The Harmony Search (HS) algorithm [
34] is a well-known population-based metaheuristic algorithm. The optimization process in HS is a mimicry of the underpinning principles of jazz music orchestra, where musicians attain a pleasant harmony through several improvisation steps. HS has been successfully applied to a wide variety of real-world optimization problems, such as system reliability, robot path planning, renewable energy systems, hyper-parameter tuning of deep neural networks, intelligent manufacturing, and credit scoring (see [
35,
36]); university timetabling, structural design, water distribution, and supply chain management (see [
37,
38]); and, music composition, Sudoku puzzle solving, tour planning, web page clustering, vehicle routing, dam scheduling, groundwater modeling, soil stability analysis, ecological conservation, heat exchanger design, transportation energy modeling, satellite heat pipe design, medical physics, medical imaging, RNA structure prediction, and image segmentation (see [
39], among others). Besides that, the implementation of HS in various parameter estimation studies indicated the potentiality of HS as an effective parameter estimation tool. Some of the notable parameter estimation problems that applied HS include parameter estimation of the nonlinear Muskingum model [
40,
41], parameter estimation in vapor-liquid equilibrium modeling [
42], parameter estimation in an electrochemical lithium-ion battery model [
43], parameter identification of synthetic gene networks [
44], and design storm estimation from probability distribution models [
45]. In addition, HS was also successfully employed in human activity pattern modeling, such as disease spread and disaster response [
46]. Hereinafter, the term HS represents the family of HS variants, while the standard HS is denoted SHS. The five primary steps in SHS, as outlined in [
47], are as follows:
The complete details of SHS are provided as in Algorithm 1. SHS is designed with fewer mathematical operations and it is relatively easy to code, but easily applicable to a wide variety of optimization problems. The advantages of HS are discussed in [
37]. Over the two decades since the inception of HS, many HS variants have been developed to date. A majority of the variants modifies the improvisation procedures either by internal modification or hybridization with other heuristics. Some of the recent comprehensive reviews on HS variants are presented in [
37,
39,
47,
48,
49]. In this study, only the internally modified HS variants were considered and selected on a minimal parameter setting requirement basis.
| Algorithm 1: Standard Harmony Search | |
| 1: Set , , , , and | |
| 2: | % generate HM |
| 3: Compute | % compute fitness |
| 4: while or (any stopping criterion) do | |
| 5: for each do | |
| 6: if then | |
| 7: where | % memory consideration |
| 8: if then | |
| 9: | % pitch adjustment |
| 10: end if | |
| 11: else | |
| 12: | % random generation |
| 13: end if | |
| 14: end for | |
| 15: if then | |
| 16: replace | |
| 17: end if | |
| 18: end while | |
2.1. Improved Harmony Search (IHS)
The Improved Harmony Search (IHS) [
50] is the prototypical HS variant. While still requiring to fine-tune the
parameter, the parameters
and
are made dynamic in this variant with the introduction of the
(
) and
(
) iterative parameters. Although IHS has shown better performance than SHS, this variant increased the burdensome process of setting suitable values for four parameters instead of just two in SHS [
51]. Nevertheless, IHS is considered to be a breakthrough that paved the way for the development of various HS variants to date.
and
are adjusted at each iteration while using:
The computational procedure of IHS is provided, as in Algorithm 2.
| Algorithm 2: Improved Harmony Search | |
| 1: Set , , using (5), using (6), and | |
| 2: | |
| 3: Compute | |
| 4: while do | |
| 5: for each do | |
| 6: if then | |
| 7: where | |
| 8: if then | |
| 9: | |
| 10: end if | |
| 11: else | |
| 12: | |
| 13: end if | |
| 14: end for | |
| 15: if then | |
| 16: replace | |
| 17: end if | |
| 18: end while | |
A year later, a new variant that was inspired by the swarm intelligence concept of the PSO was introduced by the principal developer of IHS. This variant is known as Global Harmony Search (GHS) [
52] and aims to mimic the best harmony in the HM. The parameter
is adapted from IHS, while
is removed. Thus, the pitch adjustment step in SHS is replaced with a random selection of best harmony of any decision variable from the HM, as follows:
In general, GHS is claimed to perform better than SHS and IHS, especially in high-dimensional optimization problems. However, [
53] asserted that GHS has flaws that will cause premature convergence and the name of this variant is also said to be misleading. The most serious flaw, as noted by [
51], is the frequent generation of infeasible new harmonies, whenever the upper and lower bounds of each decision variable are not identical in the given optimization problem. Hence, GHS is not considered in this paper.
