Advanced Label-Free Laser Scanning Microscopy and Its Biological Imaging Application
Abstract
1. Introduction
2. Laser Scanning Coherent Raman Scattering Microscopy (CRSM)
2.1. Laser Scanning Coherent Anti-Stokes Raman Scattering Microscopy (CARSM)
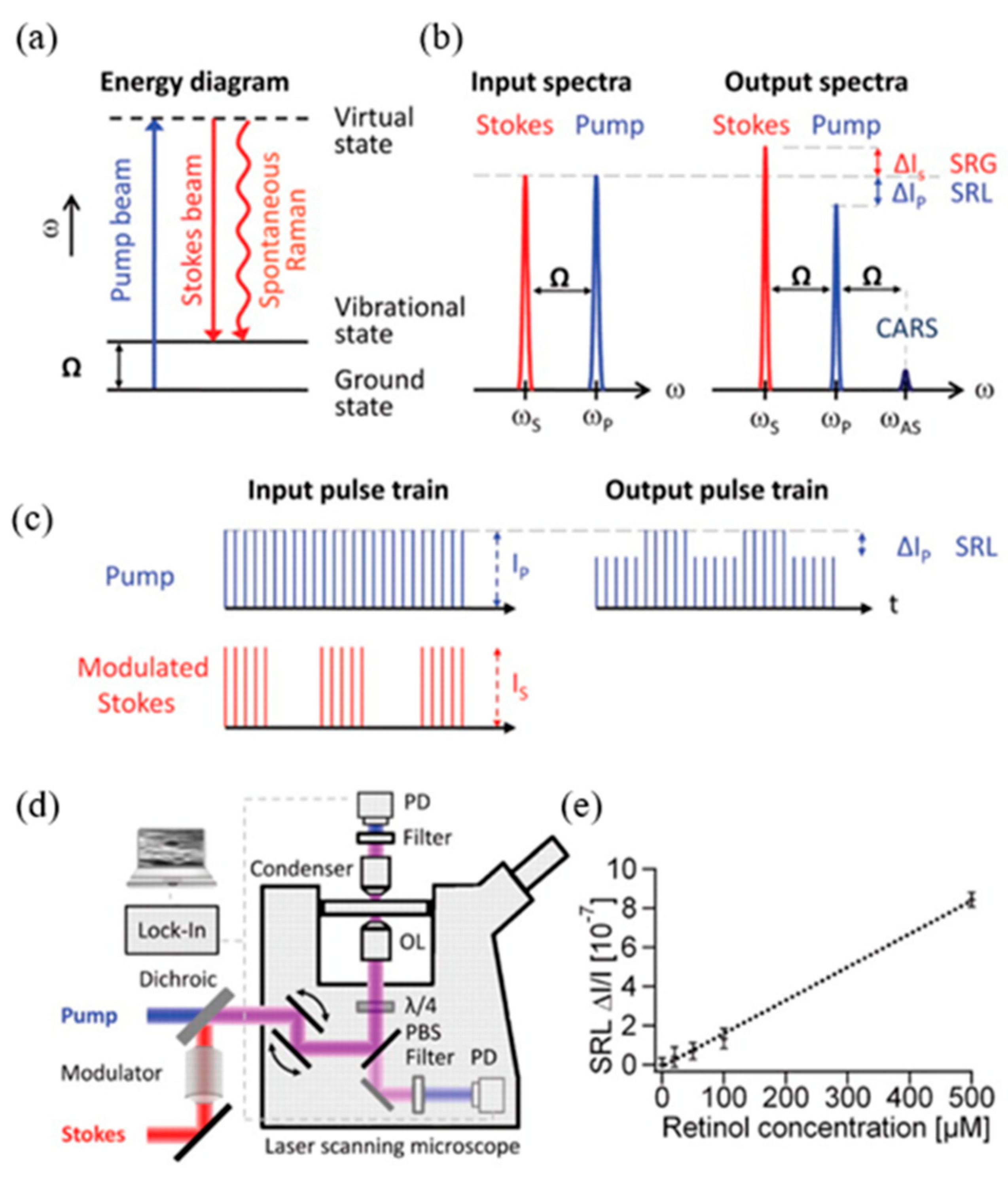
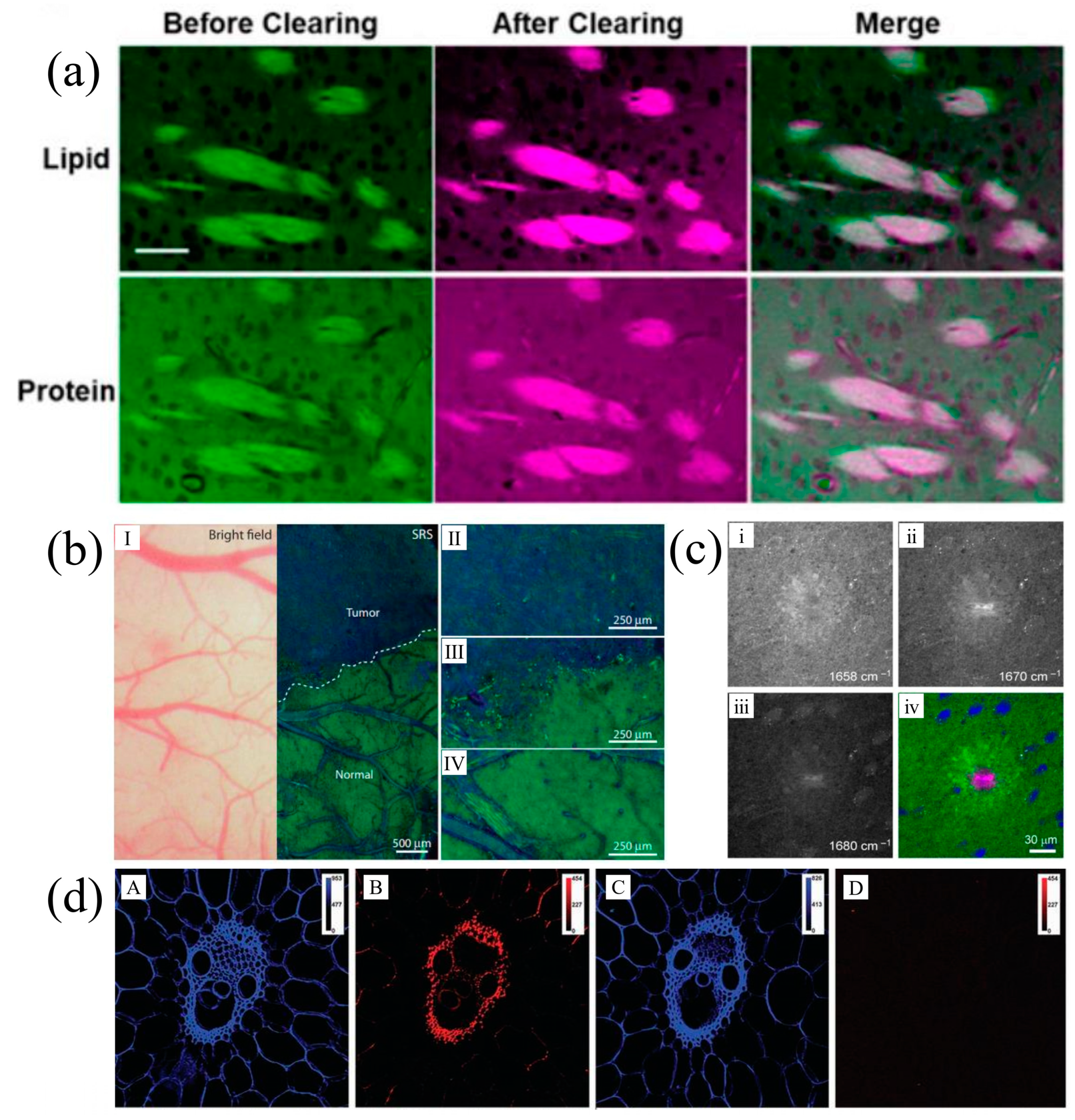
2.2. Laser Scanning Stimulated Raman Scattering Microscopy (SRSM)
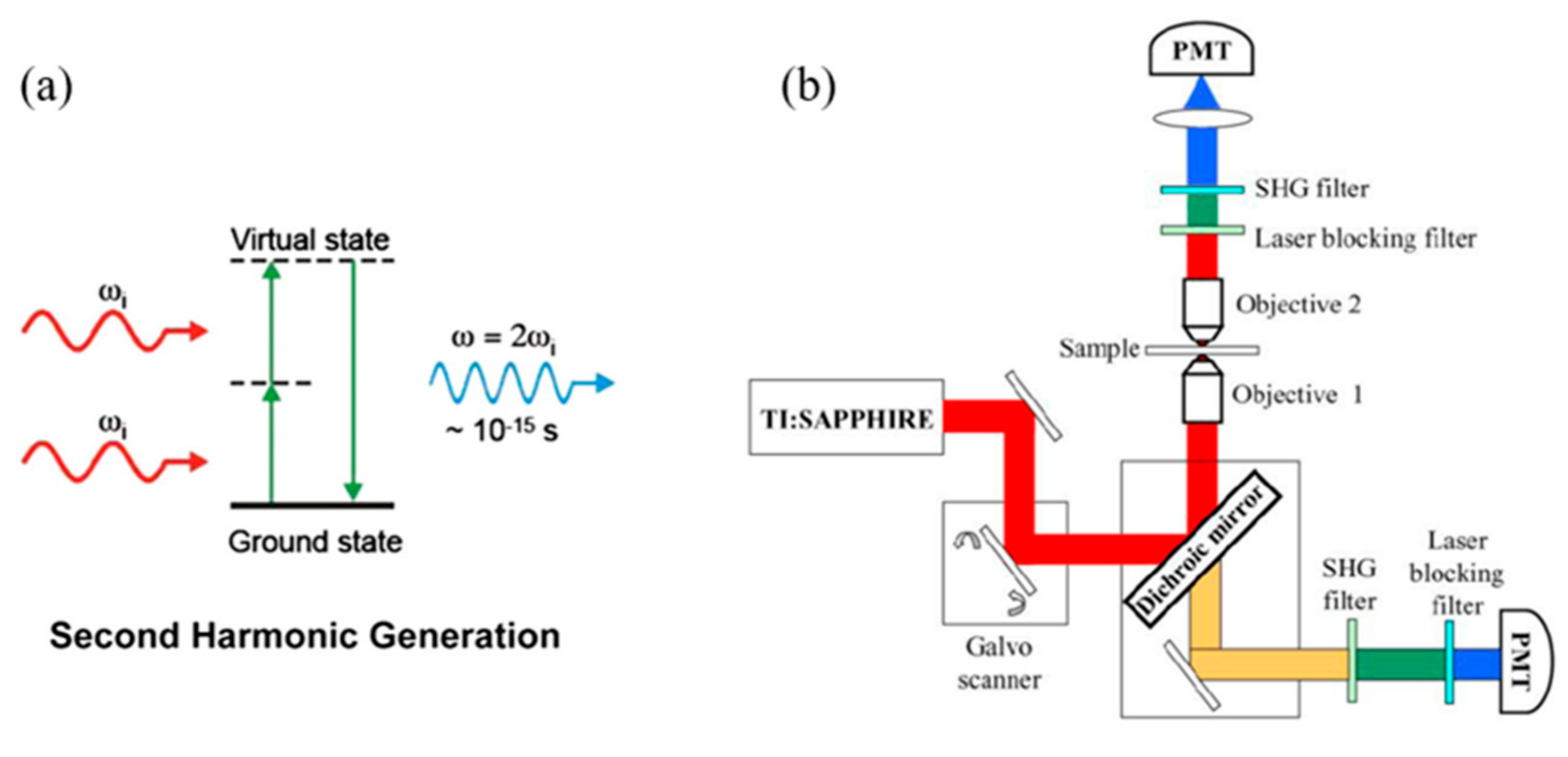
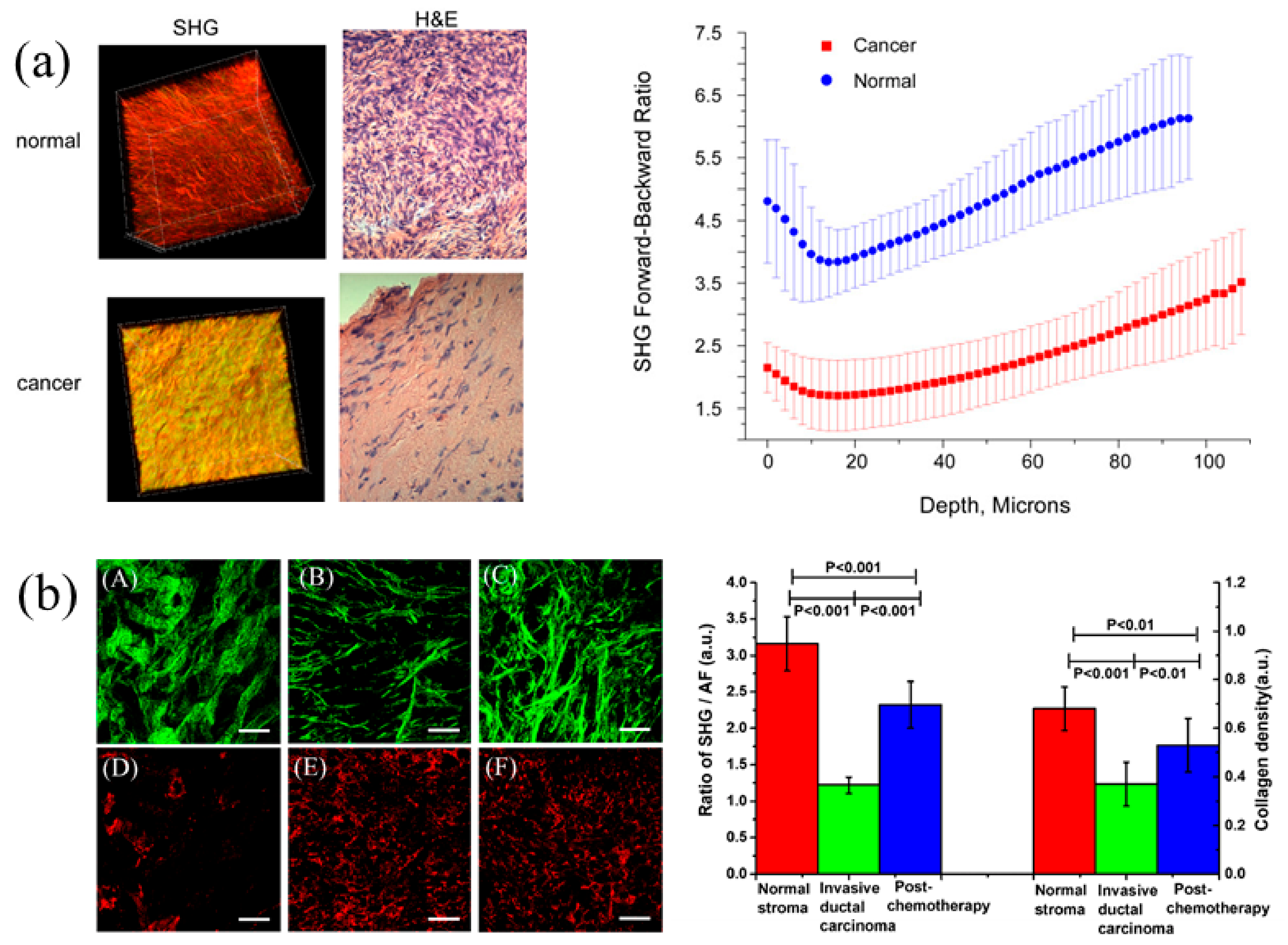
3. Second Harmonic Generation Microscopy (SHGM)
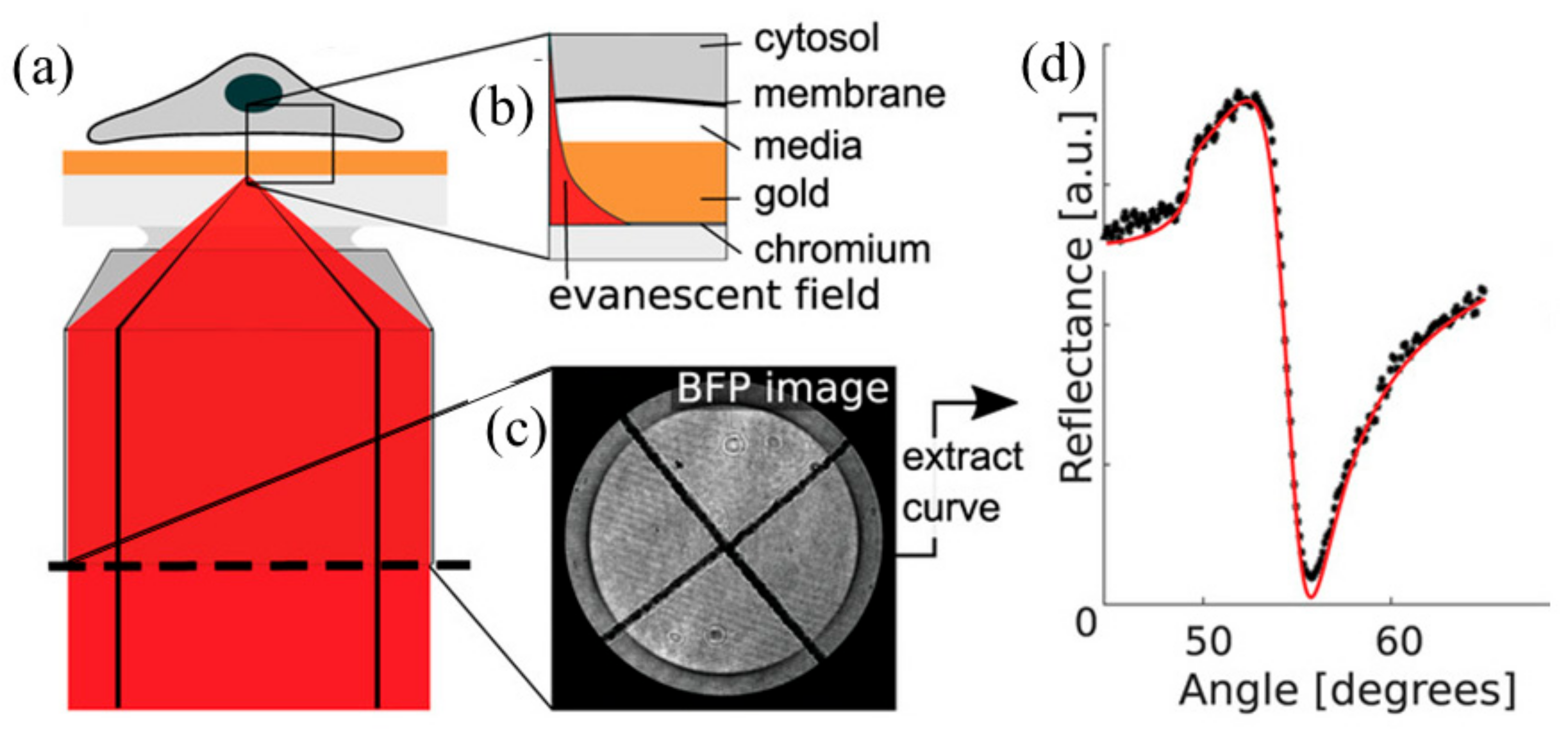
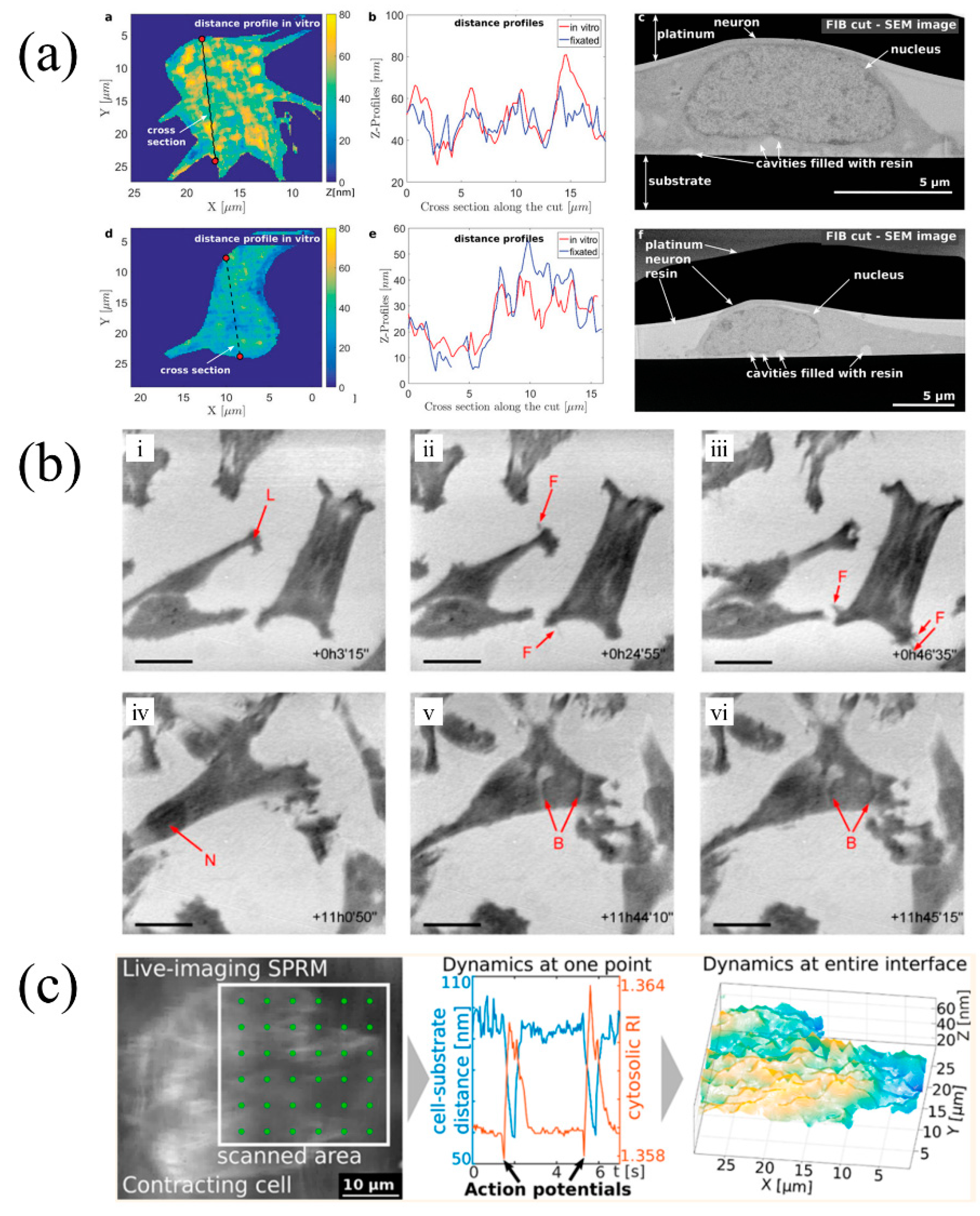
4. Scanning Localized Surface Plasmon Microscopy (SLSPM)
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Minsky, M. Memoir on Inventing the Confocal Scanning Microscope. Scanning 1988, 10, 128–138. [Google Scholar] [CrossRef]
- Hell, S.W.; Wichmann, J. Breaking the Diffraction Resolution Limit by Stimulated-Emission—Stimulated-Emission-Depletion Fluorescence Microscopy. Opt. Lett. 1994, 19, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Vicidomini, G.; Bianchini, P.; Diaspro, A. STED super-resolved microscopy. Nat. Methods 2018, 15, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Gottfert, F.; Wurm, C.A.; Mueller, V.; Berning, S.; Cordes, V.C.; Honigmann, A.; Hell, S.W. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 2013, 105, L01–L03. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Eggeling, C.; Jakobs, S.; Hell, S.W. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 17565–17569. [Google Scholar] [CrossRef]
- Keller, J.; Schonle, A.; Hell, S.W. Efficient fluorescence inhibition patterns for RESOLFT microscopy. Opt. Express 2007, 15, 3361–3371. [Google Scholar] [CrossRef]
- Frawley, A.T.; Wycisk, V.; Xiong, Y.; Galiani, S.; Sezgin, E.; Urbančič, I.; Jentzsch, A.V.; Leslie, K.G.; Eggeling, C.; Anderson, H.L. Super-resolution RESOLFT microscopy of lipid bilayers using a fluorophore-switch dyad. Chem. Sci. 2020, 11, 8955–8960. [Google Scholar] [CrossRef]
- Yamanaka, M.; Yonemaru, Y.; Kawano, S.; Uegaki, K.; Smith, N.I.; Kawata, S.; Fujita, K. Saturated excitation microscopy for sub-diffraction-limited imaging of cell clusters. J. Biomed. Opt. 2013, 18, 126002. [Google Scholar] [CrossRef]
- Nawa, Y.; Yonemaru, Y.; Kasai, A.; Oketani, R.; Hashimoto, H.; Smith, N.I.; Fujita, K. Saturated excitation microscopy using differential excitation for efficient detection of nonlinear fluorescence signals. APL Photonics 2018, 3, 080805. [Google Scholar] [CrossRef]
- Terhune, R.W.; Maker, P.D.; Savage, C.M. Measurements of Nonlinear Light Scattering. Phys. Rev. Lett. 1965, 14, 681–684. [Google Scholar] [CrossRef]
- Duncan, M.D.; Reintjes, J.; Manuccia, T.J. Scanning coherent anti-Stokes Raman microscope. Opt. Lett. 1982, 7, 350–352. [Google Scholar] [CrossRef]
- Potcoava, M.C.; Futia, G.L.; Aughenbaugh, J.; Schlaepfer, I.R.; Gibson, E.A. Raman and coherent anti-Stokes Raman scattering microscopy studies of changes in lipid content and composition in hormone-treated breast and prostate cancer cells. J. Biomed. Opt. 2014, 19, 111605. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, X.X.; Zhou, Y.; Zong, H.; Chen, M.D.; Sun, M.T. Advances in nonlinear optical microscopy for biophotonics. J. Nanophotonics 2018, 12, 033007. [Google Scholar] [CrossRef]
- Cheng, J.X.; Jia, Y.K.; Zheng, G.; Xie, X.S. Laser-scanning coherent anti-Stokes Raman scattering microscopy and applications to cell biology. Biophys. J. 2002, 83, 502–509. [Google Scholar] [CrossRef]
- Evans, C.L.; Potma, E.O.; Puoris’haag, M.; Cote, D.; Lin, C.P.; Xie, X.S. Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 16807–16812. [Google Scholar] [CrossRef]
- Kinegawa, R.; Hiramatsu, K.; Hashimoto, K.; Badarla, V.R.; Ideguchi, T.; Goda, K. High-speed broadband Fourier-transform coherent anti-stokes Raman scattering spectral microscopy. J. Raman Spectrosc. 2019, 50, 1141–1146. [Google Scholar] [CrossRef]
