Kombucha Tea as a Reservoir of Cellulose Producing Bacteria: Assessing Diversity among Komagataeibacter Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Kombucha Preparation, Sampling and Physico-Chemical Determinations
2.2. Plating and Isolation
2.3. Phenotypic Characterization
2.4. Genomic DNA Extraction, RFLP and Amplification of (GTG)5/REP-PCR and ERIC Elements
2.5. Statistics and Clustering Analysis of (GTG)5 and ERIC Rep-PCR Patterns
3. Results
3.1. Kombucha Characteristics and Isolated Strains
3.2. Diversity of Acetic Acid Bacteria from Kombucha Tea
3.3. Improving the Fingerprinting of Komagataeibacter Strains
3.4. Correlation between Strains and Cellulose Yield
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Robotti, F.; Bottan, S.; Fraschetti, F.; Mallone, A.; Pellegrini, G.; Lindenblatt, N.; Starck, C.; Falk, V.; Poulikakos, D.; Ferrari, A. A micron-scale surface topography design reducing cell adhesion to implanted materials. Sci. Rep. 2018. [Google Scholar] [CrossRef]
- Haghighi, H.; Gullo, M.; La China, S.; Pfeifer, F.; Siesler, H.W.; Licciardello, F.; Pulvirenti, A. Characterization of bio-nanocomposite films based on gelatin/polyvinyl alcohol blend reinforced with bacterial cellulose nanowhiskers for food packaging applications. Food Hydrocoll. 2020, 106454. [Google Scholar] [CrossRef]
- Barbi, S.; Taurino, C.; La China, S.; Anguluri, K.; Gullo, M.; Montorsi, M. Mechanical and structural properties of environmental green composites based on functionalized bacterial cellulose. Cellulose 2021, 1–7. [Google Scholar] [CrossRef]
- Valera, M.J.; Torija, M.J.; Mas, A. Detrimental effects of acetic acid bacteria in foods. In Acetic Acid Bacteria: Fundamentals and Food Applications, 1st ed.; Sengun, I., Ed.; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Boca Raton, FL, USA, 2017; Volume 10, pp. 299–320. [Google Scholar]
- Gullo, M.; La China, S.; Falcone, P.M.; Giudici, P. Biotechnological production of cellulose by acetic acid bacteria: Current state and perspectives. Appl. Microbiol. Biotechnol. 2018, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Son, H.J.; Heo, M.S.; Kim, Y.G.; Lee, S.J. Optimization of fermentation conditions for the production of bacterial cellulose by a newly isolated Acetobacter sp. A9 in shaking cultures. Biotechnol. Appl. Biochem. 2001, 33, 1. [Google Scholar] [CrossRef] [PubMed]
- La China, S.; Zanichelli, G.; De Vero, L.; Gullo, M. Oxidative fermentations and exopolysaccharides production by acetic acid bacteria: A mini review. Biotechnol. Lett. 2018, 40, 1289–1302. [Google Scholar] [CrossRef]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Laureys, D.; Britton, S.J.; De Clippeleer, J. Kombucha tea fermentation: A review. J. Am. Soc. Brew. Chem. 2020, 78, 165–174. [Google Scholar] [CrossRef]
- Sievers, M.; Lanini, C.; Weber, A.; Schuler-Schmid, U.; Teuber, M. Microbiology and fermentation balance in a Kombucha beverage obtained from a tea fungus fermentation. Syst. Appl. Microbiol. 1995, 18, 590–594. [Google Scholar] [CrossRef]
- Arıkan, M.; Mitchell, A.L.; Finn, R.D.; Gürel, F. Microbial composition of Kombucha determined using amplicon sequencing and shotgun metagenomics. J. Food Sci. 2020, 85, 455–464. [Google Scholar] [CrossRef]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.P.; Taillandier, P. Understanding Kombucha tea fermentation: A review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef]
