1. Introduction
Dengue is an acute febrile disease caused by dengue virus (DENV). Commonly, dengue incidences happen in tropical and subtropical countries, such as South America, Southeast Asia, etc. [
1]. Dengue incidences usually occur in the rainy season, and happen in urban and suburban areas. Based on a study that analyzed 130 countries, there were around 9221 dengue deaths per year from 1990 to 2013, with the lowest of 8277 in 1992, and the highest of 11,302 in 2010 [
2]. Dengue had made Indonesia suffered the most significant economic loss in Southeast Asia. The average annual economic burden of dengue in Indonesia was approximately USD 381.5 million. Indonesia has the highest dengue infection rate in Southeast Asia and the second-highest dengue infection rate in the world after Brazil [
3]. In 2017, there were 59,047 Dengue Hemorrhagic Fever (DHF) cases and 444 DHF-associated deaths in Indonesia, with 0.75% case fatality rate and 22.55 incidence rate per 100,000 person-years [
4]. In 2018, the national incidence of dengue was 24.75 cases per 100,000 population, resulting in 467 deaths [
5].
DENV is a member of the family Flaviviridae and genus Flavivirus [
6]. DENV is transferred by
Aedes aegypti and
Aedes albopictus female mosquitoes [
7]. Those female mosquitoes consume blood as their regular meal to mature their eggs [
8]. They fulfill their need for blood by biting humans. The incubation period of DENV is around 4 to 10 days. After the incubation period ends, an infected mosquito can transfer DENV in its lifetime [
9]. There are four serotypes of DENV, which are DENV-1, 2, 3, and 4. A DENV-infected person can be infected by one serotype of DENV or more [
10].
Dengue consists of three clinical phases, which are febrile phase (occurs on the first until the third day of fever), critical phase (occurs on the fourth until sixth day of fever), and recovery phase (occurs on the seventh day of fever or afterwards). The common clinical symptoms of dengue are as follows: during the febrile phase, the symptoms are high fever, headache, nausea, myalgia, arthralgia, malaise, retro-orbital pain, and vomiting [
11]. During the critical phase, the symptoms are thrombocytopenia, leukocytopenia, and plasma leakage, which is clinically manifested by hemoconcentration, pleural effusion, and/or ascites. Patients may also experience severe bleeding and shock [
11]. During the recovery phase, the extravasated fluid is re-absorbed into the intravascular compartment. Some patients may experience an erythematous rash. The severity level of dengue consists of Dengue Fever (DF), Dengue Hemorrhagic Fever (DHF), and Dengue Shock Syndrome (DSS) [
12]. DF is commonly classified as dengue, while DHF and DSS are commonly classified as severe dengue. The main difference between dengue and severe dengue is, patients who suffer severe dengue experience plasma leakage, while patients who suffer dengue do not. In this research, we only considered level DF and DHF. DHF itself was divided into two different levels, which were DHF grade 1 and DHF grade 2.
The gold standard to confirm a DENV infection are Reverse Transcription-Polymerase Chain Reaction (RT-PCR) test, culture of DENV, hemagglutination inhibition test, laboratory test, and tourniquet test. A suspected person can go through a laboratory test that measures hematocrit, thrombocyte (platelet count), and white blood cell (WBC). DENV infection will decrease in platelet count below 100,000 per μL between the third to eighth day onset of fever, and an increase in hematocrit of 20% or more. A positive tourniquet test also indicates that the corresponding patient might suffer dengue. Laboratory test parameters that we analyzed consist of hematocrit, hemoglobin, platelet count, WBC, monocyte, lymphocyte, and neutrophil.
In this research, we developed models that can predict the severity level of a dengue patient, based on the values of the seven laboratory test parameters. In other words, the laboratory test parameters were used as predictors to predict the severity level of a dengue patient. We used two methods, which were Artificial Neural Network (ANN) and Discriminant Analysis (DA). We decided to use ANN because ANN is one of machine learning methods that imitates the neuron structure of a human brain that has been used in many fields, including medical diagnosis prediction. Meanwhile, we decided to use DA because DA is one of dependency statistical analysis techniques in Multivariate Analysis that is used to classify an object into one of the statistically independent groups or categories. This is aligned with our purpose of this research, which was to classify data of dengue patients into one of the statistically independent severity levels of dengue.
