Identification of Co-Deregulated Genes in Urinary Bladder Cancer Using High-Throughput Methodologies
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Collection
2.2. Data Pre-Processing
2.3. Background Correction and Normalization
2.4. Differentially Expressed Genes (DEGs)
2.5. Unsupervised Classification Methods
2.6. Common Expression Patterns in UBC
2.7. Gene Ontology (GO) Enrichment
2.8. Inter-Cohort Validation of the DEGs
3. Results
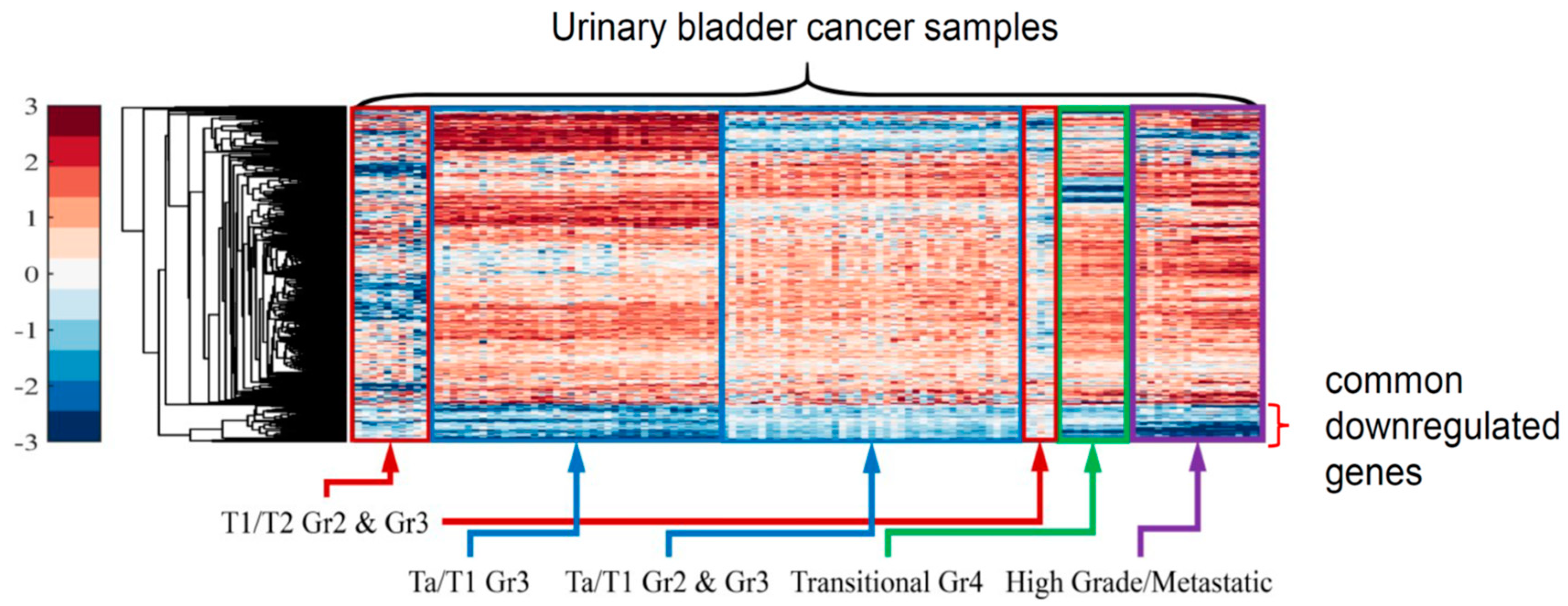
3.1. HCL of DEGs
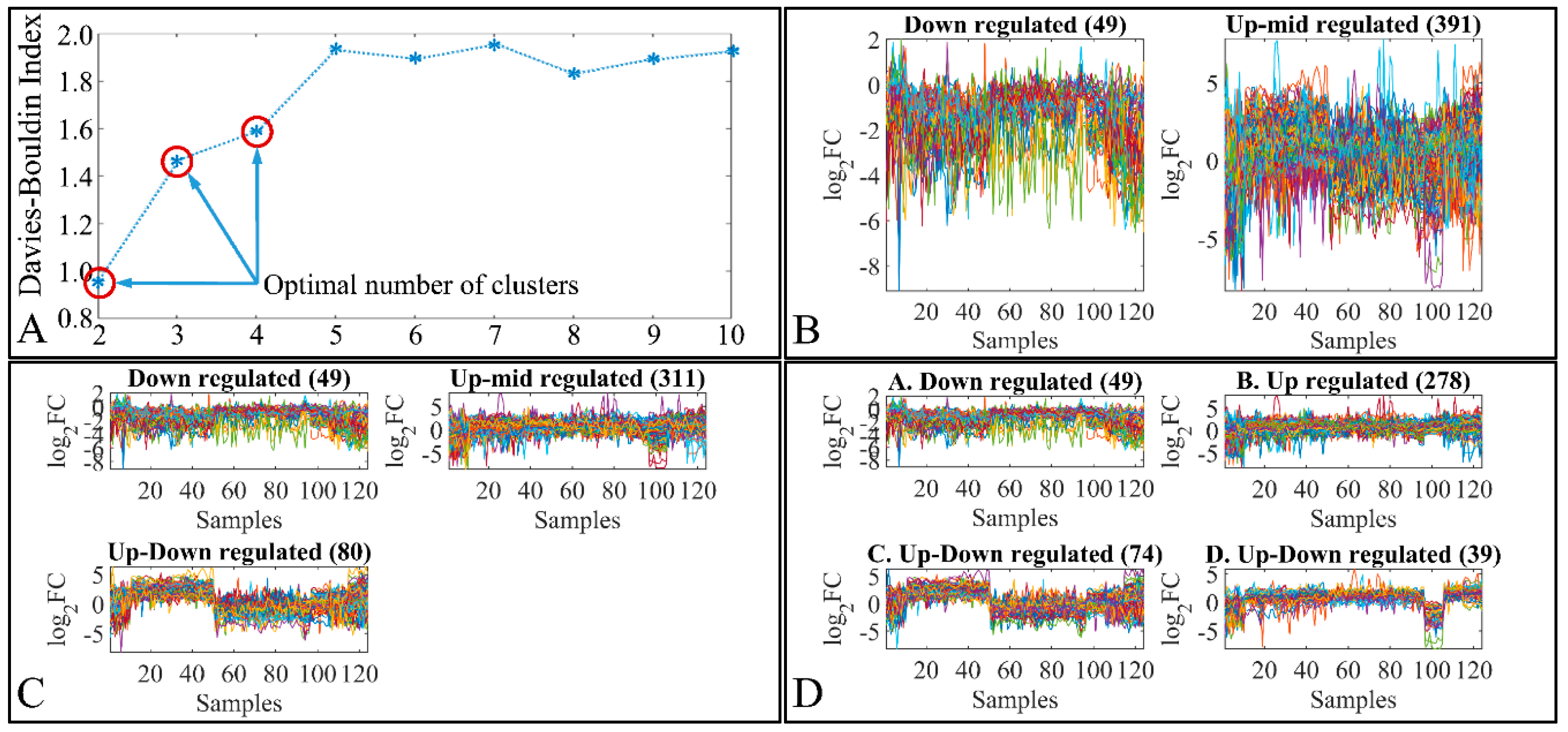
3.2. K-Means of DEGs
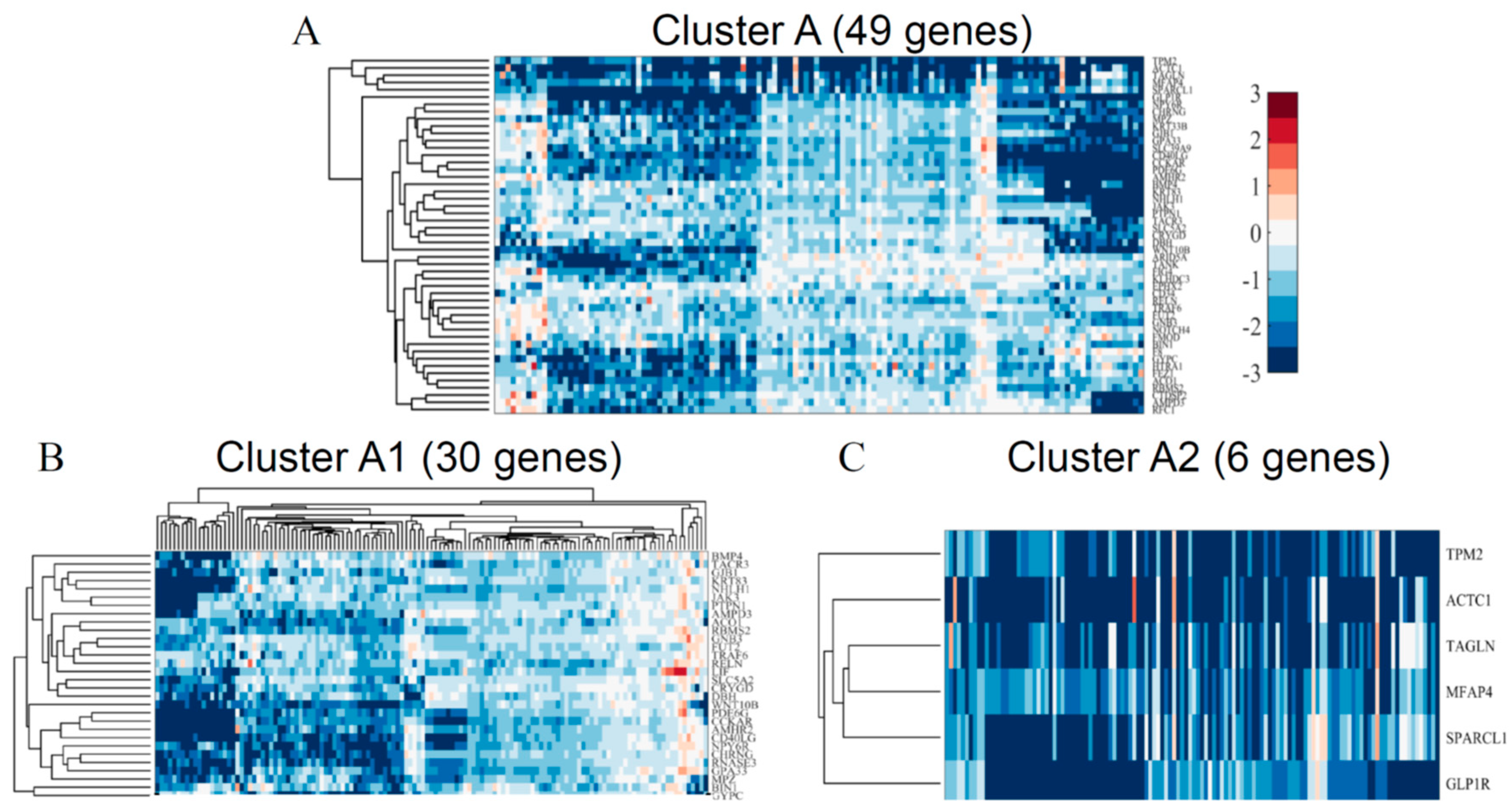
3.2.1. Globally Down-Regulated Genes
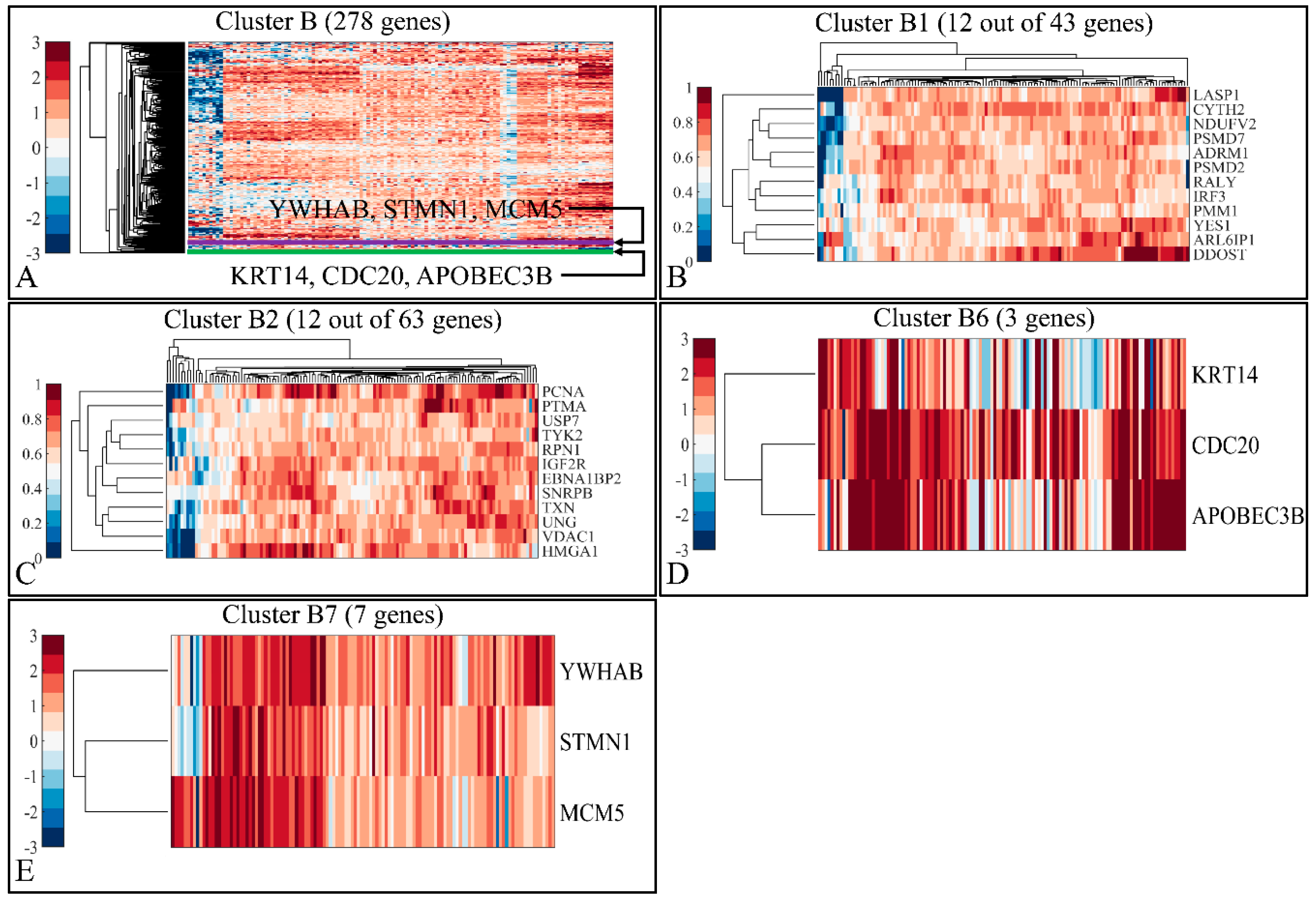
3.2.2. Globally Up-Regulated Genes
3.2.3. Cohort Validation
3.3. Functional Annotation of the DEGs
3.3.1. Functional Annotation of Globally Down-Regulated Genes
3.3.2. Functional Annotation of the Globally Up-Regulated Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bilski, K.; Dobruch, J.; Kozikowski, M.; Skrzypczyk, M.A.; Oszczudlowski, M.; Ostrowski, J. Urobiome in gender-related diversities of bladder cancer. Int. J. Mol. Sci. 2020, 21, 4488. [Google Scholar] [CrossRef] [PubMed]
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder cancer incidence and mortality: A global overview and recent trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Center, M.M.; Jemal, A.; Lortet-Tieulent, J.; Ward, E.; Ferlay, J.; Brawley, O.; Bray, F. International variation in prostate cancer incidence and mortality rates. Eur. Urol. 2012, 61, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Fidler, M.M.; Gupta, S.; Soerjomataram, I.; Ferlay, J.; Steliarova-Foucher, E.; Bray, F. Cancer incidence and mortality among young adults aged 20–39 years worldwide in 2012: A population-based study. Lancet. Oncol. 2017, 18, 1579–1589. [Google Scholar] [CrossRef]
- Babjuk, M.; Bohle, A.; Burger, M.; Capoun, O.; Cohen, D.; Comperat, E.M.; Hernandez, V.; Kaasinen, E.; Palou, J.; Roupret, M.; et al. Eau guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2016. Eur. Urol. 2017, 71, 447–461. [Google Scholar] [CrossRef]
- Dyrskjot, L.; Kruhoffer, M.; Thykjaer, T.; Marcussen, N.; Jensen, J.L.; Moller, K.; Orntoft, T.F. Gene expression in the urinary bladder: A common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004, 64, 4040–4048. [Google Scholar] [CrossRef]
- Kawahara, T.; Kojima, T.; Kandori, S.; Kurobe, M.; Yoshino, T.; Kimura, T.; Nagumo, Y.; Ishituka, R.; Mitsuzuka, K.; Narita, S.; et al. Tp53 codon 72 polymorphism is associated with fgfr3 and ras mutation in non-muscle-invasive bladder cancer. PLoS ONE 2019, 14, e0220173. [Google Scholar] [CrossRef]
- Boulalas, I.; Zaravinos, A.; Karyotis, I.; Delakas, D.; Spandidos, D.A. Activation of ras family genes in urothelial carcinoma. J. Urol. 2009, 181, 2312–2319. [Google Scholar] [CrossRef]
- Elwy, A.E.; Elsaba, T.M.; Elzaher, A.R.A.; Nassar, M.I. Prognostic value of c-myc immunohistochemical expression in muscle invasive urothelial carcinoma of the urinary bladder: A retrospective study. Asian Pac. J. Cancer Prev. Apjcp 2019, 20, 3735–3746. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.M.; Kim, J.S.; Kim, S.; Kim, K.H. Differential expression and clinicopathological significance of her2, indoleamine 2,3-dioxygenase and pd-l1 in urothelial carcinoma of the bladder. J. Clin. Med. 2020, 9, 1265. [Google Scholar] [CrossRef] [PubMed]
- Lipunova, N.; Wesselius, A.; Cheng, K.K.; van Schooten, F.J.; Cazier, J.B.; Bryan, R.T.; Zeegers, M.P. Systematic review: Genetic associations for prognostic factors of urinary bladder cancer. Biomark. Cancer 2019, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.P.B.; Almeida, T.C.; Barros, T.M.B.; Rocha, L.C.M.; Garcia, C.C.M.; da Silva, G.N. Toxicogenetic and antiproliferative effects of chrysin in urinary bladder cancer cells. Mutagenesis 2020. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhou, Q.; Wang, Z.; Zhang, H.; Liu, Z.; Huang, Q.; Wang, J.; Chang, Y.; Bai, Q.; Xia, Y.; et al. Stromal lag-3(+) cells infiltration defines poor prognosis subtype muscle-invasive bladder cancer with immunoevasive contexture. J. Immunother. Cancer 2020, 8, e000651. [Google Scholar] [CrossRef]