- Gaggìa, F.; Baffoni, L.; Galiano, M.; Nielsen, D.; Jakobsen, R.; Castro-Mejía, J.; Bosi, S.; Truzzi, F.; Musumeci, F.; Dinelli, G.; et al. Kombucha beverage from green, black and rooibos teas: A comparative study looking at microbiology, chemistry and antioxidant activity. Nutrients 2018, 11, 1. [Google Scholar] [CrossRef]
- Gullo, M.; La China, S.; Petroni, G.; Di Gregorio, S.; Giudici, P. Exploring K2G30 genome: A high bacterial cellulose producing strain in glucose and mannitol based media. Front. Microbiol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Singhsa, P.; Narain, R.; Manuspiya, H. Physical structure variations of bacterial cellulose produced by different Komagataeibacter xylinus strains and carbon sources in static and agitated conditions. Cellulose 2018, 25, 1571–1581. [Google Scholar] [CrossRef]
- Mamlouk, D.; Gullo, M. Acetic acid bacteria: Physiology and carbon sources oxidation. Indian J. Microbiol. 2013, 53, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Jayabalan, R.; Marimuthu, S.; Swaminathan, K. Changes in content of organic acids and tea polyphenols during kombucha tea fermentation. Food Chem. 2007, 102, 392–398. [Google Scholar] [CrossRef]
- Mamlouk, D. Insight Into Physiology and Functionality of Acetic Acid Bacteria Through a Multiphasic Approach. Ph.D. Thesis Dissertation, University of Modena and Reggio Emilia, Reggio Emilia, Italy, 2012. [Google Scholar]
- De Vero, L.; Boniotti, M.B.; Budroni, M.; Buzzini, P.; Cassanelli, S.; Comunian, R.; Gullo, M.; Logrieco, A.F.; Mannazzu, I.; Musumeci, R.; et al. Preservation, characterization and exploitation of microbial biodiversity: The perspective of the Italian Network of Culture Collections. Microorganisms 2019, 7, 685. [Google Scholar] [CrossRef]
- Gullo, M.; Mamlouk, D.; De Vero, L.; Giudici, P. Acetobacter pasteurianus strain AB0220: Cultivability and phenotypic stability over 9 years of preservation. Curr. Microbiol. 2012, 64, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Navarro, R.; Uchimura, T.; Komagata, K. Taxonomic heterogeneity of strains comprising Gluconacetobacter hansenii. J. Gen. Appl. Microbiol. 1999, 45, 295–300. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gullo, M.; Sola, A.; Zanichelli, G.; Montorsi, M.; Messori, M.; Giudici, P. Increased production of bacterial cellulose as starting point for scaled-up applications. Appl. Microbiol. Biotechnol. 2017, 101, 8115–8127. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Yang, Y.K.; Hwang, J.K.; Pyun, Y.R.; Kim, Y.S. Effects of pH and dissolved oxygen on cellulose production by Acetobacter xylinum BRC5 in agitated culture. J. Biosci. Bioeng. 1999, 88, 183–188. [Google Scholar] [CrossRef]
- Gullo, M.; De Vero, L.; Giudici, P. Succession of selected strains of Acetobacter pasteurianus and other acetic acid bacteria in traditional balsamic vinegar. Appl. Environ. Microbiol. 2009, 75, 2585–2589. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Schneider, M.; De Bruijn, F.J.; Lupski, J.R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef]
- Harrell, F.E., Jr.; Dupont, M.C. R Package Hmisc; CRAN; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: https://CRAN.R-project.org/package=Hmisc (accessed on 3 February 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 3 February 2020).
- Taiyun, W.; Viliam, S. R package “corrplot”: Visualization of a Correlation Matrix: Version 0.84. 2017. Available online: https://github.com/taiyun/corrplot (accessed on 3 February 2020).