We only used patients’ data on the third day onset of fever. Because on the third day, dengue patients are going to enter the critical phase. We expected that this model can help physicians to treat dengue patients early before the patients enter the critical phase, so they can have timely treatment and fatal cases or deaths can be avoided. We also used the data on the third day because there haven’t been any previous researches that used data on the third day.
The objectives of this research were to develop models that can predict the severity level of dengue using Artificial Neural Network (ANN) and Discriminant Analysis (DA), to evaluate the performances of the models, and to conclude which model has the best performance. A model with the best performance would be the proposed predictive model in this research. The predictive model will assist physicians in predicting the severity level of dengue.
3. Related Works
Abdiel E. Laureano-Rosario et al. [
13] utilized ANN, which was trained with genetic algorithm to predict dengue fever outbreak in Puerto Rico and some areas in the coast of Mexico. They concluded that the model they developed using ANN had a good predictive ability.
Jorge D. Mello-Román et al. [
14] compared two machine learning methods, which were Artificial Neural Networks multilayer perceptron (ANN-MLP) and Support Vector Machine (SVM) as the tools to assist medical diagnosis. ANN-MLP produced a better result with an average of 96% accuracy, 96% sensitivity, and 97% specificity. In conclusion, ANN-MLP could be used as a classifier to diagnose dengue infection with high accuracy, sensitivity, and specificity.
Oswaldo Santos Baquero et al. [
15] compared Seasonal Autoregressive Integrated Moving Average (SARIMA), Generalized Additive Models (GAM), Artificial Neural Networks (ANN), naïve model, and ensemble model to predict dengue cases in São Paulo for one month ahead.
Tanujit Chakraborty et al. [
16] developed a novel hybrid model, which was a combination of Autoregressive Integrated Moving Average (ARIMA) and Neural Network Autoregressive (NNAR). The model was used to analyze time series dengue data of three dengue endemic regions, which were San Juan, Iquitos, and Filipina. They concluded that the proposed hybrid model was easy to interpret, had an excellent performance in forecasting dengue epidemic for three dengue time series data from different regions, and had better forecasting accuracy compared to the other methods used in previous researches, such as traditional methods or other hybrid methods.
Siriyasatien et al. [
17] developed models that enable forecasting of outbreaks of dengue, giving medical professionals the opportunity to develop plans for handling the outbreak, well in advance. They utilized several methods, one of them is ANN. Based on their results, the ANN model had two advantages: easy to be used in incremental learning and can learn to ignore irrelevant attributes.
Yulia Resti et al. [
18] applied Quadratic Discriminant Analysis (QDA) in mapping the incidence of dengue into five areas in Palembang based on significant factors, such as age, gender, blood group, etc. The overall correct percentage of the mapping results was 66.7%.
Abdul Halim Poh et al. [
19] utilized Principal Component Analysis (PCA) and DA to predict the patients’ clinical dengue positivity. The DA method yielded accuracy between 93–98%, sensitivity between 75–89%, and specificity between 94–100%.
ANN and DA have been widely used to predict dengue incidences. That is why we decided to use both methods separately in our research to develop models that can predict the severity level of dengue based on the laboratory test results.
4. Materials and Methods
4.1. Dataset
We used dengue patients’ data from the year 2009 and 2010 to develop the models (
https://drive.google.com/drive/folders/1C1ZciLa2Cwsb1IrDpBpULP-2IZrcskBQ?usp=sharing). Many information was written in that data, such as age, gender, patients’ length of stay, clinical symptoms, laboratory test results, and patient diagnosis. The diagnoses were classified into three distinct severity levels, which were DF as the mild level, DHF grade 1 as the intermediate level, and DHF grade 2 as the severe level.