- De Martino, M.; Zhuang, D.; Klatte, T.; Rieken, M.; Roupret, M.; Xylinas, E.; Clozel, T.; Krzywinski, M.; Elemento, O.; Shariat, S.F. Impact of erbb2 mutations on in vitro sensitivity of bladder cancer to lapatinib. Cancer Biol. Ther. 2014, 15, 1239–1247. [Google Scholar] [CrossRef]
- Erben, P.; Wezel, F.; Wirtz, R.; Martini, T.; Stein, D.; Weis, C.A.; Hartmann, A.; Bolenz, C. Role of the human erbb family receptors in urothelial carcinoma of the bladder: Mrna expression status and prognostic relevance. Aktuelle Urol. 2017, 48, 356–362. [Google Scholar]
- Gunes, S.; Sullu, Y.; Yegin, Z.; Buyukalpelli, R.; Tomak, L.; Bagci, H. Erbb receptor tyrosine kinase family expression levels in urothelial bladder carcinoma. Pathol. Res. Pract. 2013, 209, 99–104. [Google Scholar] [CrossRef]
- Quackenbush, J. Computational approaches to analysis of DNA microarray data. Yearb. Med. Inform. 2006, 91–103. [Google Scholar]
- Tsai, C.A.; Chen, C.H.; Lee, T.C.; Ho, I.C.; Yang, U.C.; Chen, J.J. Gene selection for sample classifications in microarray experiments. DNA Cell Biol. 2004, 23, 607–614. [Google Scholar] [CrossRef][Green Version]
- Zaravinos, A.; Lambrou, G.I.; Boulalas, I.; Delakas, D.; Spandidos, D.A. Identification of common differentially expressed genes in urinary bladder cancer. PLoS ONE 2011, 6, e18135. [Google Scholar] [CrossRef]
- Zaravinos, A.; Lambrou, G.I.; Volanis, D.; Delakas, D.; Spandidos, D.A. Spotlight on differentially expressed genes in urinary bladder cancer. PLoS ONE 2011, 6, e18255. [Google Scholar] [CrossRef]
- Ramasamy, A.; Mondry, A.; Holmes, C.C.; Altman, D.G. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008, 5, e184. [Google Scholar] [CrossRef] [PubMed]
- Shippy, R.; Sendera, T.J.; Lockner, R.; Palaniappan, C.; Kaysser-Kranich, T.; Watts, G.; Alsobrook, J. Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations. BMC Genom. 2004, 5, 61. [Google Scholar] [CrossRef]
- Yauk, C.L.; Berndt, M.L.; Williams, A.; Douglas, G.R. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004, 32, e124. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Davies, D.L.; Bouldin, D.W. A cluster separation measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, 1, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Schmoyer, D.; Kirov, S.; Snoddy, J. Gotree machine (gotm): A web-based platform for interpreting sets of interesting genes using gene ontology hierarchies. BMC Bioinform. 2004, 5, 16. [Google Scholar]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. Webgestalt 2019: Gene set analysis toolkit with revamped uis and apis. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. Webgestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Wang, J.; Duncan, D.; Shi, Z.; Zhang, B. Web-based gene set analysis toolkit (webgestalt): Update 2013. Nucleic Acids Res. 2013, 41, W77–W83. [Google Scholar] [CrossRef]
- Zhang, B.; Kirov, S.; Snoddy, J. Webgestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef] [PubMed]
- Lambrou, G.I.; Sdraka, M.; Koutsouris, D. The “gene cube”: A novel approach to three-dimensional clustering of gene expression data. Curr. Bioinform. 2019, 14, 721–727. [Google Scholar] [CrossRef]
- Jiang, D.; Tang, C.; Zhang, A. Cluster analysis for gene expression data: A survey. IEEE Trans. Knowl. Data Eng. 2004, 16, 1370–1386. [Google Scholar] [CrossRef]
- Yang, Z.R. Machine Learning Approaches to Bioinformatics; World Scientific: London, UK, 2010; Volume 4. [Google Scholar]
- Zhang, A. Advanced Analysis of Gene Expression Microarray Data; World Scientific: London, UK, 2006. [Google Scholar]
- Madeira, S.C.; Oliveira, A.L. Biclustering algorithms for biological data analysis: A survey. IEEE/ACM Trans. Comput. Biol. Bioinform. 2004, 1, 24–45. [Google Scholar] [CrossRef] [PubMed]
- Kluger, Y.; Basri, R.; Chang, J.T.; Gerstein, M. Spectral biclustering of microarray data: Coclustering genes and conditions. Genome Res. 2003, 13, 703–716. [Google Scholar] [CrossRef]
