information using distance-based measures. Int. J. Approx. Reason. 2016, 89, 3–26. [Google Scholar] [CrossRef]
- Tran, T.H. Inconsistency measures for probabilistic knowledge bases. In Proceedings of the 2017 9th International Conference on Knowledge and Systems Engineering (KSE), Hue, Vietnam, 19–21 October 2017; pp. 148–153. [Google Scholar]
- Garbayo, L.; Zadrozny, W.; Hematialam, H. Converging in Breast Cancer Diagnostic Screening: A Computational Model Proposal. Diagnosis 2019, 6, eA60. [Google Scholar]
- American College of Obstetricians-Gynecologists. Practice Bulletin No. 122: Breast Cancer Screening. Obstet. Gynecol. 2011, 118, 372–382. [Google Scholar] [CrossRef] [PubMed]
- AAFP Policy Action. Summary of Recommendations for Clinical Preventive Services; American Academy of Family Physicians: Leawood, KS, USA, 2017. [Google Scholar]
- Wilt, T.J.; Harris, R.P.; Qaseem, A. Screening for cancer: Advice for high-value care from the American College of Physicians. Ann. Intern. Med. 2015, 162, 718–725. [Google Scholar] [CrossRef]
- Lee, C.H.; Dershaw, D.D.; Kopans, D.; Evans, P.; Monsees, B.; Monticciolo, D.; Brenner, R.J.; Bassett, L.; Berg, W.; Feig, S.; et al. Breast cancer screening with imaging: Recommendations from the Society of Breast Imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J. Am. Coll. Radiol. 2010, 7, 18–27. [Google Scholar] [CrossRef]
- Oeffinger, K.C.; Fontham, E.T.; Etzioni, R.; Herzig, A.; Michaelson, J.S.; Shih, Y.C.T.; Walter, L.C.; Church, T.R.; Flowers, C.R.; LaMonte, S.J.; et al. Breast cancer screening for women at average risk: 2015 guideline update from the American Cancer Society. JAMA 2015, 314, 1599–1614. [Google Scholar] [CrossRef]
- Jørgensen, K.J.; Bewley, S. Breast-Cancer Screening—Viewpoint of the IARC Working Group. N. Engl. J. Med. 2015, 373, 1478. [Google Scholar]
- Siu, A.L. Screening for breast cancer: US Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2016, 164, 279–296. [Google Scholar] [CrossRef]
- Kusner, M.; Sun, Y.; Kolkin, N.; Weinberger, K. From word embeddings to document distances. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 957–966. [Google Scholar]
- Monge, G. Mémoire sur la théorie des déblais et des remblais. In Histoire de l’Académie Royale des Sciences de Paris; 1781. [Google Scholar]
- Rehurek, R.; Sojka, P. Gensim—Statistical Semantics in Python; Retrieved from gensim.org; 2011; Available online: https://www.semanticscholar.org/paper/Gensim-Statistical-Semantics-in-Python-Rehurek-Sojka/b55fe23d7290f59d14e51e7813f5950f5ff08b2b (accessed on 24 February 2021).
- Tsatsaronis, G.; Balikas, G.; Malakasiotis, P.; Partalas, I.; Zschunke, M.; Alvers, M.R.; Weissenborn, D.; Krithara, A.; Petridis, S.; Polychronopoulos, D.; et al. An overview of the BioASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinform. 2015, 16, 138. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, X.; Conesa, A.; Pereira, C. GRAM-CNN: A deep learning approach with local context for named entity recognition in biomedical text. Bioinformatics 2017, 34, 1547–1554. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Peters, M.E.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. arXiv 2018, arXiv:1802.05365. [Google Scholar]
- Cer, D.; Yang, Y.; Kong, S.Y.; Hua, N.; Limtiaco, N.; John, R.S.; Constant, N.; Guajardo-Cespedes, M.; Yuan, S.; Tar, C.; et al. Universal sentence encoder. arXiv 2018, arXiv:1803.11175. [Google Scholar]
- Catherine, R.; Cohen, W. Personalized Recommendations Using Knowledge Graphs: A Probabilistic Logic Programming Approach. In Proceedings of the 10th ACM Conference on Recommender Systems, RecSys ’16; Association for Computing Machinery: New York, NY, USA, 2016; pp. 325–332. [Google Scholar]
- Mercorio, F.; Mezzanzanica, M.; Moscato, V.; Picariello, A.; Sperli, G. DICO: A graph-db framework for community detection on big scholarly data. IEEE Trans. Emerg. Top. Comput. 2019. [Google Scholar] [CrossRef]
- Nickel, M.; Murphy, K.; Tresp, V.; Gabrilovich, E. A review of relational machine learning for knowledge graphs. Proc. IEEE 2015, 104, 11–33. [Google Scholar] [CrossRef]
- Ji, S.; Pan, S.; Cambria, E.; Marttinen, P.; Yu, P.S. A survey on knowledge graphs: Representation, acquisition and applications. arXiv 2020, arXiv:2002.00388. [Google Scholar]
- Maretic, H.P.; Gheche, M.E.; Chierchia, G.; Frossard, P. GOT: An optimal transport framework for graph comparison. arXiv 2019, arXiv:1906.02085. [Google Scholar]
- Chenot, J.F.; Greitemann, B.; Kladny, B.; Petzke, F.; Pfingsten, M.; Schorr, S.G. Non-specific low back pain. Dtsch. Ärztebl. Int. 2017, 114, 883. [Google Scholar] [CrossRef]
- Mansor, M. The Malaysian LOW BACK PAIN Management Guidelines, 1st ed.; 2009; Available online: https://www.semanticscholar.org/paper/The-Malaysian-LOW-BACK-PAIN-management-Edition.-Mansor/7c8f2bbf0968f7c175754dee819f302dc8beef83 (accessed on 24 February 2021).