- Battikh, H.; Chaieb, K.; Bakhrouf, A.; Ammar, E. Antibacterial and antifungal activities of black and green kombucha teas. J. Food Biochem. 2013. [Google Scholar] [CrossRef]
- Sievers, M.; Sellmer, S.; Teuber, M. Acetobacter europaeus sp. nov., a main component of industrial vinegar fermenters in central Europe. Syst. Appl. Microbiol. 1992, 15, 386–392. [Google Scholar] [CrossRef]
- Ludwig, W.; Klenk, H.P. Overview: A phylogenetic backbone and taxonomic framework for procaryotic systematics. In Bergey’s Manual of Systematic Bacteriology, The Proteobacteria, Part A, Introductory Essays, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2005; Volume 2, pp. 49–65. [Google Scholar]
- La China, S.; Bezzecchi, A.; Moya, F.; Petroni, G.; Di Gregorio, S.; Gullo, M. Genome sequencing and phylogenetic analysis of K1G4: A new Komagataeibacter strain producing bacterial cellulose from different carbon sources. Biotechnol. Lett. 2020, 42, 807–818. [Google Scholar] [CrossRef]
- Jakmuangpak, S.; Prada, T.; Mongkolthanaruk, W.; Harnchana, V.; Pinitsoontorn, S. Engineering bacterial cellulose films by nanocomposite approach and surface modification for biocompatible triboelectric nanogenerator. ACS Appl. Electron. Mater. 2020, 2, 2498–2506. [Google Scholar] [CrossRef]
- Cleenwerck, I.; Vandemeulebroecke, K.; Janssens, D.; Swings, J. Re-examination of the genus Acetobacter, with descriptions of Acetobacter cerevisiae sp. nov. and Acetobacter malorum sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 1551–1558. [Google Scholar] [CrossRef]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef]