- Yin, L.; Huang, C.H.; Ni, J. Clustering of gene expression data: Performance and similarity analysis. BMC Bioinform. 2006, 7 (Suppl. 4), S19. [Google Scholar] [CrossRef]
- D’Haeseleer, P. How does gene expression clustering work? Nat. Biotechnol. 2005, 23, 1499–1501. [Google Scholar] [CrossRef]
- Mahanta, P.; Ahmed, H.A.; Bhattacharyya, D.K.; Kalita, J.K. Triclustering in Gene Expression Data Analysis: A Selected Survey. In Emerging Trends and Applications in Computer Science (NCETACS), Proceedings of the 2011 2nd National Conference, Meghalaya, India, 4–5 March 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 1–6. [Google Scholar]
- Zhao, L.; Zaki, M.J. Tricluster: An effective algorithm for mining coherent clusters in 3d microarray data. In Proceedings of the 2005 ACM SIGMOD International Conference on Management of Data, Baltimore, MA, USA, 14–16 June 2005; pp. 694–705. [Google Scholar]
- Bhar, A.; Haubrock, M.; Mukhopadhyay, A.; Maulik, U.; Bandyopadhyay, S.; Wingender, E. Coexpression and coregulation analysis of time-series gene expression data in estrogen-induced breast cancer cell. Algorithms Mol. Biol. AMB 2013, 8, 9. [Google Scholar] [CrossRef]
- Ciaramella, A.; Cocozza, S.; Iorio, F.; Miele, G.; Napolitano, F.; Pinelli, M.; Raiconi, G.; Tagliaferri, R. Interactive data analysis and clustering of genomic data. Neural Netw. 2008, 21, 368–378. [Google Scholar] [CrossRef]
- Gutierrez, A.D.; Rubio-Escudero, C.; Riquelme, J.C. Triclustering on temporary microarray data using the trigen algorithm. In Intelligent Systems Design and Applications (ISDA), Proceedings of the 2011 11th International Conference, Graz, Austria, 22–24 November 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 877–881. [Google Scholar]
- Araújo, R.B.; Ferreira, G.H.T.; Orair, G.H.; Meira, W., Jr.; Ferreira, R.A.C.; Neto, D.O.G.; Zaki, M.J. The partricluster algorithm for gene expression analysis. Int. J. Parallel Program. 2008, 36, 226–249. [Google Scholar] [CrossRef]
- Jiang, D.; Pei, J.; Ramanathan, M.; Tang, C.; Zhang, A. Mining coherent gene clusters from gene-sample-time microarray data. In Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Washington, DC, USA, 22–25 August 2004; ACM: Seattle, WA, USA, 2004; pp. 430–439. [Google Scholar]
- Tchagang, A.B.; Phan, S.; Famili, F.; Shearer, H.; Fobert, P.; Huang, Y.; Zou, J.; Huang, D.; Cutler, A.; Liu, Z.; et al. Mining biological information from 3d short time-series gene expression data: The optricluster algorithm. BMC Bioinform. 2012, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Mankad, S.; Michailidis, G. Biclustering three-dimensional data arrays with plaid models. J. Comput. Graph. Stat. 2014, 23, 943–965. [Google Scholar] [CrossRef][Green Version]
- Li, A.; Tuck, D. An effective tri-clustering algorithm combining expression data with gene regulation information. Gene Regul. Syst. Biol. 2009, 3, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Murty, M.N.; Flynn, P.J. Data clustering: A review. Acm Comput. Surv. (Csur) 1999, 31, 264–323. [Google Scholar] [CrossRef]
- Jain, A.K. Data clustering: 50 years beyond k-means. Pattern Recognit. Lett. 2010, 31, 651–666. [Google Scholar] [CrossRef]
- Lu, C.; Ghoman, S.K.; Cutumisu, M.; Schmölzer, G.M. Unsupervised machine learning algorithms examine healthcare providers’ perceptions and longitudinal performance in a digital neonatal resuscitation simulator. Front. Pediatrics 2020, 8, 544. [Google Scholar] [CrossRef]
- Maleki, F.; Ovens, K.; Najafian, K.; Forghani, B.; Reinhold, C.; Forghani, R. Overview of machine learning part 1: Fundamentals and classic approaches. Neuroimaging Clin. N. Am. 2020, 30, e17–e32. [Google Scholar] [CrossRef]
- Qi, G.J.; Zhang, L.; Lin, F.; Wang, X. Learning generalized transformation equivariant representations via autoencoding transformations. IEEE Trans. Pattern Anal. Mach. Intell. 2020. [Google Scholar] [CrossRef]