- Yamato, N.; Niioka, H.; Miyake, J.; Hashimoto, M. Improvement of nerve imaging speed with coherent anti-Stokes Raman scattering rigid endoscope using deep-learning noise reduction. Sci. Rep. 2020, 10, 15212. [Google Scholar] [CrossRef]
- Cheng, J.X.; Volkmer, A.; Book, L.D.; Xie, X.S. Multiplex coherent anti-stokes Raman scattering microspectroscopy and study of lipid vesicles. J. Phys. Chem. B 2002, 106, 8493–8498. [Google Scholar] [CrossRef]
- Müller, M.; Schins, J.M. Imaging the Thermodynamic State of Lipid Membranes with Multiplex CARS Microscopy. J. Phys. Chem. B 2002, 106, 3715–3723. [Google Scholar] [CrossRef]
- Petrov, G.I.; Arora, R.; Yakovlev, V.V.; Wang, X.; Sokolov, A.V.; Scully, M.O. Comparison of coherent and spontaneous Raman microspectroscopies for noninvasive detection of single bacterial endospores. Proc. Natl. Acad. Sci. USA 2007, 104, 7776–7779. [Google Scholar] [CrossRef]
- Kano, H.; Hamaguchi, H.-O. Dispersion-compensated supercontinuum generation for ultrabroadband multiplex coherent anti-Stokes Raman scattering spectroscopy. J. Raman Spectrosc. 2006, 37, 411–415. [Google Scholar] [CrossRef]
- Lee, Y.J.; Liu, Y.; Cicerone, M.T. Characterization of three-color CARS in a two-pulse broadband CARS spectrum. Opt. Lett. 2007, 32, 3370–3372. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.X.; Book, L.D.; Xie, X.S. Polarization coherent anti-Stokes Raman scattering microscopy. Opt. Lett. 2001, 26, 1341–1343. [Google Scholar] [CrossRef] [PubMed]
- Oron, D.; Dudovich, N.; Silberberg, Y. Femtosecond phase-and-polarization control for background-free coherent anti-Stokes Raman spectroscopy. Phys. Rev. Lett. 2003, 90, 213902. [Google Scholar] [CrossRef] [PubMed]
- Pestov, D.; Murawski, R.K.; Ariunbold, G.O.; Wang, X.; Zhi, M.; Sokolov, A.V.; Sautenkov, V.A.; Rostovtsev, Y.V.; Dogariu, A.; Huang, Y. Optimizing the laser-pulse configuration for coherent Raman spectroscopy. Science 2007, 316, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Camp, C.H., Jr.; Lee, Y.J.; Heddleston, J.M.; Hartshorn, C.M.; Hight Walker, A.R.; Rich, J.N.; Lathia, J.D.; Cicerone, M.T. High-Speed Coherent Raman Fingerprint Imaging of Biological Tissues. Nat. Photonics 2014, 8, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bryant, G.W.; Stranick, S.J. Superresolution four-wave mixing microscopy. Opt. Express 2012, 20, 6042–6051. [Google Scholar] [CrossRef]
- Gong, L.; Lin, J.; Hao, C.; Zheng, W.; Wu, S.Q.Y.; Teng, J.; Qiu, C.W.; Huang, Z. Supercritical focusing coherent anti-Stokes Raman scattering microscopy for high-resolution vibrational imaging. Opt. Lett. 2018, 43, 5615–5618. [Google Scholar] [CrossRef]
- Wang, D.; Fu, W.; Lei, Y.; Cai, H.; Liu, J. A theoretical study of diffraction limit breaking via coherent control of the relative phase in coherent anti-Stokes Raman scattering microscopy. Opt. Express 2019, 27, 5005–5013. [Google Scholar] [CrossRef]
- Gong, L.; Zheng, W.; Ma, Y.; Huang, Z. Higher-order coherent anti-Stokes Raman scattering microscopy realizes label-free super-resolution vibrational imaging. Nat. Photonics 2019, 14, 115–122. [Google Scholar] [CrossRef]
- Dorosz, A.; Grosicki, M.; Dybas, J.; Matuszyk, E.; Rodewald, M.; Meyer, T.; Popp, J.; Malek, K.; Baranska, M. Eosinophils and Neutrophils-Molecular Differences Revealed by Spontaneous Raman, CARS and Fluorescence Microscopy. Cells 2020, 9, 2041. [Google Scholar] [CrossRef] [PubMed]