The data was obtained from Department of Microbiology, Universitas Indonesia. The data only consists of 77 dengue patients’ data. It is very small because it is difficult to collect laboratory test results of dengue patients needed in this research. The data was split into training and testing data. We developed the models with three different data splits. That is, with the ratio of training: testing = 70%:30%, 80%:20%, and 90%:10%. Training data was used for learning, which was to fit the parameters (i.e., weights). While testing data was used to assess the (generalization) performance of the neural network [
20].
Table 1 is the summary of our data:
4.2. Discriminant Analysis (DA)
Discriminant Analysis (DA) is one of dependency statistical analysis techniques in Multivariate Analysis. It means that in DA, there are dependent and independent variables being analyzed, where the value of the dependent variable depends on the values of the independent variables. In DA, there is only one dependent variable, but there are more than one independent variables. DA can be used if the dependent variable is categorical (in nominal or ordinal scale), and the independent variables are metric (in interval or ratio scale). A categorical dependent variable means the corresponding variable consists of certain categories.
DA is used to classify an object into one of the statistically independent groups or categories based on the values of the independent variables. Classification process in DA is mutually exclusive, that is, if an object is classified into one particular group, it will not be classified into another group.
In DA, discriminant function(s) will be formulated. A discriminant function is used to classify an object into a group or category based on the value of the discriminant function itself. The values of the independent variables of an object will be inputted in the function. Then, the obtained value of the function, which is called the discriminant value, will determine in which group an object belongs. For k categories, k-1 discriminant functions have to be formulated.
Based on the amount of the categories of the dependent variable, there are two types of DA, which are two-group discriminant analysis and Multiple Discriminant Analysis (MDA). Two-group discriminant analysis is a type of DA where the dependent variable consists of two categories. While MDA is another type of DA where the dependent variable consists of more than two categories.
Based on the type of the discriminant function, there are also two types of DA, which are Linear Discriminant Analysis (LDA) and Quadratic Discriminant Analysis (QDA). LDA is a classification method used to find the optimum linear combination of features to separate two or more groups of objects [
21]. QDA can be considered as a direct extension of LDA. There are some differences between LDA and QDA. First, the discriminant function in LDA is a linear combination of the independent variables. While the discriminant function in QDA is in the form of quadratic function. Second, in LDA, every covariance matrix of every independent variable has to be the identical. While in QDA, every covariance matrix of every independent variable can be different. The third difference, the decision curve in LDA is in the form of a straight line, while the decision curve in QDA is in the form of a quadratic curve [
22].
Assume that observations
of a group
are random vectors of size
sampled from a Gaussian distribution
, for all
. With a Bayesian approach, an object
can be classified into group
that reach the maximum value of the posterior density function
where
is the density function of a Gaussian vector and
is the probability of an object belongs to group
. Formula (1) is the Quadratic Discriminant Analysis (QDA) formula. Linear Discriminant Analysis (LDA) has a similar formula. The only difference is in LDA, the covariance matrices
are assumed to be identical for all classes [
23].
4.3. Artificial Neural Network (ANN)
Artificial Neural Network (ANN) is a simple imitation of neuron structure of a human brain [
24]. Similar to human brain, ANN is capable to analyze incomplete or unclear information, and furthermore, evaluate them. ANN imitates human brain in processing input signals and translating it into output signals. ANN is also capable to learn from data without any assumptions of certain functions.
ANN is a part of Artificial Intelligence, along with Support Vector Machines, Expert Systems, and Fuzzy Logic. ANN consists of processing units which are called artificial neuron. Artificial neurons try to imitate the structure and behavior of biological neurons. A neuron can consist of more than one input (dendrite), but commonly consists of only one output (synapsis through axon).
A neuron has a function which determines the activation of the neuron itself. That function is called an activation function. An activation function processes input signals that have been combined together, then transforms them into an output signal. Mathematically, the procedure of signal processing can be expressed as follows:
where
is the output signal,
is the activation function,
is the input variable, and
is the weight that is given to each of the input variable [
25].