- Rabaglino, M.B.; Kadarmideen, H.N. Machine learning approach to integrated endometrial transcriptomic datasets reveals biomarkers predicting uterine receptivity in cattle at seven days after estrous. Sci. Rep. 2020, 10, 16981. [Google Scholar] [CrossRef]
- Breheny, P.; Stromberg, A.; Lambert, J. P-value histograms: Inference and diagnostics. High-Throughput 2018, 7, 23. [Google Scholar] [CrossRef]
- Dimitri, G.M.; Spasov, S.; Duggento, A.; Passamonti, L.; Lio, P.; Toschi, N. Unsupervised stratification in neuroimaging through deep latent embeddings. In Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Proceedings of the IEEE Engineering in Medicine and Biology Society, Annual International Conference, Milano, Italy, 20 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1568–1571. [Google Scholar]
- Praiss, A.M.; Huang, Y.; St Clair, C.M.; Tergas, A.I.; Melamed, A.; Khoury-Collado, F.; Hou, J.Y.; Hu, J.; Hur, C.; Hershman, D.L.; et al. Using machine learning to create prognostic systems for endometrial cancer. Gynecol. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Gole, R.; Ghosh, S.; Basu, A. Alternative techniques for breast tumour detection using ultrasound. In Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Proceedings of the IEEE Engineering in Medicine and Biology Society, Annual International Conference, Milano, Italy, 20 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 2047–2050. [Google Scholar]
- Zhu, Z.; Cao, Y.; Qin, C.; Rao, Y.; Ni, D.; Wang, Y. Unsupervised 3d end-to-end deformable network for brain mri registration. In Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Proceedings of the IEEE Engineering in Medicine and Biology Society, Annual International Conference, Milano, Italy, 20 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1355–1359. [Google Scholar]
- Arbelaitz, O.; Gurrutxaga, I.; Muguerza, J.; Perez, J.M.; Perona, I. An extensive comparative study of cluster validity indices. Pattern Recognit. 2013, 46, 243–256. [Google Scholar] [CrossRef]
- Paul, D.; Ghorai, S.; Dinesh, U.S.; Shetty, P.; Chattopadhyay, S.; Santra, M.K. Cdc20 directs proteasome-mediated degradation of the tumor suppressor smar1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis. 2017, 8, e2882. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, Y.; Lee, J.H.; Kim, Y.S. High expression of spindle assembly checkpoint proteins cdc20 and mad2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. Int. J. Pathol. 2013, 463, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, G.; Li, J.; Li, J.; Ruan, N.; Ma, L.; Han, X.; Wei, Y.; Li, L.; Zhang, H.; et al. Screening and identification of key biomarkers for bladder cancer: A study based on tcga and geo data. Biomed Res. Int. 2020, 2020, 8283401. [Google Scholar] [CrossRef] [PubMed]
- Kidokoro, T.; Tanikawa, C.; Furukawa, Y.; Katagiri, T.; Nakamura, Y.; Matsuda, K. Cdc20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 2008, 27, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Wan, L.; Zhou, X.; Wang, Z.; Wei, W. Targeting cdc20 as a novel cancer therapeutic strategy. Pharmacol. Ther. 2015, 151, 141–151. [Google Scholar] [CrossRef]
- Dudderidge, T.; Stockley, J.; Nabi, G.; Mom, J.; Umez-Eronini, N.; Hrouda, D.; Cresswell, J.; McCracken, S.R.C. A novel, non-invasive test enabling bladder cancer detection in urine sediment of patients presenting with haematuria-a prospective multicentre performance evaluation of adxbladder. Eur. Urol. Oncol. 2020, 3, 42–46. [Google Scholar] [CrossRef]
- Stoeber, K.; Swinn, R.; Prevost, A.T.; de Clive-Lowe, P.; Halsall, I.; Dilworth, S.M.; Marr, J.; Turner, W.H.; Bullock, N.; Doble, A.; et al. Diagnosis of genito-urinary tract cancer by detection of minichromosome maintenance 5 protein in urine sediments. J. Natl. Cancer Instig. 2002, 94, 1071–1079. [Google Scholar] [CrossRef]
- Kelly, J.D.; Dudderidge, T.J.; Wollenschlaeger, A.; Okoturo, O.; Burling, K.; Tulloch, F.; Halsall, I.; Prevost, T.; Prevost, A.T.; Vasconcelos, J.C.; et al. Bladder cancer diagnosis and identification of clinically significant disease by combined urinary detection of mcm5 and nuclear matrix protein 22. PLoS ONE 2012, 7, e40305. [Google Scholar] [CrossRef] [PubMed]