- Marques, E.L. The Treatment of Low Back Pain and Scientific Evidence; 2006; Available online: https://www.intechopen.com/books/low-back-pain/the-treatment-of-low-back-pain-scientific-evidence (accessed on 24 February 2021).
- de Campos, T.F. Low back pain and sciatica in over 16s: Assessment and management NICE Guideline [NG59]. J. Physiother. 2017, 63, 120. [Google Scholar] [CrossRef]
- NSW Agency for Clinical Innovation. Management of People with Acute Low Back Pain: Model of Care; NSW Agency for Clinical Innovation: Chatswood, Australia, 2016. [Google Scholar]
- Qaseem, A.; Wilt, T.J.; McLean, R.M.; Forciea, M.A. Noninvasive treatments for acute, subacute, and chronic low back pain: A clinical practice guideline from the American College of Physicians. Ann. Intern. Med. 2017, 166, 514–530. [Google Scholar] [CrossRef]
- Toward Optimized Practice Low Back Pain Working Group. Evidence-Informed Primary Care Management of Low Back Pain; Toward Optimized Practice: Edmonton, AB, Canada, 2015. [Google Scholar]
- Stochkendahl, M.J.; Kjaer, P.; Hartvigsen, J.; Kongsted, A.; Aaboe, J.; Andersen, M.; Andersen, M.Ø; Fournier, G.; Højgaard, B.; Jensen, M.B.; et al. National Clinical Guidelines for non-surgical treatment of patients with recent onset low back pain or lumbar radiculopathy. Eur. Spine J. 2018, 27, 60–75. [Google Scholar] [CrossRef]
- Van Wambeke, P.; Desomer, A.; Ailiet, L.; Berquin, A.; Dumoulin, C.; Depreitere, B.; Dewachter, J.; Dolphens, M.; Forget, P.; Fraselle, V.; et al. Low Back Pain and Radicular Pain: Assessment and Management; KCE Report; Belgian Health Care Knowledge Centre: Brussels, Belgium, 2017; Volume 287. [Google Scholar]
- Padwal, R.S.; Hemmelgarn, B.R.; McAlister, F.A.; McKay, D.W.; Grover, S.; Wilson, T.; Penner, B.; Burgess, E.; Bolli, P.; Hill, M.; et al. The 2007 Canadian Hypertension Education Program recommendations for the management of hypertension: Part 1—Blood pressure measurement, diagnosis and assessment of risk. Can. J. Cardiol. 2007, 23, 529–538. [Google Scholar] [CrossRef][Green Version]
- Pickering, T.G.; Miller, N.H.; Ogedegbe, G.; Krakoff, L.R.; Artinian, N.T.; Goff, D. Call to action on use and reimbursement for home blood pressure monitoring: A joint scientific statement from the American Heart Association, American Society of Hypertension, and Preventive Cardiovascular Nurses Association. Hypertension 2008, 52, 10–29. [Google Scholar] [CrossRef]
- Malachias, M.; Gomes, M.; Nobre, F.; Alessi, A.; Feitosa, A.; Coelho, E. 7th Brazilian guideline of arterial hypertension: Chapter 2-diagnosis and classification. Arq. Bras. Cardiol. 2016, 107, 7–13. [Google Scholar]
- Lindsay, P.; Gorber, S.C.; Joffres, M.; Birtwhistle, R.; McKay, D.; Cloutier, L. Recommendations on screening for high blood pressure in Canadian adults. Can. Fam. Physician 2013, 59, 927–933. [Google Scholar]
- Hartle, A.; McCormack, T.; Carlisle, J.; Anderson, S.; Pichel, A.; Beckett, N.; Woodcock, T.; Heagerty, A. The measurement of adult blood pressure and management of hypertension before elective surgery: Joint Guidelines from the Association of Anaesthetists of Great Britain and Ireland and the British Hypertension Society. Anaesthesia 2016, 71, 326–337. [Google Scholar] [CrossRef]
- Flynn, J.T.; Kaelber, D.C.; Baker-Smith, C.M.; Blowey, D.; Carroll, A.E.; Daniels, S.R.; de Ferranti, S.D.; Dionne, J.M.; Falkner, B.; Flinn, S.K.; et al. Clinical practice guideline for screening and management of high blood pressure in children and adolescents. Pediatrics 2017, 140, e20171904. [Google Scholar] [CrossRef] [PubMed]
- James, P.A.; Oparil, S.; Carter, B.L.; Cushman, W.C.; Dennison-Himmelfarb, C.; Handler, J.; Lackland, D.T.; LeFevre, M.L.; MacKenzie, T.D.; Ogedegbe, O.; et al. 2014 evidence-based guideline for the management of high blood pressure in adults: Report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA 2014, 311, 507–520. [Google Scholar] [CrossRef]
- Padwal, R.S.; Hemmelgarn, B.R.; Khan, N.A.; Grover, S.; McKay, D.W.; Wilson, T.; Penner, B.; Burgess, E.; McAlister, F.A.; Bolli, P.; et al. The 2009 Canadian Hypertension Education Program recommendations for the management of hypertension: Part 1—Blood pressure measurement, diagnosis and assessment of risk. Can. J. Cardiol. 2009, 25, 279–286. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).