There’s a lot of types of activation functions, which are linear, sigmoid, step, ramp, hyperbolic tangent, etc. The most frequently used activation function is sigmoid function. Hyperbolic tangent function has a similar shape to sigmoid function, but its value lies between −1 to +1, unlike sigmoid which the values lies between 0 to 1.
ANN can be widely used in different scopes of problems, such as finding new features of an object, and classifying or predicting an object/event using huge sets of data. Some of the fields where ANN is frequently used are medical diagnosis prediction, character recognition, speech recognition, human face recognition, signature verification application, etc.
ANN has a different way of work with normal computers in many ways. ANN has strengths compared to normal computer programs. Some of the strengths of ANN are as follows: (i) ANN is an adaptive learning method. It imitates human brain on how to do its task while learning, even with different types of inputs; (ii) ANN can organize itself while learning; (iii) ANN works parallelly like human brain; (iv) ANN has a high fault tolerance. It is able to work even on fuzzy, noisy, and incomplete data; and (v) ANN is applicable to classify data, recognize patterns, and any other tasks that involve obscure data.
4.4. Confusion Matrix and Performance Measurement
Confusion matrix is a matrix used to evaluate the performance of classifier models in general. For binary classification problems, confusion matrix size 2 × 2 is used. However, in real life problems, classification can involve more than two classes or categories. These problems are considered as multi-class classification problems. For a multi-class classification problem, confusion matrix size 3 × 3 is used. Principally, 3 × 3 confusion matrix is similar to 2 × 2 confusion matrix. The general form of 3 × 3 confusion matrix is as follows:
where the columns denote actual cases, the rows denote predicted cases.
For category A,
True Positive (TP):
True Negative (TN):
False Positive (FP):
False Negative (FN):
Calculations for other categories are similar.
In this research, our problem was a multi-class classification problem, because the severity level consisted of three distinct categories, and we aimed to classify each dengue patient into one of those level. So, we used 3 × 3 confusion matrix and these performance measurements below:
Accuracy: This metric measures the overall performance of the model. Generally, accuracy is the proportion of true results among the total number of cases examined.