- Nuriya, M.; Yoneyama, H.; Takahashi, K.; Leproux, P.; Couderc, V.; Yasui, M.; Kano, H. Characterization of Intra/Extracellular Water States Probed by Ultrabroadband Multiplex Coherent Anti-Stokes Raman Scattering (CARS) Spectroscopic Imaging. J. Phys. Chem. A 2019, 123, 3928–3934. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, E.R.; Corwin, E.I.; Greenblatt, N.A.; Ashmore, J.; Wang, D.Y.; Dinsmore, A.D.; Cheng, J.X.; Xie, X.S.; Hutchinson, J.W.; Weitz, D.A. Flow and fracture in drying nanoparticle suspensions. Phys. Rev. Lett. 2003, 91, 224501. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Yang, W.Y.; Xie, X.S. CARS Microscopy Lights up Lipids in Living Cells. Biophotonics Int. 2004, 11, 44–47. [Google Scholar]
- Potma, E.O.; Xie, X.S. Direct visualization of lipid phase segregation in single lipid bilayers with coherent anti-Stokes Raman scattering microscopy. Chemphyschem 2005, 6, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Jurna, M.; Korterik, J.P.; Otto, C.; Offerhaus, H.L. Shot noise limited heterodyne detection of CARS signals. Opt. Express 2007, 15, 15207–15213. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, H.; Takamuku, S.; Iiyama, A.; Inukai, J. Dynamic Distribution of Chemical States of Water Inside a Nafion Membrane in a Running Fuel Cell Monitored by Operando Time-Resolved CARS Spectroscopy. J. Phys. Chem. C 2020, 124, 19508–19513. [Google Scholar] [CrossRef]
- Roy, S.; Gord, J.R.; Patnaik, A.K. Recent advances in coherent anti-Stokes Raman scattering spectroscopy: Fundamental developments and applications in reacting flows. Prog. Energy Combust. Sci. 2010, 36, 280–306. [Google Scholar] [CrossRef]
- Koivistoinen, J.; Myllyperkio, P.; Pettersson, M. Time-Resolved Coherent Anti-Stokes Raman Scattering of Graphene: Dephasing Dynamics of Optical Phonon. J. Phys. Chem. Lett. 2017, 8, 4108–4112. [Google Scholar] [CrossRef]
- Ling, J.; Miao, X.; Sun, Y.; Feng, Y.; Zhang, L.; Sun, Z.; Ji, M. Vibrational Imaging and Quantification of Two-Dimensional Hexagonal Boron Nitride with Stimulated Raman Scattering. ACS Nano 2019, 13, 14033–14040. [Google Scholar] [CrossRef]
- Li, R.; Wang, L.; Mu, X.; Chen, M.; Sun, M. Nonlinear optical characterization of porous carbon materials by CARS, SHG and TPEF. Spectrochim. Acta A 2019, 214, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Virga, A.; Ferrante, C.; Batignani, G.; De Fazio, D.; Nunn, A.D.G.; Ferrari, A.C.; Cerullo, G.; Scopigno, T. Coherent anti-Stokes Raman spectroscopy of single and multi-layer graphene. Nat. Commun. 2019, 10, 3658. [Google Scholar] [CrossRef] [PubMed]
- Freudiger, C.W.; Min, W.; Saar, B.G.; Lu, S.; Holtom, G.R.; He, C.; Tsai, J.C.; Kang, J.X.; Xie, X.S. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008, 322, 1857–1861. [Google Scholar] [CrossRef]
- Eckhardt, G.; Hellwarth, R.W.; McClung, F.J.; Schwarz, S.E.; Weiner, D.; Woodbury, E.J. Stimulated Raman Scattering from Organic Liquids. Phys. Rev. Lett. 1962, 9, 455–457. [Google Scholar] [CrossRef]
- Hellwarth, R.W. Theory of Stimulated Raman Scattering. Phys. Rev. 1963, 130, 1850–1852. [Google Scholar] [CrossRef]
- Ploetz, E.; Laimgruber, S.; Berner, S.; Zinth, W.; Gilch, P. Femtosecond stimulated Raman microscopy. Appl. Phys. B 2007, 87, 389–393. [Google Scholar] [CrossRef]
- Liao, C.-S.; Huang, K.-C.; Hong, W.; Chen, A.J.; Karanja, C.; Wang, P.; Eakins, G.; Cheng, J.-X. Stimulated Raman spectroscopic imaging by microsecond delay-line tuning. Optica 2016, 3, 1377–1380. [Google Scholar] [CrossRef]
- Silva, W.R.; Graefe, C.T.; Frontiera, R.R. Toward Label-Free Super-Resolution Microscopy. ACS Photonics 2015, 3, 79–86. [Google Scholar] [CrossRef]
- Bi, Y.; Yang, C.; Chen, Y.; Yan, S.; Yang, G.; Wu, Y.; Zhang, G.; Wang, P. Near-resonance enhanced label-free stimulated Raman scattering microscopy with spatial resolution near 130 nm. Light Sci. Appl. 2018, 7, 81. [Google Scholar] [CrossRef]