2.3. Self-Adaptive Global Best Harmony Search (SGHS)
The Self-Adaptive Global Best Harmony Search (SGHS) [
55] aims to improve the GHS [
52] in terms of avoiding getting trapped at local optima. In this approach,
and
are dynamically adjusted to a suitable range after a number of iterations by tracking their previous values that allowed for the replacement of new harmony in HM. Further,
and
are assumed to be normally distributed where
and
. The initial values of
and
are set at 0.98 and 0.9, respectively. Subsequently, SGHS begins with
and
values being generated from the Normal distribution. During each iteration, the values of
and
that corresponds to a replacement of new harmony in HM is recorded until a number of solutions are generated within the specified learning period
. Once
is reached, the recorded
and
values in previous iterations are averaged to obtain new
and
to be used in upcoming iterations. This process is repeated until the termination criterion is satisfied. As for
, the values are dynamically adapted, as follows:
SGHS is outlined, as in Algorithm 4.
| Algorithm 4: Self-Adaptive Global Best Harmony Search | |
| 1: Set , , , using (8), , and | |
| 2: | |
| 3: Compute | |
| 4: Initialize solution counter |
| 5: Generate and based on and |
| 6: while do | |
| 7: for each do | |
| 8: if then | |
| 9: where | |
| 10: if then | |
| 11: | |
| 12: end if | |
| 13: else | |
| 14: | |
| 15: end if | |
| 16: end for | |
| 17: if then | |
| 18: replace | |
| 19: record the values of and | |
| 20: end if | |
| 21: if then | |
| 22: recompute and by averaging the recorded values of and | |
| 23: reset | |
| 24: else | |
| 25: | |
| 26: end if | |
| 27: end while | |
2.5. Novel Self-Adaptive Harmony Search (NSHS)
The Novel Self-Adaptive Harmony Search (NSHS) is a HS variant that was developed by [
51], being inspired by the defects that the creator found in SHS and other variants, namely IHS [
50], GHS [
52], SAHS [
53], Dynamic Local Harmony Search (DLHS) [
56], and SGHS [
55]. In NSHS, the
parameter is constructed based on the dimension of the optimization problem to be solved,
With reference to Equation (
10),
is set to be directly proportional to
n, in order to use the HM more frequently, and it lies in the interval
. The parameter
is removed. Furthermore, a dynamic fine-tuned
is introduced and it depends on the standard deviation
S of the objective function,
.
diminishes in stages according to the iteration number
t, while increasing with a larger range of decision variables. The improvisation step in NSHS generates a new harmony within the narrow range of
based on conditions of
and
fstd. Algorithm 6 outlines the computational procedure of NSHS.