Accuracy can be expressed as follows:
Accuracy can also be calculated from the elements of the 3 × 3 confusion matrix using the formula below:
where
denotes an element of the confusion matrix,
denotes the number of rows of the confusion matrix, and
denotes the number of columns of the confusion matrix. Note that the numerator of the Formula (3) is the sum of the main diagonal elements of the confusion matrix, and the denominator is the sum of all of the elements of the confusion matrix.
- 2
Sensitivity: this metric measures the proportion of the True Positive (
TP) cases among the total number of positive cases. The formula to calculate
sensitivity is as follows:
- 3
Specificity: This metric measures the proportion of the True Negative (
TN) cases among the total number of negative cases. The formula to calculate
sensitivity is as follows:
5. Results and Discussion
5.1. Model Construction
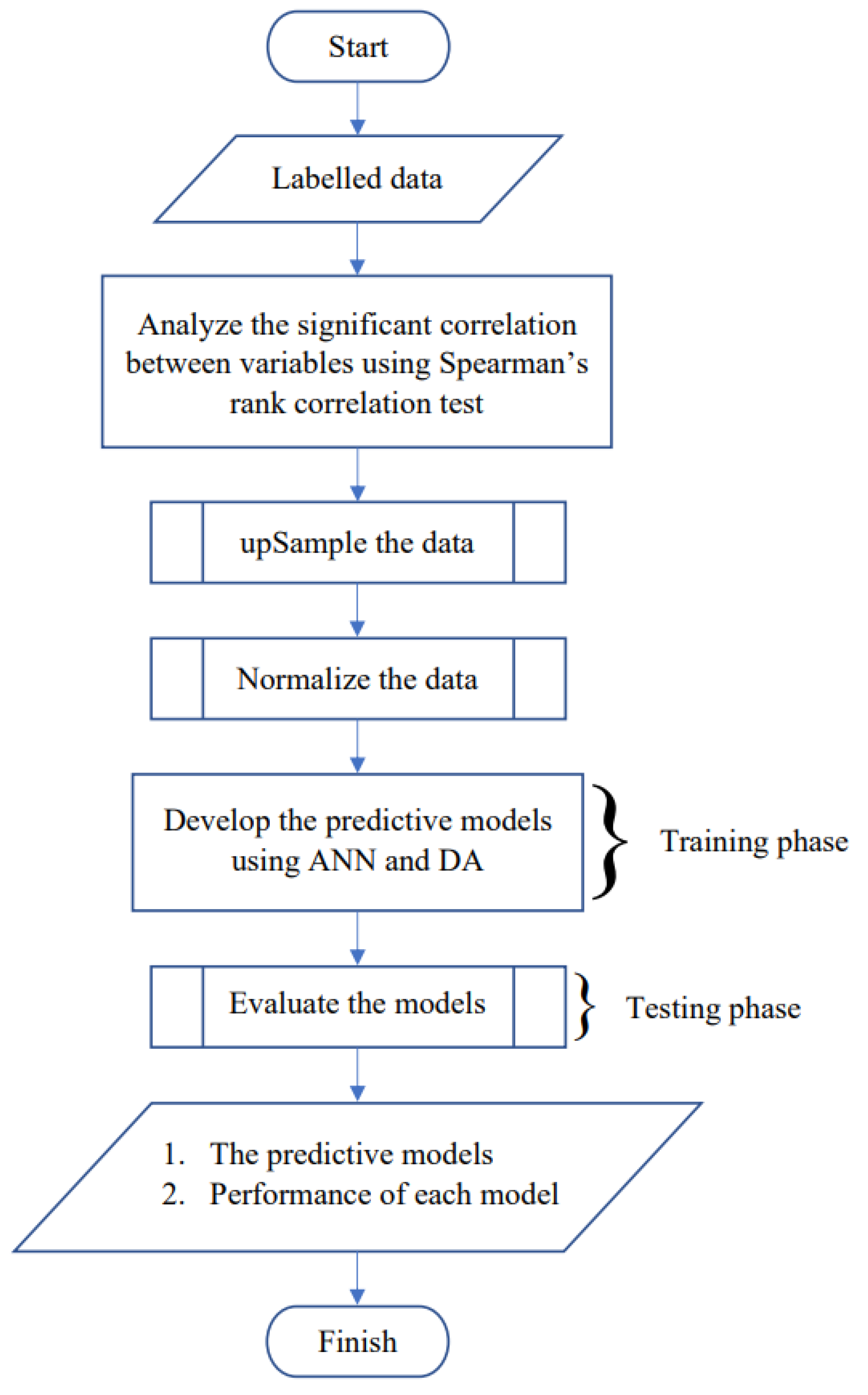
As mentioned previously, we developed models to predict the severity level of dengue with ANN and DA. The models will be able to predict the severity level of dengue based on the laboratory test results of the corresponding patients. The accuracy of both models developed with ANN and DA would be evaluated, and the one with the higher accuracy would be the proposed model. For short, we mentioned the models as “predictive models” in
Figure 1.
By inputting the values of hematocrit, hemoglobin, platelet count, WBC, monocyte, lymphocyte, and neutrophil of the corresponding patient into the predictive model, the model would be able to process those values and predict the severity level of the dengue patient. In other words, the input variables of the model were the independent variables of this research, which were the seven laboratory test parameters. While the output variable was the dependent variable of this research, which was the severity level of dengue.
Before we developed the model, we conducted Spearman’s rank correlation test. This correlation test was conducted for two purposes:
To examine whether there is a significant correlation between the independent variables. To be able to use DA, there has to be no multicollinearities between the independent variables. This means all of the independent variables are not correlated. If there’s a couple of correlated independent variables, one of them has to be eliminated.
To conduct feature selection. This means to select only the necessary independent variables to develop the models.