- Gong, L.; Zheng, W.; Ma, Y.; Huang, Z. Saturated Stimulated-Raman-Scattering Microscopy for Far-Field Superresolution Vibrational Imaging. Phys. Rev. Appl. 2019, 11, 034041. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, C.; Lin, P.; Huang, K.C.; Liang, J.; Tian, J.; Cheng, J.X. Volumetric chemical imaging by stimulated Raman projection microscopy and tomography. Nat. Commun. 2017, 8, 15117. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Shi, L.; Shen, Y.; Zhao, Z.; Guzman, A.; Kaufman, L.J.; Wei, L.; Min, W. Volumetric chemical imaging by clearing-enhanced stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 6608–6617. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Shi, L.; Min, W. Biological imaging of chemical bonds by stimulated Raman scattering microscopy. Nat. Methods 2019, 16, 830–842. [Google Scholar] [CrossRef]
- Bae, K.; Zheng, W.; Huang, Z. Spatial light-modulated stimulated Raman scattering (SLM-SRS) microscopy for rapid multiplexed vibrational imaging. Theranostics 2019, 10, 312–322. [Google Scholar] [CrossRef]
- Freudiger, C.W.; Pfannl, R.; Orringer, D.A.; Saar, B.G.; Ji, M.; Zeng, Q.; Ottoboni, L.; Wei, Y.; Waeber, C.; Sims, J.R.; et al. Multicolored stain-free histopathology with coherent Raman imaging. Lab. Investig. 2012, 92, 1492–1502. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Li, J.; Lee, S.Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef]
- Lu, F.K.; Basu, S.; Igras, V.; Hoang, M.P.; Ji, M.; Fu, D.; Holtom, G.R.; Neel, V.A.; Freudiger, C.W.; Fisher, D.E.; et al. Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E5902. [Google Scholar] [CrossRef]
- Ji, M.; Orringer, D.A.; Freudiger, C.W.; Ramkissoon, S.; Liu, X.; Lau, D.; Golby, A.J.; Norton, I.; Hayashi, M.; Agar, N.Y.; et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci. Transl. Med. 2013, 5, 201ra119. [Google Scholar] [CrossRef]
- Ji, M.; Arbel, M.; Zhang, L.; Freudiger, C.W.; Hou, S.S.; Lin, D.; Yang, X.; Bacskai, B.J.; Xie, X.S. Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy. Sci. Adv. 2018, 4, eaat7715. [Google Scholar] [CrossRef]
- Saar, B.G.; Zeng, Y.; Freudiger, C.W.; Liu, Y.S.; Himmel, M.E.; Xie, X.S.; Ding, S.Y. Label-free, real-time monitoring of biomass processing with stimulated Raman scattering microscopy. Angew. Chem. Int. Ed. Engl. 2010, 49, 5476–5479. [Google Scholar] [CrossRef]
- Ding, S.Y.; Liu, Y.S.; Zeng, Y.; Himmel, M.E.; Baker, J.O.; Bayer, E.A. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 2012, 338, 1055–1060. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, Y.; Dake, F.; Kajiyama, S.; Fukui, K.; Itoh, K. Analysis and experimental assessment of the sensitivity of stimulated Raman scattering microscopy. Opt. Express 2009, 17, 3651–3658. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, Y.; Umemura, W.; Otsuka, Y.; Satoh, S.; Hashimoto, H.; Sumimura, K.; Nishizawa, N.; Fukui, K.; Itoh, K. High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nat. Photonics 2012, 6, 845–851. [Google Scholar] [CrossRef]
- Nandakumar, P.; Kovalev, A.; Volkmer, A. Vibrational imaging based on stimulated Raman scattering microscopy. New J. Phys. 2009, 11, 033026. [Google Scholar] [CrossRef]
- Saar, B.G.; Freudiger, C.W.; Reichman, J.; Stanley, C.M.; Holtom, G.R.; Xie, X.S. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science 2010, 330, 1368–1370. [Google Scholar] [CrossRef]
- Pantazis, P.; Maloney, J.; Wu, D.; Fraser, S.E. Second harmonic generating (SHG) nanoprobes for in vivo imaging. Proc. Natl. Acad. Sci. USA 2010, 107, 14535–14540. [Google Scholar] [CrossRef]
- Franken, P.A.; Hill, A.E.; Peters, C.W.; Weinreich, G. Generation of Optical Harmonics. Phys. Rev. Lett. 1961, 7, 118–119. [Google Scholar] [CrossRef]
- Hellwarth, R.; Christensen, P. Nonlinear optical microscopic examination of structure in polycrystalline ZnSe. Opt. Commun. 1974, 12, 318–322. [Google Scholar] [CrossRef]