| Algorithm 5: Intelligent Tuned Harmony Search | |
| 1: Set , , using (9), and | |
| 2: | |
| 3: Compute | |
| 4: while do | |
| 5: for each do | |
| 6: if then | |
| 7: where | |
| 8: if then | |
| 9: | |
| 10: if then | %Group 1 |
| 11: if then | |
| 12: | |
| 13: else | %Group 2 |
| 14: | |
| 15: end if | |
| 16: else | |
| 17: | |
| 18: | |
| 19: | |
| 20: end if | |
| 21: | |
| 22: end if | |
| 23: else | |
| 24: | |
| 25: end if | |
| 26: end for | |
| 27: if then | |
| 28: replace | |
| 29: end if | |
| 30: end while | |
| Algorithm 6: Novel Self-Adaptive Harmony Search | |
| 1: Set , using (10), and | |
| 2: | |
| 3: Compute and corresponding | |
| 4: while do | |
| 5: for each do | |
| 6: if then | |
| 7: where | |
| 8: else | |
| 9: if then | |
| 10: | |
| 11: else | |
| 12: | |
| 13: end if | |
| 14: end if | |
| 15: if then | |
| 16: | |
| 17: else | |
| 18: | |
| 19: end if | |
| 20: end for | |
| 21: if then | |
| 22: replace | |
| 23: end if | |
| 24: end while | |
2.6. Global Dynamic Harmony Search (GDHS)
Based on IHS [
50], the Global Dynamic Harmony Search (GDHS) [
58] further improves the improvisation step of HS with dynamic parameters, as well as dynamic upper and lower bounds of the decision variables. The iterative values of
and
are made to be both decreasing and increasing in the search of global optima, as given by:
For
, the dynamic adjustment is adapted from IHS, but with a few modifications, as follows: -4.6cm0cm
and, based on Equation (
6) from IHS, the equation reduces to,
Next, a correction coefficient,
, is introduced at each iteration by:
where
j is the index of the selected harmony in the memory consideration step.
Finally, the dynamic lower and upper bounds are obtained for the random selection step. Algorithm 7 provides the computational steps of GDHS.
| Algorithm 7: Global Dynamic Harmony Search | |
| 1: Set , using (11), using (12), using (14), and | |
| 2: | |
| 3: Compute | |
| 4: while do | |
| 5: for each do | |
| 6: if | |
| 7: where | |
| 8: if | |
| 9: compute using (15) | |
| 10: | |
| 11: if | |
| 12: | |
| 13: end if | |
| 14: end if | |
| 15: else | |
| 16: and | |
| 17: | |
| 18: | |
| 19: end if | |
| 20: end for | |
| 21: if then | |
| 22: replace | |
| 23: end if | |
| 24: end while | |
2.7. Parameter Adaptive Harmony Search (PAHS)
The Parameter Adaptive Harmony Search (PAHS) [
59] focused on the modification of the improvisation step of IHS [
50]. The dynamic values of
,
, and
are generated during each iteration to ensure the global optima is achieved. The authors explored four different combinations of dynamic
and
iterative values i.e., (i) linear
and
; (ii) exponential
and linear
; (iii) linear
and exponential
; and, (iv) exponential
and
. Through computational experiments, it was concluded that linear
and exponential
yields the best performance. In this way,
gradually increases, while
exponentially decreases with respect to the iterations.
and
at each iteration are computed using:
whereas,
is adapted from IHS as it is. PAHS further aggravates the difficulty of finding suitable values as there are six parameters to be set now, rather than only four in IHS. PAHS is detailed in Algorithm 8.
| Algorithm 8: Parameter Adaptive Harmony Search | |
| 1: Set , using (16), using (17), using (6), and | |
| 2: | |
| 3: Compute | |
| 4: while do | |
| 5: for each do | |
| 6: if then | |
| 7: where | |
| 8: if then | |
| 9: | |
| 10: end if | |
| 11: else | |
| 12: | |
| 13: end if | |
| 14: end for | |
| 15: if then | |
| 16: replace | |
| 17: end if | |
| 18: end while | |
2.9. Improved Binary Global Harmony Search (IBGHS)
The Improved Binary Global Harmony Search (IBGHS) [
61] is a binary variant of the NGHS [
54] that aims to improve the two limitations of NGHS namely the local optima trap and slow convergence. The improvisation step is modified with the introduction of a control parameter
in the place of
and a linear combination of the best and worst harmonies in order to improve the global search ability and convergence speed of NGHS. Algorithm 10 provides the computational procedure of IBGHS.