- Proctor, I.; Stoeber, K.; Williams, G.H. Biomarkers in bladder cancer. Histopathology 2010, 57, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Butt, E.; Ebbing, J.; Bubendorf, L.; Ardelt, P. Influence of hematuria and infection on diagnostic accuracy of urinary lasp1: A new biomarker for bladder carcinoma. Biomark. Med. 2017, 11, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Ardelt, P.; Grünemay, N.; Strehl, A.; Jilg, C.; Miernik, A.; Kneitz, B.; Butt, E. Lasp-1, a novel urinary marker for detection of bladder cancer. Urol. Oncol. 2013, 31, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Enokida, H.; Kawakami, K.; Tatarano, S.; Uchida, Y.; Kawahara, K.; Nishiyama, K.; Seki, N.; Nakagawa, M. Functional role of lasp1 in cell viability and its regulation by micrornas in bladder cancer. Urol. Oncol. 2012, 30, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, L.; Zang, Y.; Xu, Z. Identification of core genes and key pathways via integrated analysis of gene expression and DNA methylation profiles in bladder cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 3024–3033. [Google Scholar] [CrossRef]
- Rubin, C.I.; Atweh, G.F. The role of stathmin in the regulation of the cell cycle. J. Cell. Biochem. 2004, 93, 242–250. [Google Scholar] [CrossRef]
- Biaoxue, R.; Xiguang, C.; Hua, L.; Shuanying, Y. Stathmin-dependent molecular targeting therapy for malignant tumor: The latest 5 years’ discoveries and developments. J. Transl. Med. 2016, 14, 279. [Google Scholar] [CrossRef]
- Hemdan, T.; Linden, M.; Lind, S.B.; Namuduri, A.V.; Sjostedt, E.; de Stahl, T.D.; Asplund, A.; Malmstrom, P.U.; Segersten, U. The prognostic value and therapeutic target role of stathmin-1 in urinary bladder cancer. Br. J. Cancer 2014, 111, 1180–1187. [Google Scholar] [CrossRef]
- Battista, S.; Fidanza, V.; Fedele, M.; Klein-Szanto, A.J.; Outwater, E.; Brunner, H.; Santoro, M.; Croce, C.M.; Fusco, A. The expression of a truncated hmgi-c gene induces gigantism associated with lipomatosis. Cancer Res. 1999, 59, 4793–4797. [Google Scholar]
- Chen, X.; Liu, M.; Meng, F.; Sun, B.; Jin, X.; Jia, C. The long noncoding rna hif1a-as2 facilitates cisplatin resistance in bladder cancer. J. Cell. Biochem. 2019, 120, 243–252. [Google Scholar] [CrossRef]
- Lin, R.; Shen, W.; Zhi, Y.; Zhou, Z. Prognostic value of mir-26a and hmga1 in urothelial bladder cancer. Biomed. Pharm. 2014, 68, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, H.; Hu, Z.; Mao, Y.; Xu, X.; Zhu, Y.; Xu, X.; Wu, J.; Li, S.; Mao, Q.; et al. Mir-26a inhibits proliferation and motility in bladder cancer by targeting hmga1. FEBS Lett. 2013, 587, 2467–2473. [Google Scholar] [CrossRef]
- Qin, M.M.; Chai, X.; Huang, H.B.; Feng, G.; Li, X.N.; Zhang, J.; Zheng, R.; Liu, X.C.; Pu, C. Let-7i inhibits proliferation and migration of bladder cancer cells by targeting hmga1. BMC Urol. 2019, 19, 53. [Google Scholar] [CrossRef]
- Iida, K.; Naiki, T.; Naiki-Ito, A.; Suzuki, S.; Kato, H.; Nozaki, S.; Nagai, T.; Etani, T.; Nagayasu, Y.; Ando, R.; et al. Luteolin suppresses bladder cancer growth via regulation of mechanistic target of rapamycin pathway. Cancer Sci. 2020, 111, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.S.; Jou, Y.C.; Tsai, H.T.; Shiau, A.L.; Wu, C.L.; Tzai, T.S. Prothymosin-α enhances phosphatase and tensin homolog expression and binds with tripartite motif-containing protein 21 to regulate kelch-like ech-associated protein 1/nuclear factor erythroid 2-related factor 2 signaling in human bladder cancer. Cancer Sci. 2019, 110, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.B.; Zhou, L.B.; Wang, H.F.; Li, G.; Xie, Q.P.; Hu, B. Loss of igf2r indicates a poor prognosis and promotes cell proliferation and tumorigenesis in bladder cancer via akt signaling pathway. Neoplasma 2020, 67, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Nord, H.; Segersten, U.; Sandgren, J.; Wester, K.; Busch, C.; Menzel, U.; Komorowski, J.; Dumanski, J.P.; Malmström, P.U.; de Ståhl, T.D. Focal amplifications are associated with high grade and recurrences in stage ta bladder carcinoma. Int. J. Cancer 2010, 126, 1390–1402. [Google Scholar] [CrossRef]