- Švec, P.; Vancanneyt, M.; Seman, M.; Snauwaert, C.; Lefebvre, K.; Sedláček, I.; Swings, J. Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiol. Lett. 2005, 247, 59–63. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Vuyst, L.; Camu, N.; De Winter, T.; Vandemeulebroecke, K.; Van De Perre, V.; Vancanneyt, M.; De Vos, P.; Cleenwerck, I. Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int. J. food Microbiol. 2007. [Google Scholar] [CrossRef]
- Cleenwerck, I.; De Wachter, M.; González, Á.; De Vuyst, L.; De Vos, P. Differentiation of species of the family Acetobacteraceae by AFLP DNA fingerprinting: Gluconacetobacter kombuchae is a later heterotypic synonym of Gluconacetobacter hansenii. Int. J. Syst. Evol. Microbiol. 2009, 59, 1771–1786. [Google Scholar] [CrossRef] [PubMed]
- Azuma, Y.; Hosoyama, A.; Matsutani, M.; Furuya, N.; Horikawa, H.; Harada, T.; Hirakawa, H.; Kuhara, S.; Matsushita, K.; Fujita, N.; et al. Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucleic Acids Res. 2009, 37, 5768–5783. [Google Scholar] [CrossRef]
- Matsutani, M.; Nishikura, M.; Saichana, N.; Hatano, T.; Masud-Tippayasak, U.; Theergool, G.; Yakushi, T.; Matsushita, K. Adaptive mutation of Acetobacter pasteurianus SKU1108 enhances acetic acid fermentation ability at high temperature. J. Biotechnol. 2013, 165, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Pothimon, R.; Gullo, M.; La China, S.; Thompson, A.K.; Krusong, W. Conducting High acetic acid and temperature acetification processes by Acetobacter pasteurianus UMCC 2951. Process Biochem. 2020, 98, 41–50. [Google Scholar] [CrossRef]
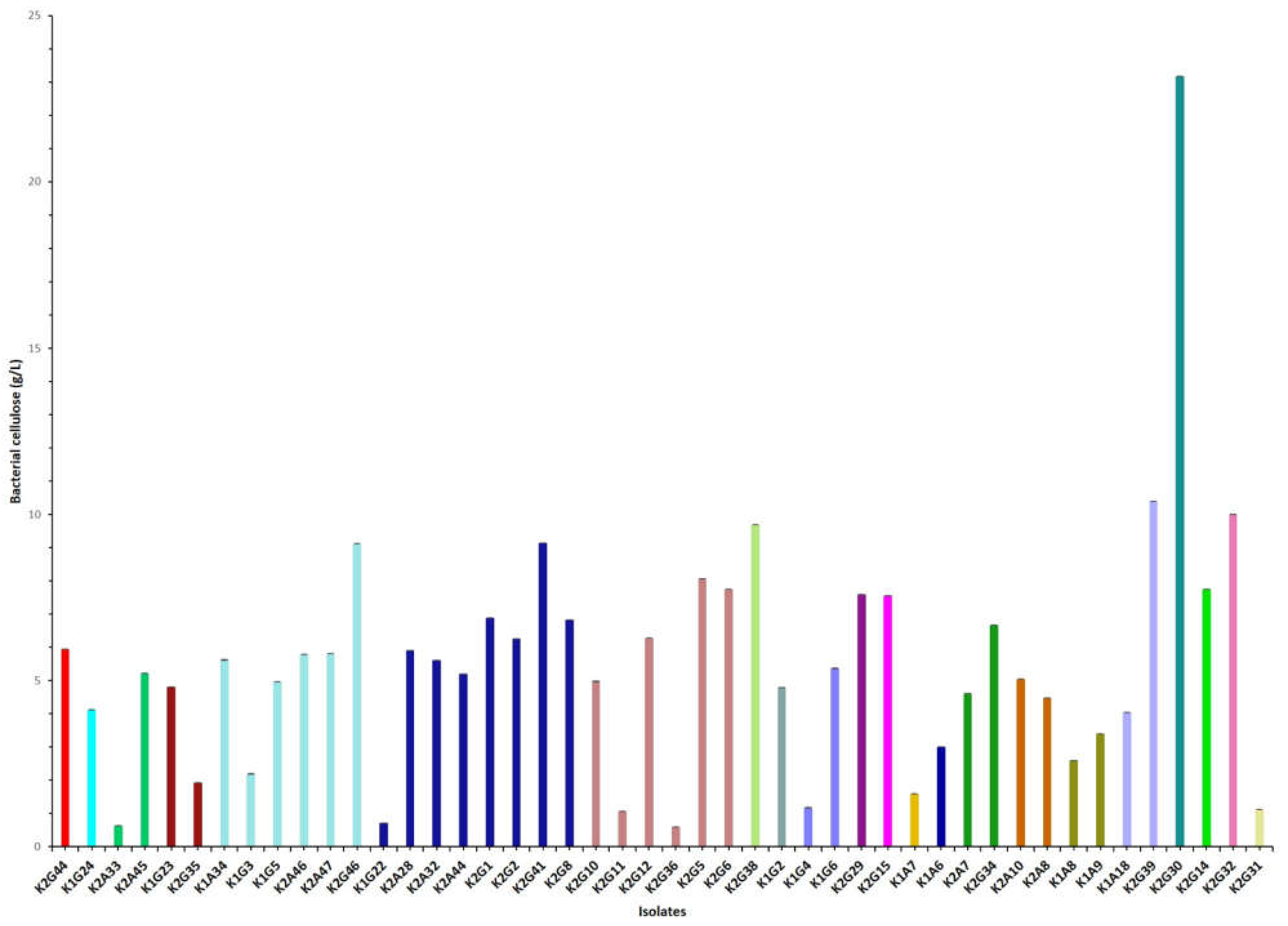

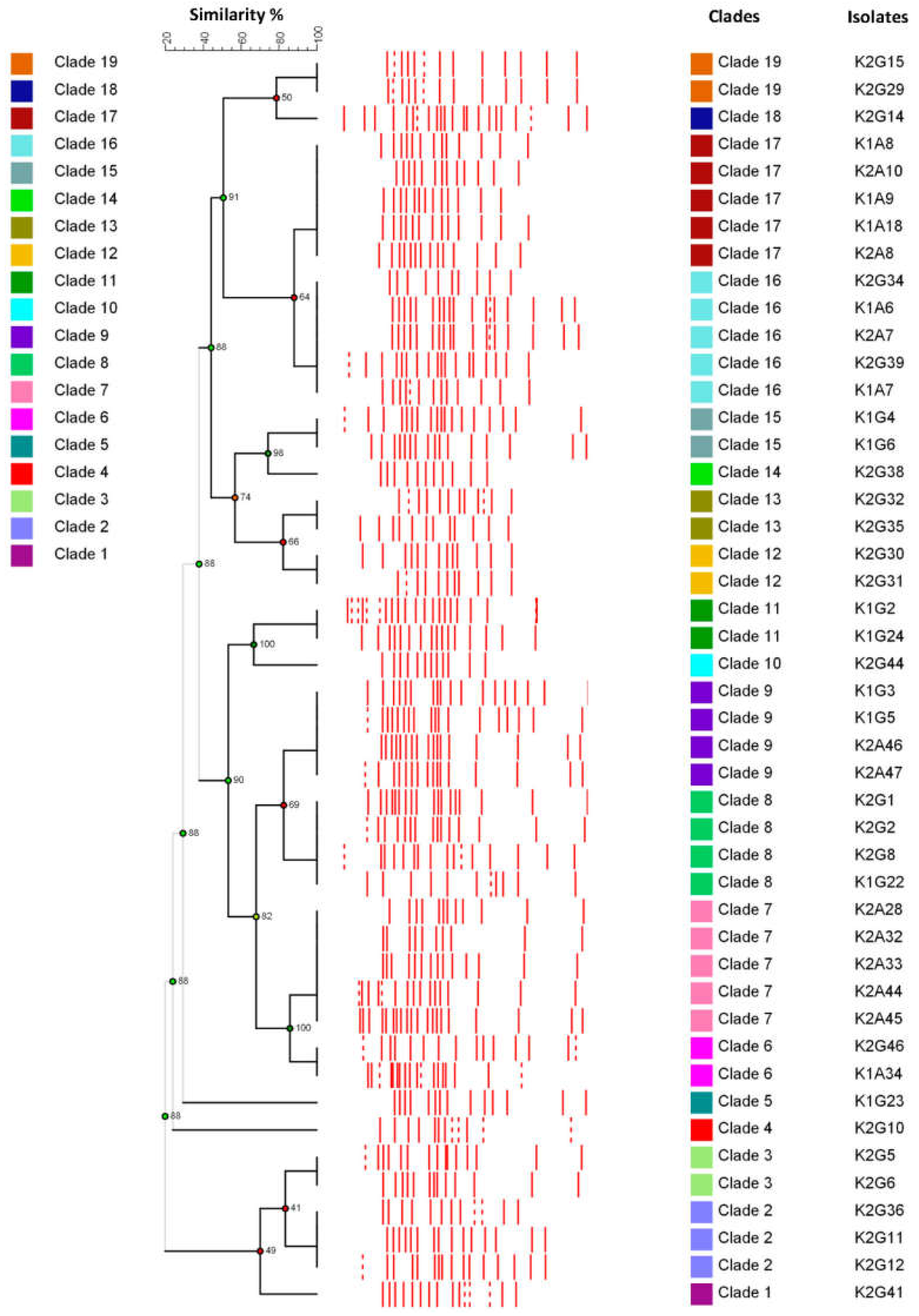