- Sivaguru, M.; Durgam, S.; Ambekar, R.; Luedtke, D.; Fried, G.; Stewart, A.; Toussaint, K.C., Jr. Quantitative analysis of collagen fiber organization in injured tendons using Fourier transform-second harmonic generation imaging. Opt. Express 2010, 18, 24983–24993. [Google Scholar] [CrossRef]
- Freund, I.; Deutsch, M. Second-harmonic microscopy of biological tissue. Opt. Lett. 1986, 11, 94. [Google Scholar] [CrossRef]
- Bouevitch, O.; Lewis, A.; Pinevsky, I.; Wuskell, J.P.; Loew, L.M. Probing membrane potential with nonlinear optics. Biophys. J. 1993, 65, 672–679. [Google Scholar] [CrossRef]
- Campagnola, P.J.; Wei, M.-D.; Lewis, A.; Loew, L.M. High-Resolution Nonlinear Optical Imaging of Live Cells by Second Harmonic Generation. Biophys. J. 1999, 77, 3341–3349. [Google Scholar] [CrossRef]
- Bianchini, P.; Diaspro, A. Three-dimensional (3D) backward and forward second harmonic generation (SHG) microscopy of biological tissues. J. Biophoton. 2008, 1, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Tan, C.Z.; Cheng, C.A.; Hung, J.T.; Chen, S.Y. Improving resolution of second harmonic generation microscopy via scanning structured illumination. Biomed. Opt. Express 2018, 9, 6081–6090. [Google Scholar] [CrossRef]
- Hashimoto, M.; Niioka, H.; Ashida, K.; Yoshiki, K.; Araki, T. High-sensitivity and high-spatial-resolution imaging of self-assembled monolayer on platinum using radially polarized beam excited second-harmonic-generation microscopy. Appl. Phys. Express 2015, 8, 112401. [Google Scholar] [CrossRef]
- Bautista, G.; Huttunen, M.J.; Makitalo, J.; Kontio, J.M.; Simonen, J.; Kauranen, M. Second-harmonic generation imaging of metal nano-objects with cylindrical vector beams. Nano Lett. 2012, 12, 3207–3212. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, B.; Liu, P.; Liu, J.; Tan, J. Calculations of second harmonic generation with radially polarized excitations by elliptical mirror focusing. J. Microsc. 2019, 273, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Tian, N.; Fu, L.; Gu, M. Resolution and contrast enhancement of subtractive second harmonic generation microscopy with a circularly polarized vortex beam. Sci. Rep. 2015, 5, 13580. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, C.J.R.; Castello, M.; Tortarolo, G.; Deguchi, T.; Koho, S.V.; Vicidomini, G.; Diaspro, A. Pixel reassignment in image scanning microscopy: A re-evaluation. J. Opt. Soc. Am. A 2020, 37, 154–162. [Google Scholar] [CrossRef]
- Wang, W.; Wu, B.; Zhang, B.; Zhang, Z.; Li, X.; Zheng, S.; Fan, Z.; Tan, J. Second harmonic generation microscopy using pixel reassignment. J. Microsc. 2021, 281, 97–105. [Google Scholar] [CrossRef]
- Chen, X.; Nadiarynkh, O.; Plotnikov, S.; Campagnola, P.J. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 2012, 7, 654–669. [Google Scholar] [CrossRef] [PubMed]
- Keikhosravi, A.; Bredfeldt, J.S.; Sagar, A.K.; Eliceiri, K.W. Second-harmonic generation imaging of cancer. Methods Cell Biol. 2014, 123, 531–546. [Google Scholar]
- Ducourthial, G.; Affagard, J.S.; Schmeltz, M.; Solinas, X.; Lopez-Poncelas, M.; Bonod-Bidaud, C.; Rubio-Amador, R.; Ruggiero, F.; Allain, J.M.; Beaurepaire, E.; et al. Monitoring dynamic collagen reorganization during skin stretching with fast polarization-resolved second harmonic generation imaging. J. Biophotonics 2019, 12, e201800336. [Google Scholar] [CrossRef]
- Nadiarnykh, O.; LaComb, R.B.; Brewer, M.A.; Campagnola, P.J. Alterations of the extracellular matrix in ovarian cancer studied by Second Harmonic Generation imaging microscopy. BMC Cancer 2010, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, Y.; Tang, Q.; Li, Z.; Horng, H.; Li, J.; Wu, Z.; Chen, Y.; Li, H. Quantitative evaluation of redox ratio and collagen characteristics during breast cancer chemotherapy using two-photon intrinsic imaging. Biomed. Opt. Express 2018, 9, 1375–1388. [Google Scholar] [CrossRef]