- Morra, F.; Merolla, F.; Criscuolo, D.; Insabato, L.; Giannella, R.; Ilardi, G.; Cerrato, A.; Visconti, R.; Staibano, S.; Celetti, A. Ccdc6 and usp7 expression levels suggest novel treatment options in high-grade urothelial bladder cancer. J. Exp. Clin. Cancer Res. 2019, 38, 90. [Google Scholar] [CrossRef]
- Varol, N.; Konac, E.; Bilen, C.Y. Does wnt/β-catenin pathway contribute to the stability of dnmt1 expression in urological cancer cell lines? Exp. Biol. Med. 2015, 240, 624–630. [Google Scholar] [CrossRef]
- Agarwal, P.; Sen, A.K.; Bhardwaj, M.; Dinand, V.; Ahuja, A.; Sood, R. Study of proliferating cell nuclear antigen expression and angiogenesis in urothelial neoplasms: Correlation with tumor grade and stage. Urol. Ann. 2018, 10, 209–214. [Google Scholar] [CrossRef]
- Almeida, T.C.; Guerra, C.C.C.; De Assis, B.L.G.; de Oliveira, R.D.A.S.; Garcia, C.C.M.; Lima, A.A.; da Silva, G.N. Antiproliferative and toxicogenomic effects of resveratrol in bladder cancer cells with different tp53 status. Environ. Mol. Mutagenesis 2019, 60, 740–751. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, P.; Wang, S.; Li, J.; Ou, T.; Zeng, X. Ciz1 knockdown suppresses the proliferation of bladder cancer cells by inducing apoptosis. Gene 2019, 719, 143946. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Deng, Z.; Zhou, Z.; Jiang, C.Y.; Zhao, R.Z.; Sun, F.; Cui, D.; Bei, X.Y.; Yang, B.Y.; Sun, Q.; et al. Qki-6 inhibits bladder cancer malignant behaviours through down-regulating e2f3 and nf-κb signalling. J. Cell. Mol. Med. 2019, 23, 6578–6594. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; He, Z.; Lei, H.; Chen, Y.; Lu, Z.; Zeng, G.; Wang, H. Identification of differentially expressed genes and biological pathways in bladder cancer. Mol. Med. Rep. 2018, 17, 6425–6434. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, B.; Sivakumar, A.; Sundaresan, S. Diindolylmethane and lupeol modulates apoptosis and cell proliferation in n-butyl-n-(4-hydroxybutyl) nitrosamine initiated and dimethylarsinic acid promoted rat bladder carcinogenesis. Pathol. Oncol. Res. 2016, 22, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, F.T.; Chade, D.C.; Reis, S.T.; Piantino, C.; Dall’Oglio, M.F.; Srougi, M.; Leite, K.R. Curcumin, but not prima-1, decreased tumor cell proliferation in the syngeneic murine orthotopic bladder tumor model. Clinics 2011, 66, 2121–2124. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Xing, S.; Lin, C.; Zhang, X.; Fu, M.; Liang, X.; Zeng, F.; Lu, G.; Wu, M. Bladder cancer therapy using combined proliferating cell nuclear antigen antisense oligonucleotides and recombinant adenovirus p53. Chin. Med. J. 2003, 116, 1860–1863. [Google Scholar]
- Burns, M.B.; Temiz, N.A.; Harris, R.S. Evidence for apobec3b mutagenesis in multiple human cancers. Nat. Genet. 2013, 45, 977–983. [Google Scholar] [CrossRef]
- Christofi, T.; Zaravinos, A. Rna editing in the forefront of epitranscriptomics and human health. J. Transl. Med. 2019, 17, 319. [Google Scholar] [CrossRef]
- Yu, Q.; Chen, D.; König, R.; Mariani, R.; Unutmaz, D.; Landau, N.R. Apobec3b and apobec3c are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 2004, 279, 53379–53386. [Google Scholar] [CrossRef]
- Delebecque, F.; Suspène, R.; Calattini, S.; Casartelli, N.; Saïb, A.; Froment, A.; Wain-Hobson, S.; Gessain, A.; Vartanian, J.P.; Schwartz, O. Restriction of foamy viruses by apobec cytidine deaminases. J. Virol. 2006, 80, 605–614. [Google Scholar] [CrossRef]
- Zielonka, J.; Bravo, I.G.; Marino, D.; Conrad, E.; Perković, M.; Battenberg, M.; Cichutek, K.; Münk, C. Restriction of equine infectious anemia virus by equine apobec3 cytidine deaminases. J. Virol. 2009, 83, 7547–7559. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Bishop, K.N.; Sheehy, A.M.; Craig, H.M.; Petersen-Mahrt, S.K.; Watt, I.N.; Neuberger, M.S.; Malim, M.H. DNA deamination mediates innate immunity to retroviral infection. Cell 2003, 113, 803–809. [Google Scholar] [CrossRef]