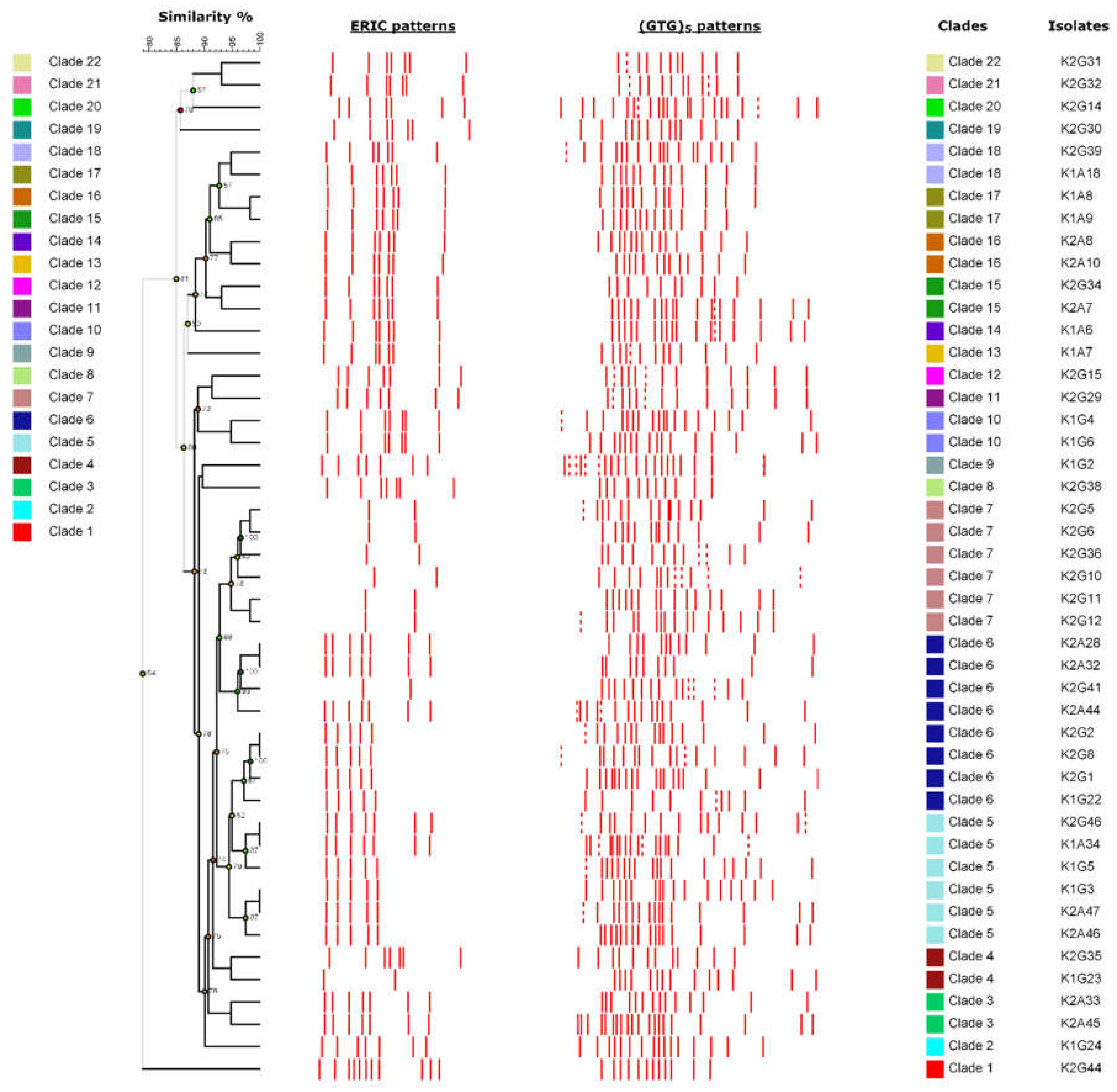
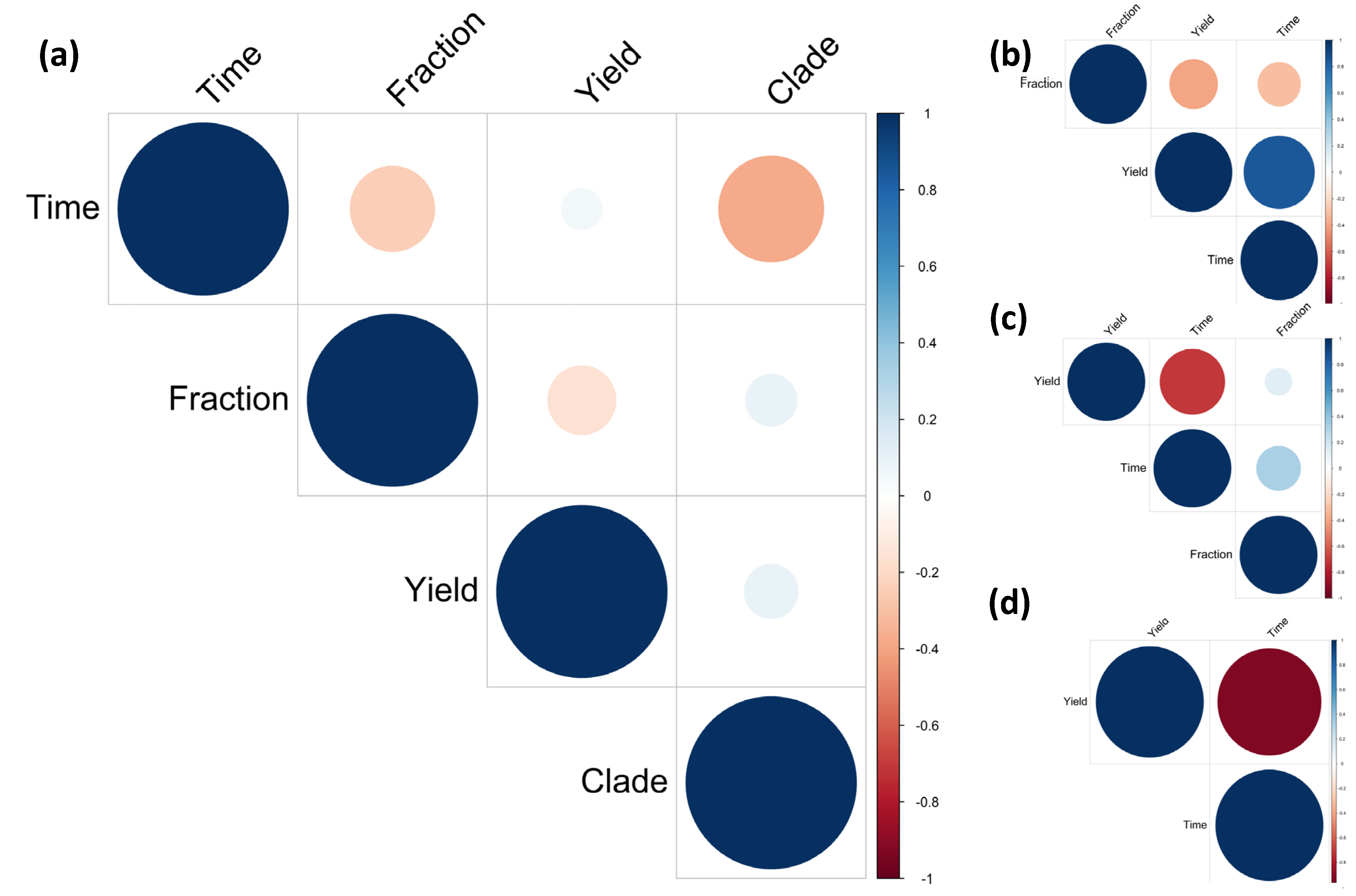
| * Strain | Sample | Time | BC (g/L) | Colony Morphology | Liquid Growth | AluI (pb) | Group | RsaI (bp) | Group | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| K1A18 | liquid | 6 | 4.0448 ± 0.0008 | green cellulosic | cellulose layer | - | - | 120, 400, 425, 550 | A | This study |
| K1A34 | liquid | 12 | 5.6214 ± 0.0010 | green with light cellulosic edge | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K1A6 | liquid | 0 | 3.0024 ± 0.0009 | cellulosic small | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K1A7 | liquid | 0 | 1.5886 ± 0.0004 | cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K1A8 | liquid | 0 | 2.5886 ± 0.0002 | cellulosic | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K1A9 | liquid | 0 | 3.4014 ± 0.0004 | green cellulosic | cellulose layer | - | - | - | - | This study |
| K1G2 (=UMCC 2964) | liquid | P1 | 4.7895 ± 0.0014 | white cellulosic edge | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K1G22 | liquid | 12 | 0.7057 ± 0.0003 | cellulose light | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K1G23 | liquid | 12 | 4.8122 ± 0.0003 | dark cellulose | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K1G24 | liquid | 12 | 4.1210 ± 0.0001 | white cellulose | only cellulose deposit | - | - | 120, 400, 425, 550 | A | [20] |
| K1G3 | liquid | P1 | 2.1857 ± 0.0052 | light beige | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K1G4 | liquid | 0 | 1.1738 ± 0.0032 | cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K1G5 | liquid | 0 | 4.9590 ± 0.0027 | cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K1G6 | liquid | 0 | 5.3676 ± 0.0025 | white cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2A10 (=UMCC 2965) | liquid | 0 | 5.0405 ± 0.0008 | cellulosic green small | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2A28 | liquid | 6 | 5.8962 ± 0.0007 | green cellulosic | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2A32 | pellicle | 6 | 5.6143 ± 0.0001 | green cellulosic | cellulose layer | - | - | - | - | This study |
| K2A33 | liquid | 6 | 0.6414 ± 0.0005 | green cellulosic | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2A44 | liquid | 12 | 5.1971 ± 0.0002 | small green | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2A45 | pellicle | 12 | 5.2133 ± 0.0004 | light edge | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2A46 | liquid | 12 | 5.7833 ± 0.0001 | dark center light edge | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2A47 | pellicle | 12 | 5.8019 ± 0.0001 | blue medium displayed | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2A7 | liquid | 0 | 4.6076 ± 0.0007 | green cellulosic small | cellulose layer | 150, 450, 800 | 1 | 400, 450, 550 | B | This study |
| K2A8 | liquid | 0 | 4.4781 ± 0.0004 | green cellulosic small | cellulose layer | - | - | - | - | This study |
| K2G1 | liquid | P2 | 6.8843 ± 0.0004 | cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | This study |
| K2G10 | liquid | 0 | 4.9752 ± 0.0016 | cellulosic small | cellulose layer/deposit | 150, 450, 800 | 1 | 400, 450, 550 | B | [20] |