- Tsafas, V.; Gavgiotaki, E.; Tzardi, M.; Tsafa, E.; Fotakis, C.; Athanassakis, I.; Filippidis, G. Polarization-dependent second-harmonic generation for collagen-based differentiation of breast cancer samples. J. Biophotonics 2020, 13, e202000180. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, D.; Cisek, R.; Joseph, A.; Golaraei, A.; Mirsanaye, K.; Krouglov, S.; Asa, S.L.; Wilson, B.C.; Barzda, V. Characterization of Pancreatic Cancer Tissue Using Multiphoton Excitation Fluorescence and Polarization-Sensitive Harmonic Generation Microscopy. Front. Oncol. 2019, 9, 272. [Google Scholar] [CrossRef]
- Olivier, N.; Luengo-Oroz, M.A.; Duloquin, L.; Faure, E.; Savy, T.; Veilleux, I.; Solinas, X.; Debarre, D.; Bourgine, P.; Santos, A.; et al. Cell Lineage Reconstruction of Early Zebrafish Embryos Using Label-Free Nonlinear Microscopy. Science 2010, 329, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Zeitoune, A.A.; Luna, J.S.; Salas, K.S.; Erbes, L.; Cesar, C.L.; Andrade, L.A.; Carvahlo, H.F.; Bottcher-Luiz, F.; Casco, V.H.; Adur, J. Epithelial Ovarian Cancer Diagnosis of Second-Harmonic Generation Images: A Semiautomatic Collagen Fibers Quantification Protocol. Cancer Inform. 2017, 16, 1176935117690162. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Y.; Chen, Y.; Gao, Q.; Peng, D.; Lin, H.; Zhan, Z.; Liu, Z.; Zhuo, S. Rapid identification of human ovarian cancer in second harmonic generation images using radiomics feature analyses and tree-based pipeline optimization tool. J. Biophotonics 2020, 13, e202000050. [Google Scholar] [CrossRef]
- Wen, B.; Campbell, K.R.; Tilbury, K.; Nadiarnykh, O.; Brewer, M.A.; Patankar, M.; Singh, V.; Eliceiri, K.W.; Campagnola, P.J. 3D texture analysis for classification of second harmonic generation images of human ovarian cancer. Sci. Rep. 2016, 6, 35734. [Google Scholar] [CrossRef] [PubMed]
- Nylander, C.; Liedberg, B.; Lind, T. Gas detection by means of surface plasmon resonance. Sens. Actuator 1982, 3, 79–88. [Google Scholar] [CrossRef]
- Liedberg, B.; Nylander, C.; Lunström, I. Surface plasmon resonance for gas detection and biosensing. Sens. Actuator 1983, 4, 299–304. [Google Scholar] [CrossRef]
- Liedberg, B.; Nylander, C.; Lundström, I. Biosensing with surface plasmon resonance—How it all started. Biosens. Bioelectron. 1995, 10, i–ix. [Google Scholar] [CrossRef]
- Watanabe, K.; Matsuura, K.; Kawata, F.; Nagata, K.; Ning, J.; Kano, H. Scanning and non-scanning surface plasmon microscopy to observe cell adhesion sites. Biomed. Opt. Express 2012, 3, 354–359. [Google Scholar] [CrossRef]
- Kano, H.; Knoll, W. A scanning microscope employing localized surface-plasmon-polaritons as a sensing probe. Opt. Commun. 2000, 182, 11–15. [Google Scholar] [CrossRef]
- Homola, J.; Yee, S.S.; Gauglitz, G. Surface plasmon resonance sensors: Review. Sens. Actuators B Chem. 1999, 54, 3–15. [Google Scholar] [CrossRef]
- Yeatman, E.M. Resolution and sensitivity in surface plasmon microscopy and sensing. Biosens. Bioelectron. 1996, 11, 635–649. [Google Scholar] [CrossRef]
- Kano, H.; Mizuguchi, S.; Kawata, S. Excitation of surface-plasmon polaritons by a focused laser beam. J. Opt. Soc. Am. B 1998, 15, 1381–1386. [Google Scholar] [CrossRef]
- Kano, H.; Knoll, W. Locally excited surface-plasmon-polaritons for thickness measurement of LBK films. Opt. Commun. 1998, 153, 235–239. [Google Scholar] [CrossRef]
- Watanabe, K.; Horiguchi, N.; Kano, H. Optimized measurement probe of the localized surface plasmon microscope by using radially polarized illumination. Appl. Opt. 2007, 46, 4985–4990. [Google Scholar] [CrossRef] [PubMed]
- Kreysing, E.; Seyock, S.; Hassani, H.; Brauweiler-Reuters, E.; Neumann, E.; Offenhäusser, A. Correlating Surface Plasmon Resonance Microscopy of Living and Fixated Cells with Electron Microscopy Allows for Investigation of Potential Preparation Artifacts. Adv. Mater. Interfaces 2020, 7, 1901991. [Google Scholar] [CrossRef]