First, we conducted a two-ways hypothesis test, where the hypotheses were as follows:
H0: , which means there is no significant correlation between variable xi and xj, where i, j = 1, …, 7, i j. H1: , which means there is a significant correlation between variable xi and xj.
We used a significance level of 0.05. Below were the results of the correlation test.
The elements of
Table 2 were the correlation coefficients between the corresponding independent variables. The starred coefficients mean that there is a significant correlation between the corresponding independent variables, or in other words, the corresponding independent variables were correlated. The correlated independent variables were as follows:
Hemoglobin and hematocrit
Hemoglobin and platelet count
Hematocrit and platelet count
WBC and platelet count
Neutrophil and lymphocyte
Neutrophil and monocyte
Therefore, we eliminated some of the independent variables which were correlated with more than one other independent variables. The eliminated independent variables were hematocrit, platelet count, and neutrophil. Furthermore, we only used the remaining four independent variables to develop the models – hemoglobin, WBC, lymphocyte, and monocyte.
Then, we applied the upSample technique because the data that had been used in this research was imbalanced. upSampling means doing a sampling with replacement, in a random manner, to the data in the minority category until the sample size equals the size of the majority category. upSample is one of the techniques to handle imbalanced dataset. An imbalanced dataset is usually upSampled before it is used to develop the model, so the developed model will tend to have higher accuracy. If the dataset is not upSampled and is directly used to develop a model, it is unlikely to yield a model with high accuracy. The dataset used in our research was imbalanced, in the sense that the classes are not represented equally. Before the data was upSampled, it consisted of 77 cases. From the 77 cases, 38 cases were from category DF, 28 cases were from category DHF grade 1, and 11 cases were from category DHF grade 2. It is clear that the majority category was DF with a size of 38, and the minority category was DHF grade 2 with size 11. After the dataset was upSampled, each category has the size 38. So, in total, the upSampled data consisted of 38 × 3 = 114 cases.
After we applied the upSample technique, we standardized the data. We only standardized the data of the independent variables, which were the seven laboratory test parameters. We didn’t standardize the data of the dependent variable, which was the severity level of dengue, because we wanted the real values of the dependent variable. Data of the independent variables was standardized because the values varied greatly and had different units. It was also standardized in order to develop better models compared to models developed using unstandardized data. After the data was standardized, we started to develop the models. We developed twelve models in total. Six models were developed with ANN, and the other six were developed with DA. We developed the models using R programming version 3.6.3. We decided to use R because it has some advantages, such as: (i) it has data science engine, specifically the statistics and machine learning packages, and (ii) it is an open-source language programming, so it is accessible by anyone.
5.2. Predictive Models Developed Using DA
We developed two DA models. The first model that we developed was Linear Discriminant Analysis (LDA) model, and the second one was Quadratic Discriminant Analysis (QDA) model. Below were the table of performance measurements of both models.
Based on
Table 3, it is shown that the LDA model with 70% training data yielded the highest accuracy (45.45%), sensitivity (45.73%), and specificity (72.89%).
- 2.
QDA
Based on
Table 4, it is shown that the QDA model with 80% training data yielded the highest accuracy (60.87%) and sensitivity (59.72%).
Based on the obtained results, QDA model with 80% training data yielded the highest accuracy (60.87%) and sensitivity (59.72%). Therefore, QDA model with 80% training data was the proposed predictive model so far. This model was then compared with the other predictive models developed using ANN.
5.3. Predictive Models Developed Using ANN
We also developed the predictive models using ANN—next will be called ANN model for short. We developed the ANN models using R programming with the nnet package. The nnet trains feed-forward neural networks with traditional backpropagation algorithm [
26].
Feed-Forward Neural Networks (FFNN) is a type of ANN that doesn’t have any feedback connection from the output to the input [
27]. In FFNN, neurons of the previous layer are entirely connected to the consecutive layer, but there are no intra-layer connections [
28]. FFNN is a supervised learning method that is utilized for classification problems and consists of input, hidden, and output layers [
29].