| K2G11 | liquid | 0 | 1.0638 ± 0.0003 | white cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G12 | liquid | 0 | 6.2805 ± 0.0001 | small cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G14 | liquid | 0 | 7.7414 ± 0.0005 | white cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G15 | liquid | 0 | 7.5448 ± 0.0001 | creamy | cellulose layer/deposit | 150, 450, 800 | 1 | 400, 450, 550 | A | [20] |
| K2G2 | pellicle | P2 | 6.2548 ± 0.0004 | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] | |
| K2G29 (=UMCC 2967) | pellicle | 6 | 7.5848 ± 0.0002 | cellulosic | cellulose layer | 150, 450, 800 | 1 | 400, 450, 550 | B | [20] |
| K2G30 | pellicle | 6 | 23.1829 ± 0.0001 | cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 400, 450, 550 | B | [20] |
| K2G31 | pellicle | 6 | 1.1267 ± 0.0007 | cellulosic | cellulose layer/deposit | - | - | - | - | [20] |
| K2G32 (=UMCC 2968) | liquid | 6 | 9.9976 ± 0.0003 | cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G34 | liquid | 6 | 6.6614 ± 0.0001 | cellulosic | cellulose layer | - | - | - | - | [20] |
| K2G35 | liquid | 6 | 1.9314 ± 0.0005 | cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G36 | liquid | 6 | 0.5929 ± 0.0004 | yellow cellulosic | cellulose layer | 150, 450, 800 | 1 | 400, 450, 550 | B | [20] |
| K2G38 (=UMCC 2969) | liquid | 6 | 9.6957 ± 0.0009 | cellulosic | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G39 (=UMCC 2970) | liquid | 6 | 10.3957 ± 0.0001 | cellulosic | cellulose | 150, 450, 800 | 1 | 400, 450, 550 | B | [20] |
| K2G41 (=UMCC 2971) | liquid | 6 | 9.1443 ± 0.0003 | cellulosic | cellulose layer | 150, 450, 800 | 1 | 400, 450, 550 | B | [20] |
| K2G44 (=UMCC 2972) | pellicle | 12 | 5.9500 ± 0.0005 | dark brown cellulose | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G46 | liquid | 12 | 9.1195 ± 0.0007 | brown cellulose | cellulose layer/deposit | - | - | - | - | [20] |