- Zou, J.; Wang, C.; Ma, X.; Wang, E.; Peng, G. Apobec3b, a molecular driver of mutagenesis in human cancers. Cell Biosci. 2017, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Kuong, K.J.; Loeb, L.A. Apobec3b mutagenesis in cancer. Nat. Genet. 2013, 45, 964–965. [Google Scholar] [CrossRef] [PubMed]
- Periyasamy, M.; Singh, A.K.; Gemma, C.; Kranjec, C.; Farzan, R.; Leach, D.A.; Navaratnam, N.; Palinkas, H.L.; Vertessy, B.G.; Fenton, T.R.; et al. P53 controls expression of the DNA deaminase apobec3b to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res. 2017, 45, 11056–11069. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Shirakawa, K.; Yokoyama, M.; Fukuda, H.; Sarca, A.D.; Koyabu, S.; Yamazaki, H.; Kazuma, Y.; Matsui, H.; Maruyama, W.; et al. Protein kinase a inhibits tumor mutator apobec3b through phosphorylation. Sci. Rep. 2019, 9, 8307. [Google Scholar] [CrossRef]
- Vasudevan, A.A.J.; Kreimer, U.; Schulz, W.A.; Krikoni, A.; Schumann, G.G.; Haussinger, D.; Munk, C.; Goering, W. Apobec3b activity is prevalent in urothelial carcinoma cells and only slightly affected by line-1 expression. Front. Microbiol. 2018, 9, 2088. [Google Scholar] [CrossRef]
- Paraskevopoulou, V.; Papafotiou, G.; Klinakis, A. Krt14 marks bladder progenitors. Cell Cycle 2016, 15, 3161–3162. [Google Scholar] [CrossRef]
- Papafotiou, G.; Paraskevopoulou, V.; Vasilaki, E.; Kanaki, Z.; Paschalidis, N.; Klinakis, A. Krt14 marks a subpopulation of bladder basal cells with pivotal role in regeneration and tumorigenesis. Nat. Commun. 2016, 7, 11914. [Google Scholar] [CrossRef]
- Volkmer, J.P.; Sahoo, D.; Chin, R.K.; Ho, P.L.; Tang, C.; Kurtova, A.V.; Willingham, S.B.; Pazhanisamy, S.K.; Contreras-Trujillo, H.; Storm, T.A.; et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc. Natl. Acad. Sci. USA 2012, 109, 2078–2083. [Google Scholar] [CrossRef] [PubMed]



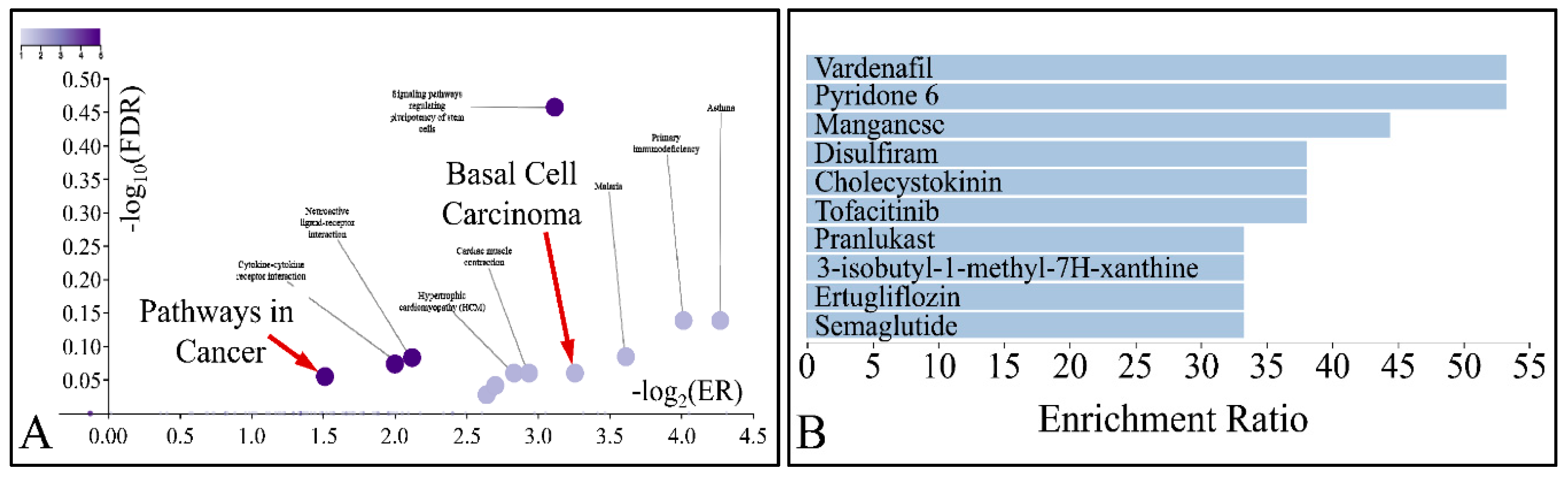

| Inv. | Gene Name | Cluster | n | (%) |
|---|---|---|---|---|
| 1 | ACO1 | A1 | 128 | 97.71% |
| 2 | CCKAR | A1 | 128 | 97.71% |
| 3 | JAK3 | A1 | 128 | 97.71% |
| 4 | GLP1R | A2 | 127 | 96.95% |
| 5 | RNASE3 | A1 | 127 | 96.95% |
| 6 | GJB1 | A1 | 126 | 96.18% |
| 7 | KRT83 | A1 | 126 | 96.18% |
| 8 | MPZ | A1 | 126 | 96.18% |
| 9 | PDE6G | A1 | 126 | 96.18% |
| 10 | TPM2 | A2 | 126 | 96.18% |
| 11 | AMHR2 | A1 | 125 | 95.42% |
| 12 | CD40LG | A1 | 125 | 95.42% |
| 13 | GYPC | A1 | 125 | 95.42% |
| 14 | PTPN1 | A1 | 125 | 95.42% |
| 15 | SLC5A2 | A1 | 124 | 94.66% |
| 16 | WNT10B | A1 | 124 | 94.66% |
| 17 | MFAP4 | A2 | 123 | 93.89% |
| 18 | TAGLN | A2 | 123 | 93.89% |
| 19 | AMPD3 | A1 | 122 | 93.13% |
| 20 | CHRNG | A1 | 122 | 93.13% |
| 21 | GNB3 | A1 | 122 | 93.13% |
| 22 | NHLH1 | A1 | 122 | 93.13% |
| 23 | ACTC1 | A2 | 121 | 92.37% |
| 24 | DBH | A1 | 121 | 92.37% |
| 25 | FUT2 | A1 | 121 | 92.37% |
| 26 | NPY6R | A1 | 121 | 92.37% |
| 27 | TACR3 | A1 | 121 | 92.37% |
| 28 | BMP4 | A1 | 120 | 91.60% |
| 29 | GPA33 | A1 | 120 | 91.60% |
| 30 | RBMS2 | A1 | 120 | 91.60% |
| 31 | RELN | A1 | 120 | 91.60% |
| 32 | TRAF6 | A1 | 120 | 91.60% |
| 33 | BIN1 | A1 | 119 | 90.84% |
| 34 | CRYGD | A1 | 119 | 90.84% |
| 35 | LIF | A1 | 119 | 90.84% |
| 36 | SPARCL1 | A2 | 117 | 89.31% |
| Inv. | Gene Name | Cluster | n | (%) |
|---|---|---|---|---|
| 1 | YES1 | B1 | 125 | 95.42% |
| 2 | PMM1 | B1 | 123 | 93.89% |
| 3 | CDC20 | B6 | 123 | 93.89% |
| 4 | IRF3 | B1 | 122 | 93.13% |
| 5 | MCM5 | B7 | 122 | 93.13% |
| 6 | ARL6IP1 | B1 | 121 | 92.37% |
| 7 | CYTH2 | B1 | 121 | 92.37% |
| 8 | LASP1 | B1 | 121 | 92.37% |
| 9 | PSMD2 | B1 | 121 | 92.37% |
| 10 | PSMD7 | B1 | 121 | 92.37% |
| 11 | DDOST | B1 | 120 | 91.60% |
| 12 | NDUFV2 | B1 | 120 | 91.60% |
| 13 | RALY | B1 | 120 | 91.60% |
| 14 | STMN1 | B7 | 120 | 91.60% |
| 15 | ADRM1 | B1 | 119 | 90.84% |
| 16 | HMGA1 | B2 | 119 | 90.84% |
| 17 | RPN1 | B2 | 119 | 90.84% |