- Kreysing, E.; Hassani, H.; Hampe, N.; Offenhausser, A. Nanometer-Resolved Mapping of Cell-Substrate Distances of Contracting Cardiomyocytes Using Surface Plasmon Resonance Microscopy. ACS Nano 2018, 12, 8934–8942. [Google Scholar] [CrossRef] [PubMed]
- Hassani, H.; Kreysing, E. Noninvasive measurement of the refractive index of cell organelles using surface plasmon resonance microscopy. Opt. Lett. 2019, 44, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Yang, P.T.; Wu, T.H.; Lu, Y.L.; Gu, F.; Sung, K.B.; Lin, C.W. Characteristic investigation of scanning surface plasmon microscopy for nucleotide functionalized nanoarray. Opt. Express 2015, 23, 20104–20114. [Google Scholar] [CrossRef] [PubMed]
- Berguiga, L.; Orobtchouk, R.; Elezgaray, J.; Arneodo, A.; Argoul, F. High-resolution-scanning waveguide microscopy: Spatial refractive index and topography quantification. Opt. Lett. 2017, 42, 2523–2526. [Google Scholar] [CrossRef]
- Somekh, M.G.; Liu, S.G.; Velinov, T.S.; See, C.W. Optical V(z) for high-resolution 2pi surface plasmon microscopy. Opt. Lett. 2000, 25, 823–825. [Google Scholar] [CrossRef]
- Zhang, B.; Pechprasarn, S.; Zhang, J.; Somekh, M.G. Confocal surface plasmon microscopy with pupil function engineering. Opt. Express 2012, 20, 7388–7397. [Google Scholar] [CrossRef]
- Watanabe, K.; Miyazaki, R.; Terakado, G.; Okazaki, T.; Morigaki, K.; Kano, H. High resolution imaging of patterned model biological membranes by localized surface plasmon microscopy. Appl. Opt. 2010, 49, 887–891. [Google Scholar] [CrossRef]
- Giebel, K.; Bechinger, C.; Herminghaus, S.; Riedel, M.; Leiderer, P.; Weiland, U.; Bastmeyer, M. Imaging of cell/substrate contacts of living cells with surface plasmon resonance microscopy. Biophys. J. 1999, 76 Pt 1, 509–516. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Y.; Wang, S.; Nagaraj, V.J.; Liu, Q.; Wu, J.; Tao, N. Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nat. Chem. 2012, 4, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.W.; Yang, Y.; Wang, W.; Wang, S.; Gao, M.; Wu, J.; Tao, N. Plasmonic-Based Electrochemical Impedance Imaging of Electrical Activities in Single Cells. Angew. Chem. Int. Ed. 2017, 56, 8855–8859. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Lakowicz, J.R.; Streppa, L.; Berguiga, L.; Boyer Provera, E.; Ratti, F.; Goillot, E.; Martinez Torres, C.; Schaeffer, L.; Elezgaray, J.; et al. Tracking in real time the crawling dynamics of adherent living cells with a high resolution surface plasmon microscope. Plasmon. Biol. Med. XIII 2016, 9724, 97240G. [Google Scholar]
- Berguiga, L.; Streppa, L.; Boyer-Provera, E.; Martinez-Torres, C.; Schaeffer, L.; Elezgaray, J.; Arneodo, A.; Argoul, F. Time-lapse scanning surface plasmon microscopy of living adherent cells with a radially polarized beam. Appl. Opt. 2016, 55, 1216–1227. [Google Scholar] [CrossRef] [PubMed]
- Toma, K.; Kano, H.; Offenhausser, A. Label-free measurement of cell-electrode cleft gap distance with high spatial resolution surface plasmon microscopy. ACS Nano 2014, 8, 12612–12619. [Google Scholar] [CrossRef] [PubMed]
- Icha, J.; Weber, M.; Waters, J.C.; Norden, C. Phototoxicity in live fluorescence microscopy, and how to avoid it. Bioessays 2017, 39, 1700003–1700015. [Google Scholar] [CrossRef]
- Combs, C.A. Comparing phototoxicity during the development of a zebrafish craniofacial bone using confocal and light sheet fluorescence microscopy techniques. J. Biophotonics 2013, 6, 920–927. [Google Scholar]
- Fu, Y.; Wang, H.; Shi, R.; Cheng, J.X. Characterization of photodamage in coherent anti-Stokes Raman scattering microscopy. Opt. Express 2006, 14, 3942–3951. [Google Scholar] [CrossRef]
- Tong, L.; Lu, Y.; Lee, R.J.; Cheng, J.X. Imaging receptor-mediated endocytosis with a polymeric nanoparticle-based coherent anti-stokes Raman scattering probe. J. Phys. Chem. B 2007, 111, 9980. [Google Scholar] [CrossRef]