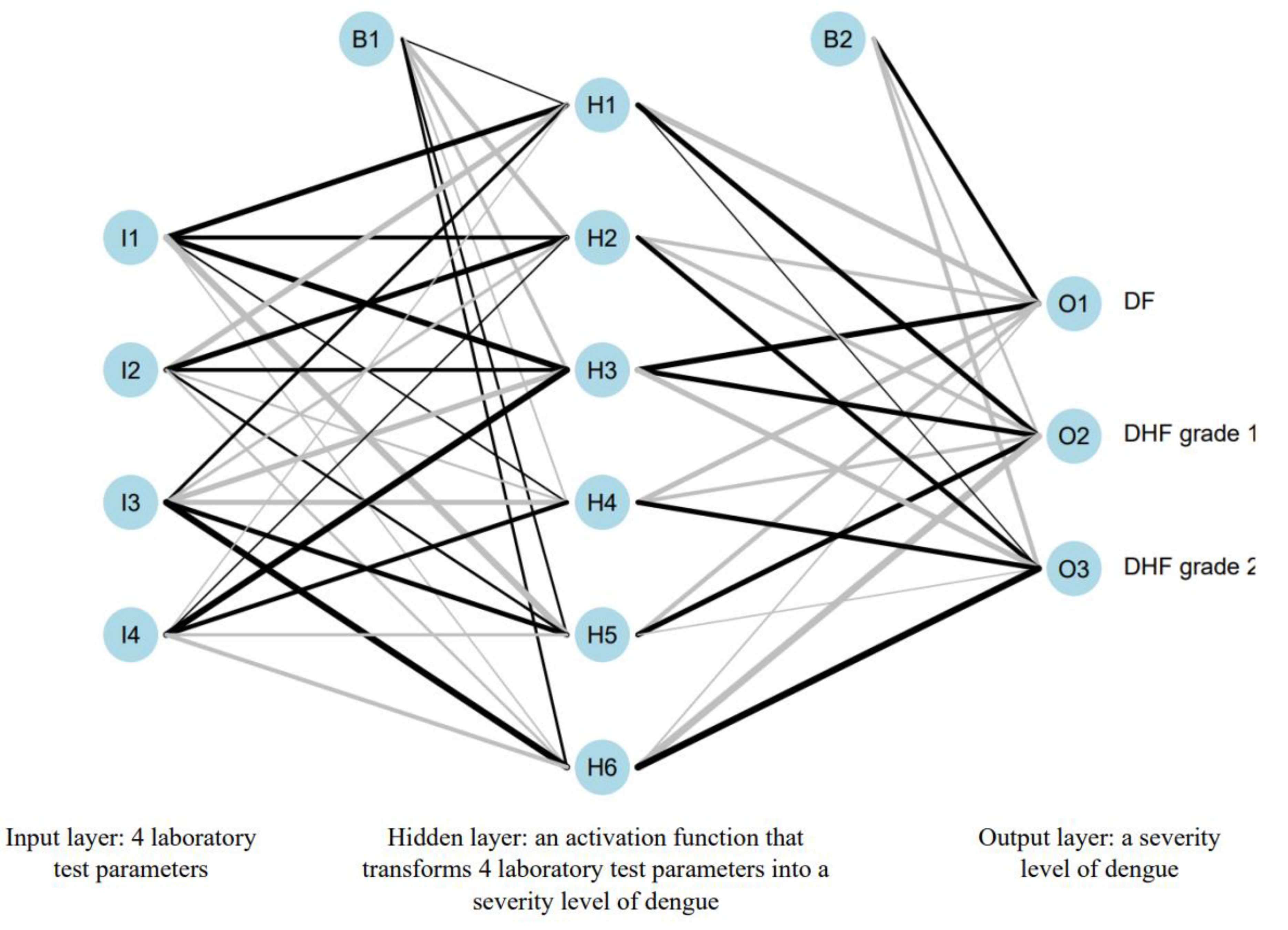
Our ANN architecture consisted of one input layer, one hidden layer, and one output layer. There were four neurons in input layer that represented hemoglobin, WBC, lymphocyte, and monocyte.