| Algorithm 10: Improved Binary Global Harmony Search | |
| 1: Set , , , , , and | |
| 2: | |
| 3: Compute | |
| 4: while do | |
| 5: for each do | |
| 6: if then | %control |
| 7: | |
| 8: | |
| 9: | %position updating |
| 10: if then | %genetic mutation |
| 11: | |
| 12: else | |
| 13: | |
| 14: if then | |
| 15: | |
| 16: | |
| 17: end if | |
| 18: end if | |
| 19: end for | |
| 20: if then | |
| 21: replace | |
| 22: end if | |
| 23: end while | |
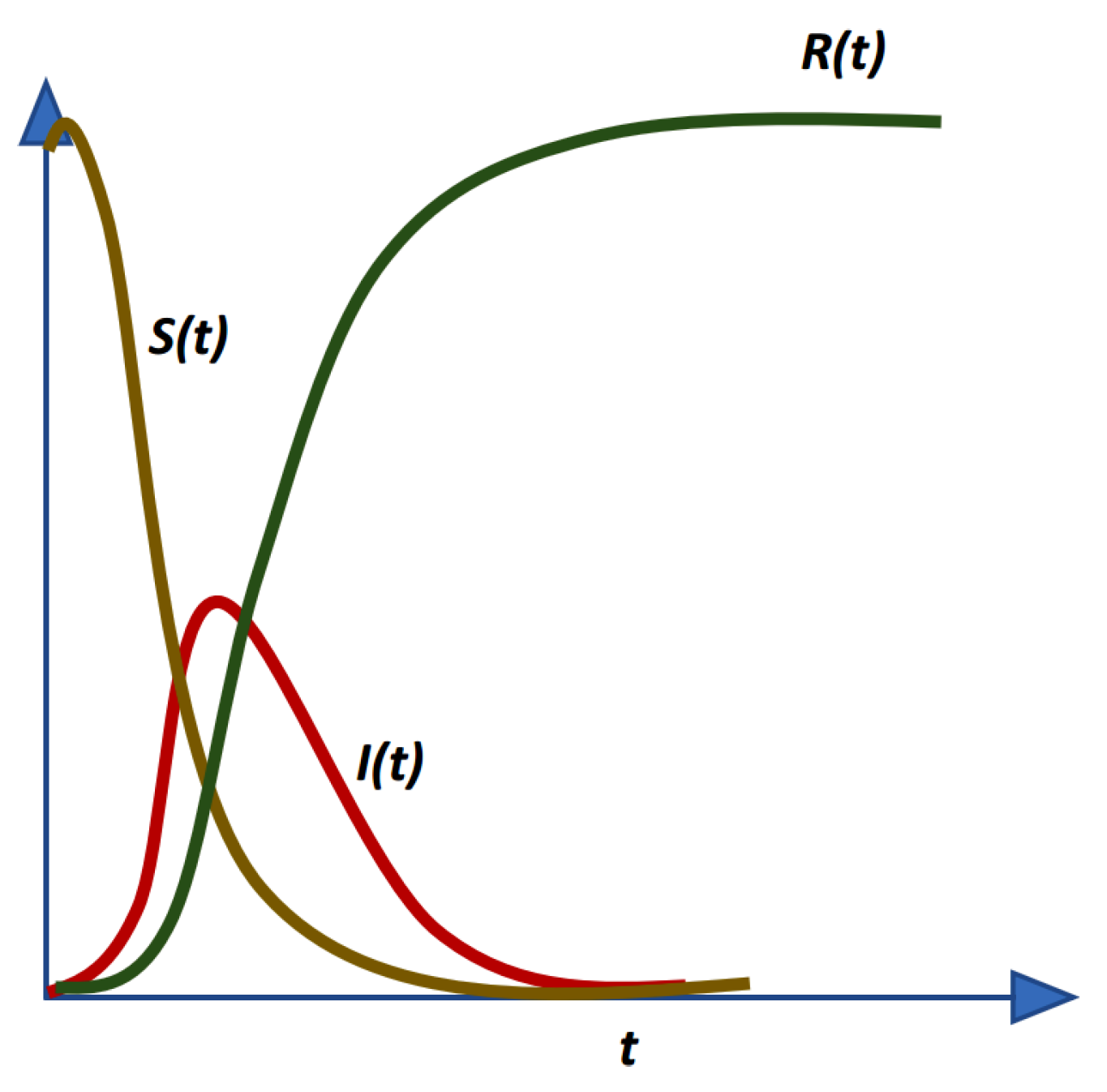
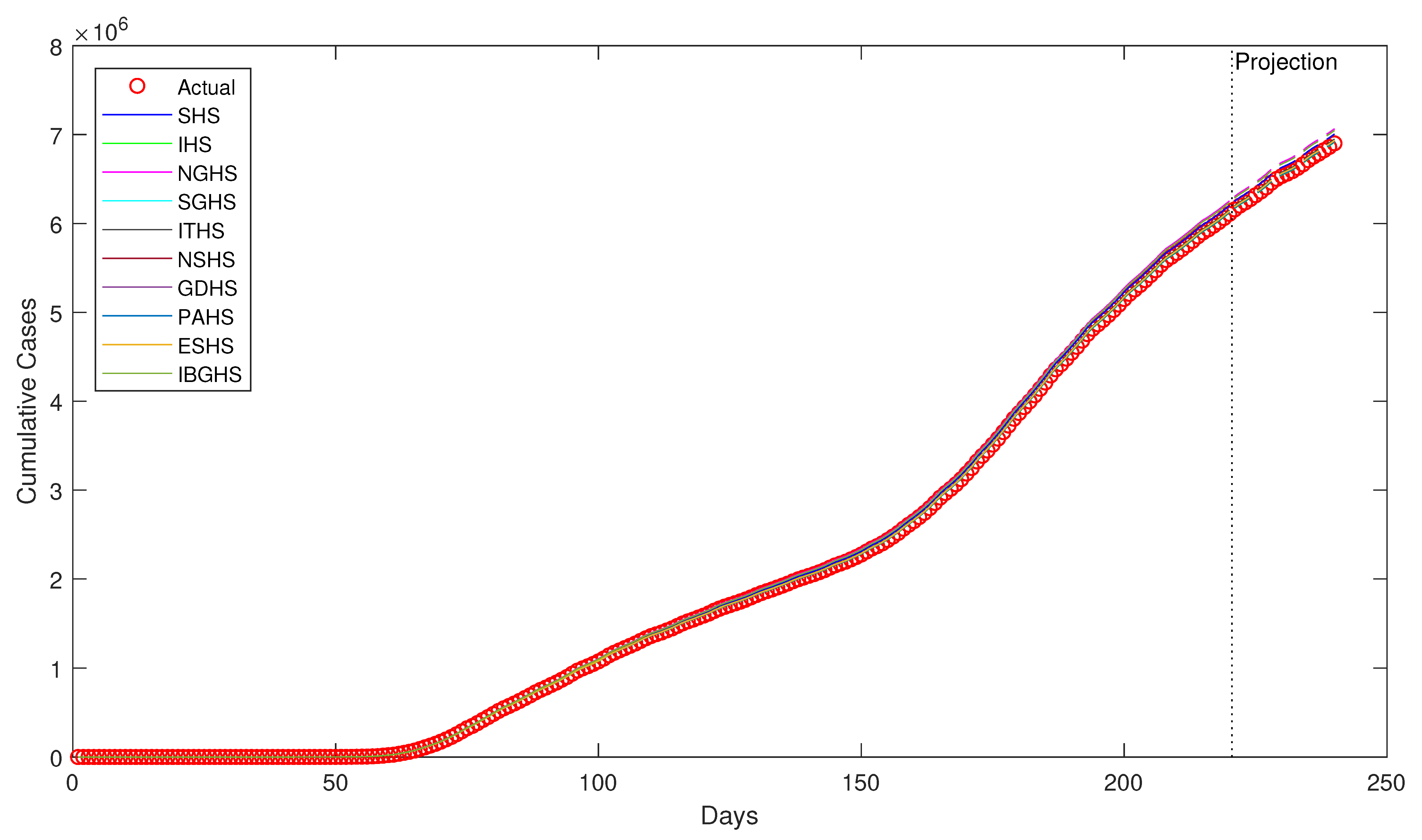
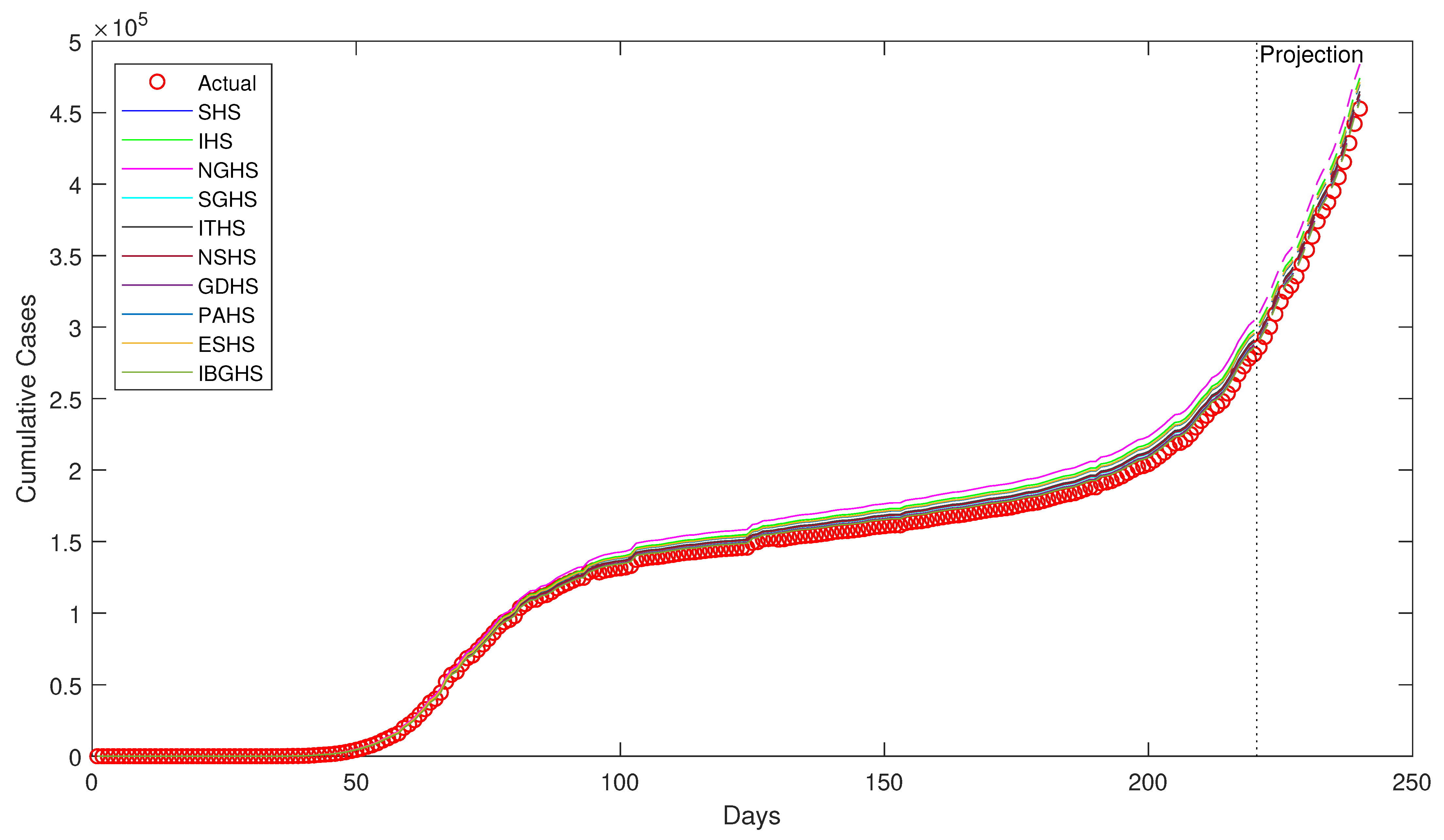
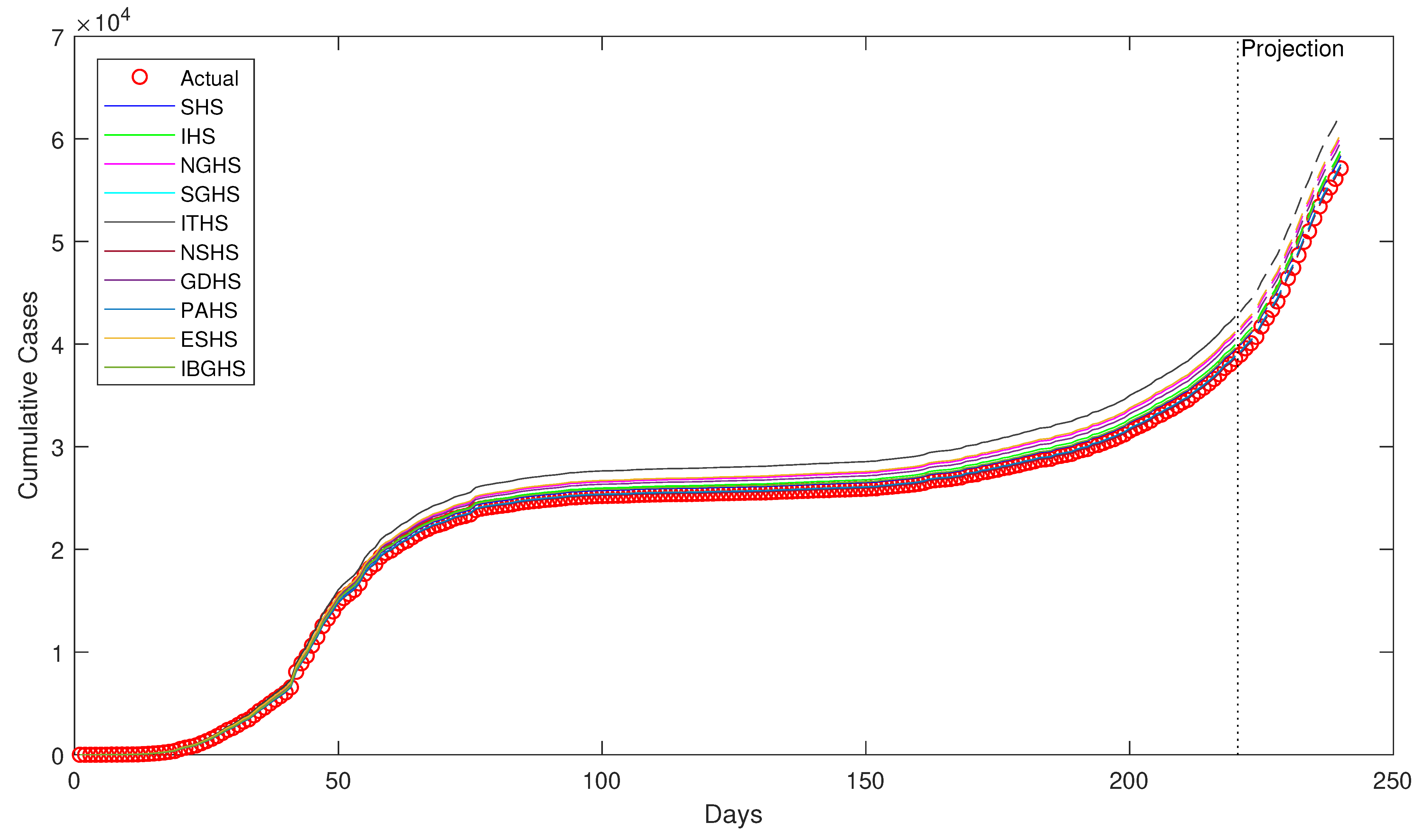
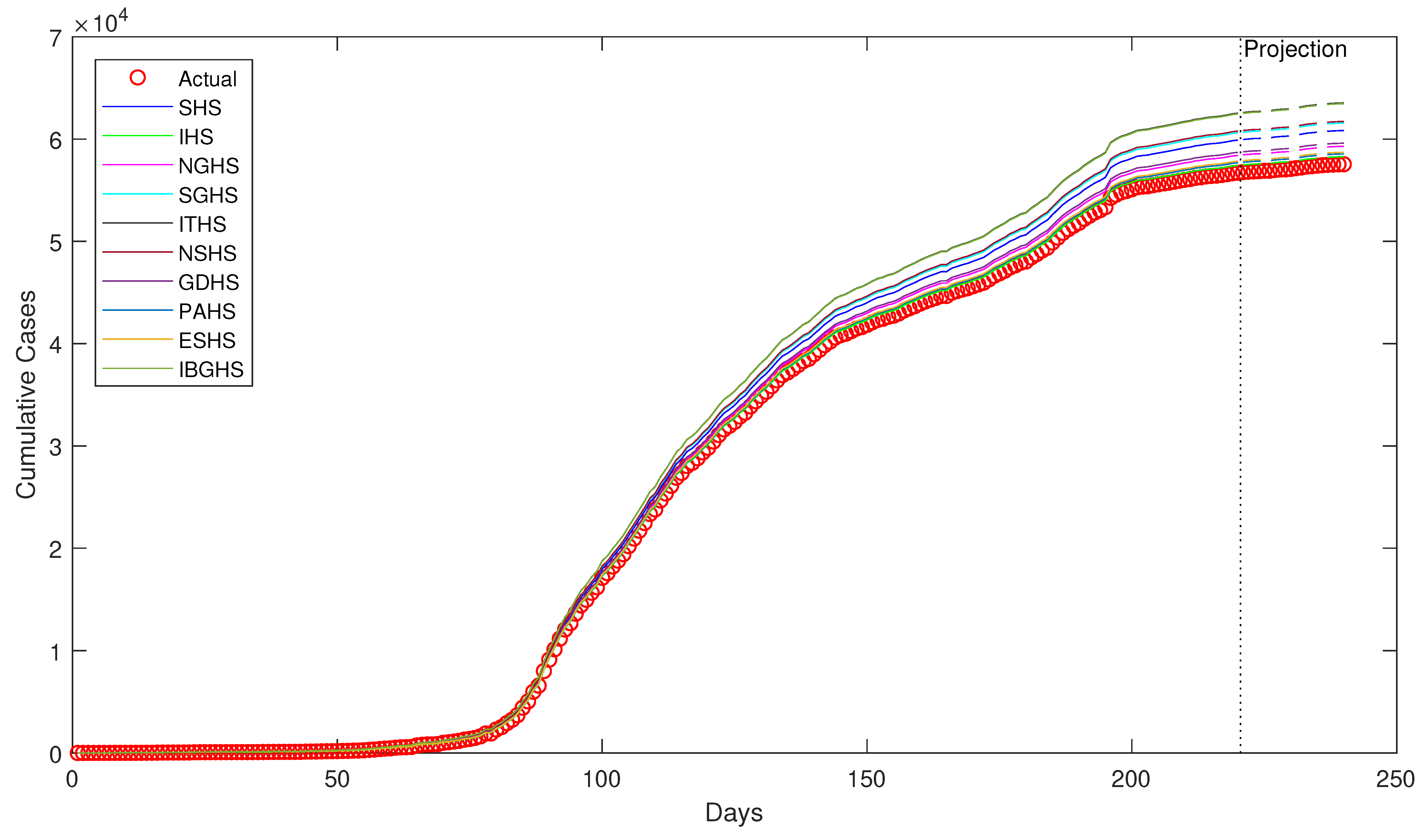
Despite being successfully applied in various fields, to the best of our knowledge, HS algorithms have yet to be applied in epidemiology, particularly in epidemiological modeling. Thus, in this study, ten variants of HS algorithm is proposed to be applied in order to estimate the epidemiological parameters of interest in the prototypical compartmental epidemiological SIR model and compare the estimation performance of each algorithm.