| 18 | TXN | B2 | 119 | 90.84% |
| 19 | TYK2 | B2 | 119 | 90.84% |
| 20 | UNG | B2 | 119 | 90.84% |
| 21 | EBNA1BP2 | B2 | 118 | 90.08% |
| 22 | IGF2R | B2 | 118 | 90.08% |
| 23 | PCNA | B2 | 118 | 90.08% |
| 24 | PTMA | B2 | 118 | 90.08% |
| 25 | SNRPB | B2 | 118 | 90.08% |
| 26 | USP7 | B2 | 118 | 90.08% |
| 27 | VDAC1 | B2 | 118 | 90.08% |
| 28 | YWHAB | B7 | 118 | 90.08% |
| 29 | APOBEC3B | B6 | 106 | 80.92% |
| 30 | KRT14 | B6 | 96 | 73.28% |
| ID | Source | Term | Term ID | Padj | Genes |
|---|---|---|---|---|---|
| 1 | GO:MF | protein binding | GO:0005515 | 0.009515 | YES1,PMM1,CDC20,IRF3,MCM5,ARL6IP1,CYTH2,LASP1,PSMD2,PSMD7,DDOST,NDUFV2,RALY,STMN1,ADRM1,HMGA1,RPN1,TXN,TYK2,UNG,EBNA1BP2,IGF2R,PCNA,PTMA,SNRPB,USP7,VDAC1,YWHAB,APOBEC3B,KRT14 |
| 2 | GO:MF | dolichyl-diphosphooligosaccharide-protein glycotransferase activity | GO:0004579 | 0.019061 | DDOST,RPN1 |
| 3 | GO:MF | oligosaccharyl transferase activity | GO:0004576 | 0.025388 | DDOST,RPN1 |
| 4 | GO:MF | protein-containing complex binding | GO:0044877 | 0.044508 | CDC20,LASP1,ADRM1,UNG,PCNA,SNRPB,VDAC1,YWHAB,KRT14 |
| 5 | GO:BP | viral process | GO:0016032 | 0.021014 | IRF3,HMGA1,UNG,IGF2R,PCNA,USP7,VDAC1,YWHAB,APOBEC3B |
| GO:BP | interspecies interaction between organisms | GO:0044419 | 0.031201 | IRF3,PSMD2,PSMD7,STMN1,HMGA1,TYK2,UNG,IGF2R,PCNA,USP7,VDAC1,YWHAB,APOBEC3B | |
| 6 | GO:BP | symbiotic process | GO:0044403 | 0.034472 | IRF3,HMGA1,UNG,IGF2R,PCNA,USP7,VDAC1,YWHAB,APOBEC3B |
| 7 | GO:CC | proteasome regulatory particle | GO:0005838 | 0.001241 | PSMD2,PSMD7,ADRM1 |
| 8 | GO:CC | cytosol | GO:0005829 | 0.001839 | YES1,PMM1,CDC20,IRF3,MCM5,ARL6IP1,CYTH2,PSMD2,PSMD7,STMN1,ADRM1,HMGA1,RPN1,TXN,TYK2,PTMA,SNRPB,USP7,YWHAB,KRT14 |
| GO:CC | proteasome accessory complex | GO:0022624 | 0.001847 | PSMD2,PSMD7,ADRM1 | |
| 9 | GO:CC | catalytic complex | GO:1902494 | 0.007993 | CDC20,PSMD2,PSMD7,DDOST,NDUFV2,RALY,ADRM1,RPN1,PCNA,SNRPB |
| 10 | GO:CC | intracellular organelle | GO:0043229 | 0.035435 | YES1,CDC20,IRF3,MCM5,ARL6IP1,CYTH2,LASP1,PSMD2,PSMD7,DDOST,NDUFV2,RALY,STMN1,ADRM1,HMGA1,RPN1,TXN,TYK2,UNG,EBNA1BP2,IGF2R,PCNA,PTMA,SNRPB,USP7,VDAC1,YWHAB,APOBEC3B,KRT14 |
| 11 | GO:CC | proteasome complex | GO:0000502 | 0.036772 | PSMD2,PSMD7,ADRM1 |
| 12 | GO:CC | endopeptidase complex | GO:1905369 | 0.038428 | PSMD2,PSMD7,ADRM1 |
| 13 | GO:CC | oligosaccharyltransferase complex | GO:0008250 | 0.042715 | DDOST,RPN1 |
| 14 | GO:CC | intracellular membrane-bounded organelle | GO:0043231 | 0.043905 | YES1,CDC20,IRF3,MCM5,ARL6IP1,CYTH2,PSMD2,PSMD7,DDOST,NDUFV2,RALY,ADRM1,HMGA1,RPN1,TXN,TYK2,UNG,EBNA1BP2,IGF2R,PCNA,PTMA,SNRPB,USP7,VDAC1,YWHAB,APOBEC3B,KRT14 |
| 15 | KEGG | Proteasome | KEGG:03050 | 0.022187 | PSMD2,PSMD7,ADRM1 |
| 16 | KEGG | Cell cycle | KEGG:04110 | 0.031652 | CDC20,MCM5,PCNA,YWHAB |
| 17 | REAC | Ub-specific processing proteases | REAC:R-HSA-5689880 | 0.002821 | CDC20,PSMD2,PSMD7,ADRM1,USP7,VDAC1 |
| 18 | WP | Cell Cycle | WP:WP179 | 0.029044 | CDC20,MCM5,PCNA,YWHAB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lambrou, G.I.; Vichos, K.; Koutsouris, D.; Zaravinos, A. Identification of Co-Deregulated Genes in Urinary Bladder Cancer Using High-Throughput Methodologies. Appl. Sci. 2021, 11, 1785. https://doi.org/10.3390/app11041785
Lambrou GI, Vichos K, Koutsouris D, Zaravinos A. Identification of Co-Deregulated Genes in Urinary Bladder Cancer Using High-Throughput Methodologies. Applied Sciences. 2021; 11(4):1785. https://doi.org/10.3390/app11041785
Chicago/Turabian StyleLambrou, George I., Kleanthis Vichos, Dimitrios Koutsouris, and Apostolos Zaravinos. 2021. "Identification of Co-Deregulated Genes in Urinary Bladder Cancer Using High-Throughput Methodologies" Applied Sciences 11, no. 4: 1785. https://doi.org/10.3390/app11041785
APA StyleLambrou, G. I., Vichos, K., Koutsouris, D., & Zaravinos, A. (2021). Identification of Co-Deregulated Genes in Urinary Bladder Cancer Using High-Throughput Methodologies. Applied Sciences, 11(4), 1785. https://doi.org/10.3390/app11041785