| K2G5 (=UMCC 2966) | liquid | P2 | 8.0605 ± 0.0003 | white cellulosic | cellulose layer | - | - | 120, 400, 425, 550 | A | [20] |
| K2G6 | liquid | P2 | 7.7533 ± 0.0003 | white cellulosic | cellulose layer/deposit | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
| K2G8 | liquid | 0 | 6.8343 ± 0.0002 | white cellulosic | cellulose layer | 150, 450, 800 | 1 | 120, 400, 425, 550 | A | [20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La China, S.; De Vero, L.; Anguluri, K.; Brugnoli, M.; Mamlouk, D.; Gullo, M. Kombucha Tea as a Reservoir of Cellulose Producing Bacteria: Assessing Diversity among Komagataeibacter Isolates. Appl. Sci. 2021, 11, 1595. https://doi.org/10.3390/app11041595
La China S, De Vero L, Anguluri K, Brugnoli M, Mamlouk D, Gullo M. Kombucha Tea as a Reservoir of Cellulose Producing Bacteria: Assessing Diversity among Komagataeibacter Isolates. Applied Sciences. 2021; 11(4):1595. https://doi.org/10.3390/app11041595
Chicago/Turabian StyleLa China, Salvatore, Luciana De Vero, Kavitha Anguluri, Marcello Brugnoli, Dhouha Mamlouk, and Maria Gullo. 2021. "Kombucha Tea as a Reservoir of Cellulose Producing Bacteria: Assessing Diversity among Komagataeibacter Isolates" Applied Sciences 11, no. 4: 1595. https://doi.org/10.3390/app11041595
APA StyleLa China, S., De Vero, L., Anguluri, K., Brugnoli, M., Mamlouk, D., & Gullo, M. (2021). Kombucha Tea as a Reservoir of Cellulose Producing Bacteria: Assessing Diversity among Komagataeibacter Isolates. Applied Sciences, 11(4), 1595. https://doi.org/10.3390/app11041595