There was only one hidden layer in this ANN architecture. The number of hidden layers and neurons in each hidden layer were obtained through the iteration process until the accuracy could not be furtherly improved [
30]. The number of hidden layers is arbitrary. However, one hidden layer is commonly used for simple problems. The numbers of input and output neurons are determined according to the problem, while the number of hidden neurons was well optimized [
20]. The problem in this research could be considered as a simple problem because it only consisted of four independent variables and the purpose was related to medical diagnosis, one of the common purposes of using ANN. That was why one hidden layer was enough for this research. Meanwhile, to ease the iteration process, we decided to use the rule of thumb in determining the hidden neurons. Based on the research conducted by Panchal and Panchal [
31], one of the rules in determining the number of hidden neurons is “the number of hidden neurons should be 2/3 of the input layer size, plus the size of the output layer”.
We followed this rule when developing the ANN model. So, the number of hidden neurons is:
Meanwhile, the number of input neurons was 4 because there were four remaining laboratory test parameters. The number of the output neuron was 3 because it represented three categories of severity level. The output that would come out was the probability values from each neuron that would sum up to 1. The probability value means the probability of a patient to suffer from dengue in a corresponding level. For example, the output value from neuron DF is 0.3, from DHF grade 1 is also 0.3, and from DHF grade 2 is 0.4. So, the patient is suffering dengue level DHF grade 2 because the probability value is the highest compared to the other two.
Figure 2 was the architecture of the developed ANN predictive model.
The activation function is frequently a bounded nondecreasing, differentiable, and nonlinear function such as the hyperbolic tangent or the logistic function [
26]. We tried to used logistic and hyperbolic tangent as the activation functions. First, we developed an ANN model with logistic activation function. Then, we developed another ANN model with hyperbolic tangent activation function.
Logistic function may have different forms, but the one used in R programming—the one we used in this research—is the standard logistic function, which is also known as sigmoid function. The formula of the logistic function is as follows
where
is the value of the
th independent variable [
32].
Meanwhile, hyperbolic tangent function, which is also known as tanh function, has the formula as follows
where
is the value of the
th independent variable [
33].
We provided the biases and weights of our ANN models, so our developed ANN models can be useful for other researchers who want to conduct similar researches. We displayed them in tables so it would be easier to read. Below is the annotation of the notations in the tables below:
Bi–Hj: weight from Bi to Hj, where i = 1, 2 and j = 1, …, 6.
Ik–Hj: weight from Ik to Hj, where k = 1, …, 4.
Bi–Ol: weight from Bi to Ol, where l = 1, 2, 3.
Hj–Ol: weight from Hj to Ol.
Table 5 and
Table 6 were the tables of the biases and weights of our ANN models.
- 1.
ANN model developed using logistic activation function
- 2.
ANN model developed using hyperbolic tangent (tanh) activation function
Below were the tables of performance measurements of both models.
Based on
Table 7, it is shown that the ANN model with 70% training data yielded the highest accuracy (90.91%), sensitivity (91.11%), and specificity (95.51%).
- 2
ANN model developed using hyperbolic tangent (tanh) activation function
Based on
Table 8, it is shown that the ANN model with 70% training data yielded the highest accuracy (90.91%), sensitivity (91.11%), and specificity (95.51%).
5.4. The Proposed Predictive Model
Based on the obtained results, both ANN models developed using logistic and hyperbolic tangent activation function with 70% training data yielded the highest accuracy (90.91%), sensitivity (91.11%), and specificity (95.51%). These models also have the highest accuracy, sensitivity, and specificity compared to the previously proposed model, QDA model with 80% training data. Therefore, both ANN models developed using logistic and tanh activation function with 70% training data were the proposed predictive models in this research. We displayed the proposed predictive models’ performances in
Table 9.