- De Oliveira, M.A.; Smith, Z.J.; Knorr, F.; De Araujo, R.E.; Wachsmann-Hogiu, S. Long term Raman spectral study of power-dependent photodamage in red blood cells. Appl. Phys. Lett. 2014, 104, 1517. [Google Scholar] [CrossRef]
- Lombardini, A.; Berto, P.; Duboisset, J.; Andresen, E.R.; Heuke, S.; Büttner, E.; Rimke, I.; Vergnole, S.; Shinkar, V.; de Bettignies, P.; et al. Background-suppressed SRS fingerprint imaging with a fully integrated system using a single optical parametric oscillator. Opt. Express 2020, 28, 14490–14502. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.W.; Liu, T.M.; Sun, C.K.; Lin, C.Y.; Tsai, H.J. Real-time second-harmonic-generation microscopy based on a 2-GHz repetition rate Ti:sapphire laser. Opt. Express 2003, 11, 933. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.W.; Chen, I.H.; Liu, T.M.; Chen, P.C.; Sun, C.K.; Lin, B.L. Multimodal nonlinear spectral microscopy based on a femtosecond Cr: Forsterite laser. Opt. Lett. 2001, 26, 1909–1911. [Google Scholar] [CrossRef] [PubMed]
- Sarri, B.; Canonge, R.; Audier, X.; Simon, E.; Wojak, J.; Caillol, F.; Cador, C.; Marguet, D.; Poizat, F.; Giovannini, M.; et al. Fast stimulated Raman and second harmonic generation imaging for intraoperative gastro-intestinal cancer detection. Sci. Rep. 2019, 9, 10052. [Google Scholar] [CrossRef]
- Xiong, H.; Shi, L.; Wei, L.; Shen, Y.; Long, R.; Zhao, Z.; Min, W. Stimulated Raman Excited Fluorescence Spectroscopy and Imaging. Nat. Photonics 2019, 13, 412–417. [Google Scholar] [CrossRef]
- Adams, W.R.; Mehl, B.; Leiser, E.; Wang, M.; Mahadevan-Jansen, A. Multimodal Nonlinear Optical and Thermal Imaging Platform for Label-Free Characterization of Biological Tissue. bioRxiv 2020, 023820. [Google Scholar] [CrossRef]


| Type | Work Principle | Work Wavelength (nm) | Repetition Rate | Laser Power | Lateral Resolution (nm) | Axial Resolution | Ref. | |
|---|---|---|---|---|---|---|---|---|
| CRS | CARSM | supercritical focusing CARS | 835 (pump) 1107 (Stokes) | 76 MHz | 3 mW (pump) 10 mW (Stokes) | 295 | 3.32 μm | [28] |
| coherent controlling of the relative phase | 785 (Stokes) 727.8 (pump) | - | - | 130 | - | [29] | ||
| higher-order CARS | 1041 (Stokes) 680–1300 (pump) | 80 MHz | 2.0 × 109–3.6 × 1012 W/cm2 (pump) 1.8 × 109–4.7 × 1012 W/cm2 (Stokes) | 196 | - | [30] | ||
| SRSM | near-resonance enhancement | 450 | 80 MHz | 4.2 mW (pump) 2.6 mW (Stokes) (brain tissue) | 130 | - | [49] | |
| Saturated SRS | - | - | 227 × 109 W/cm2 (pump) 214 × 109 W/cm2 (Stokes) | 255 | 3 μm | [50] | ||
| SHGM | structured illumination | 1064 | 40 MHz | 30 mW | 235 | 693 nm | [74] | |
| subtractive imaging | 800 | 80 MHz | - | 210 | - | [78] | ||
| pixel reassignment | 840 | - | - | 300 | - | [80] | ||
| SLSPM | scanning localized SPP | 632.8 | - | - | 186 | 2.3 nm | [103] | |
| scanning localized SPP | 632.8 | - | - | 170 | 0.33 nm | [109] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Lu, X.; Huang, C. Advanced Label-Free Laser Scanning Microscopy and Its Biological Imaging Application. Appl. Sci. 2021, 11, 1002. https://doi.org/10.3390/app11031002
Wang X, Lu X, Huang C. Advanced Label-Free Laser Scanning Microscopy and Its Biological Imaging Application. Applied Sciences. 2021; 11(3):1002. https://doi.org/10.3390/app11031002
Chicago/Turabian StyleWang, Xue, Xinchao Lu, and Chengjun Huang. 2021. "Advanced Label-Free Laser Scanning Microscopy and Its Biological Imaging Application" Applied Sciences 11, no. 3: 1002. https://doi.org/10.3390/app11031002
APA StyleWang, X., Lu, X., & Huang, C. (2021). Advanced Label-Free Laser Scanning Microscopy and Its Biological Imaging Application. Applied Sciences, 11(3), 1002. https://doi.org/10.3390/app11031002








