Comparison of Genotoxicity and Pulmonary Toxicity Study of Modified SiO2 Nanomaterials
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Animals
2.3. In Vitro Genotoxicity Study
2.3.1. Ames Test
2.3.2. Chromosomal Aberration (CA) Test
2.4. In Vivo Genotoxicity and Acute Pulmonary Toxicity Test
2.4.1. Acute Pulmonary Toxicity Test (1 D)
2.4.2. Rat Peripheral Blood Micronucleus Test and Acute Pulmonary Toxicity Test (14 D)
2.5. Statistical Analysis
3. Results
3.1. Ames Test
3.2. CA Test
3.3. MN Test
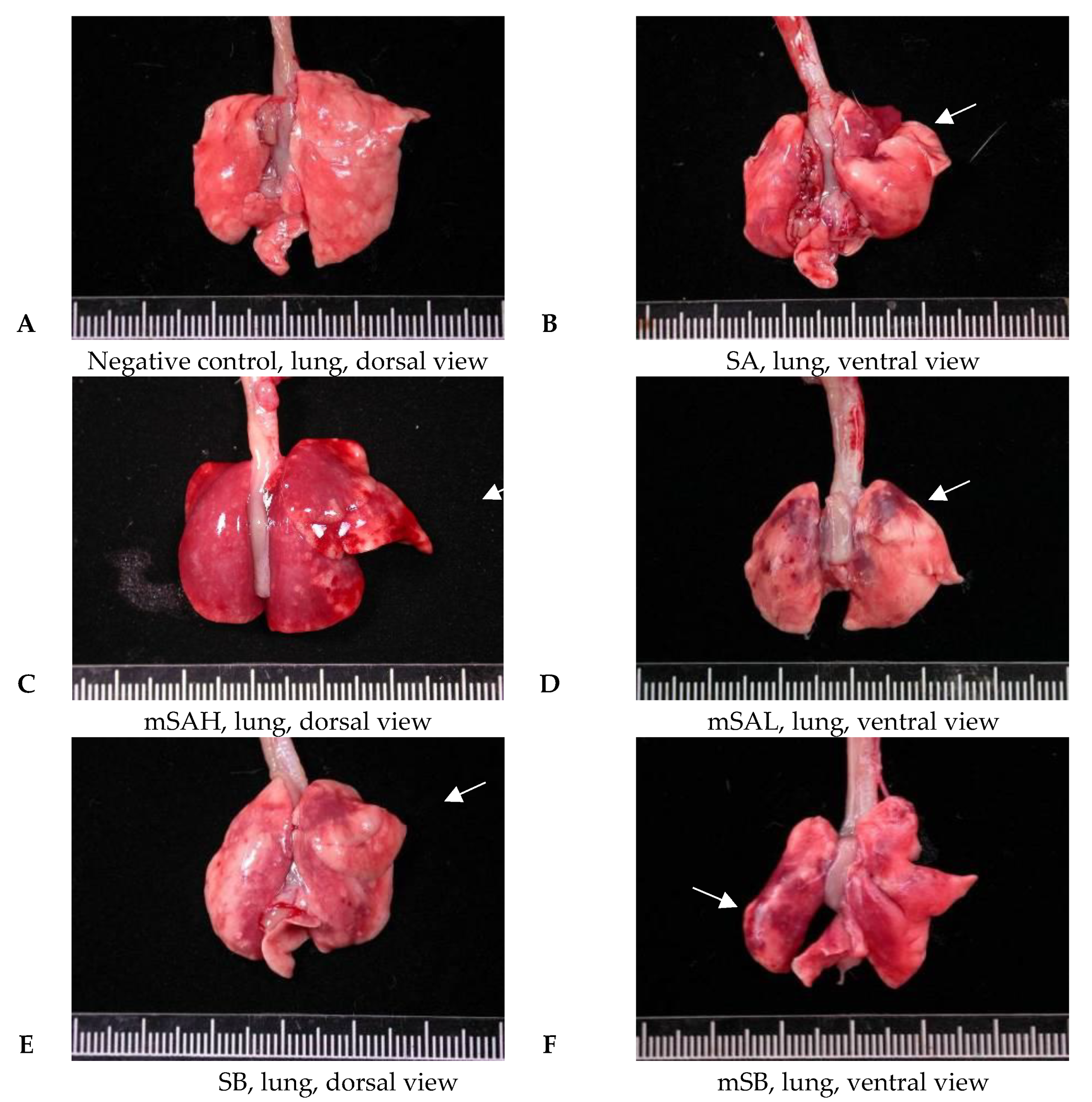
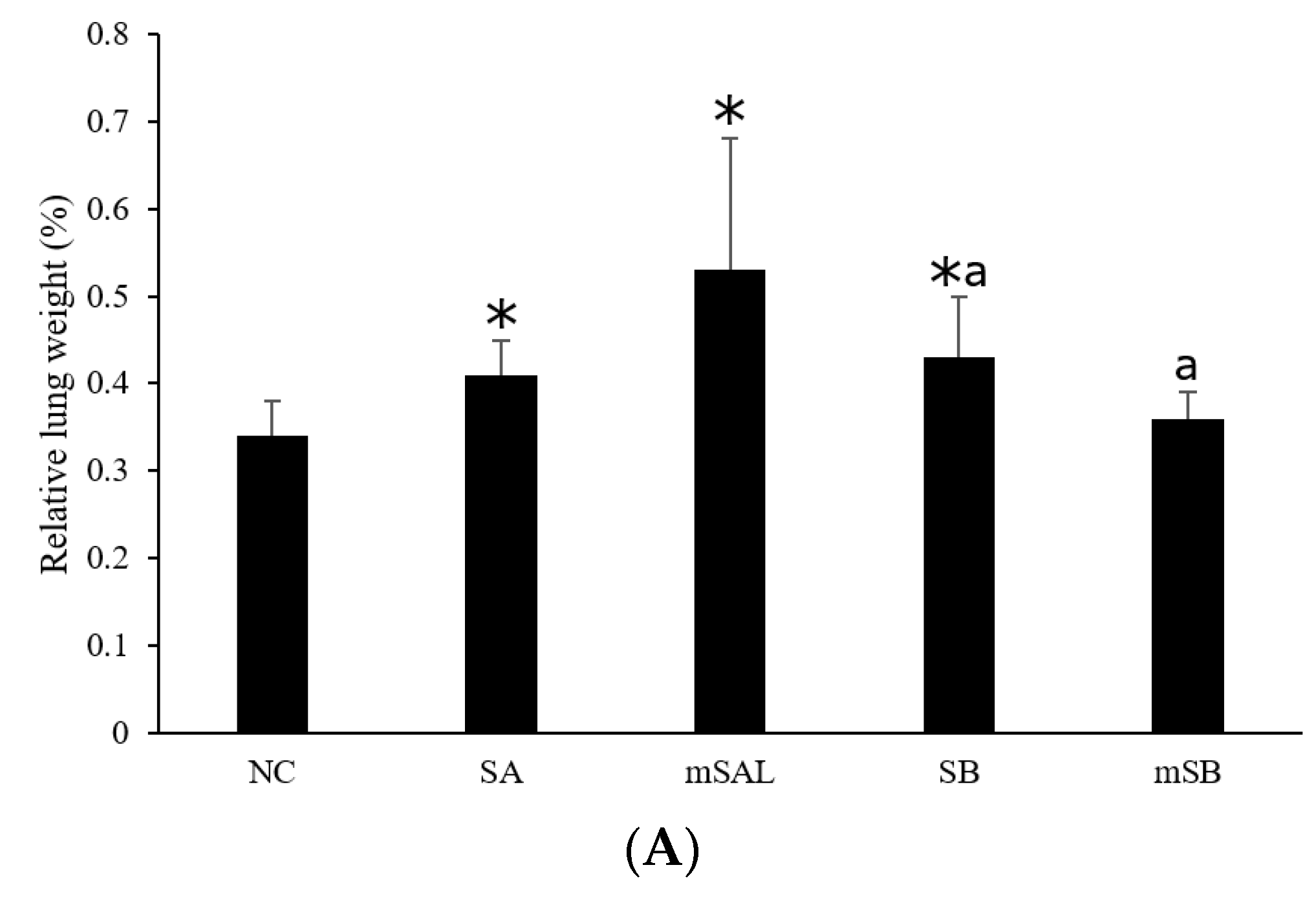
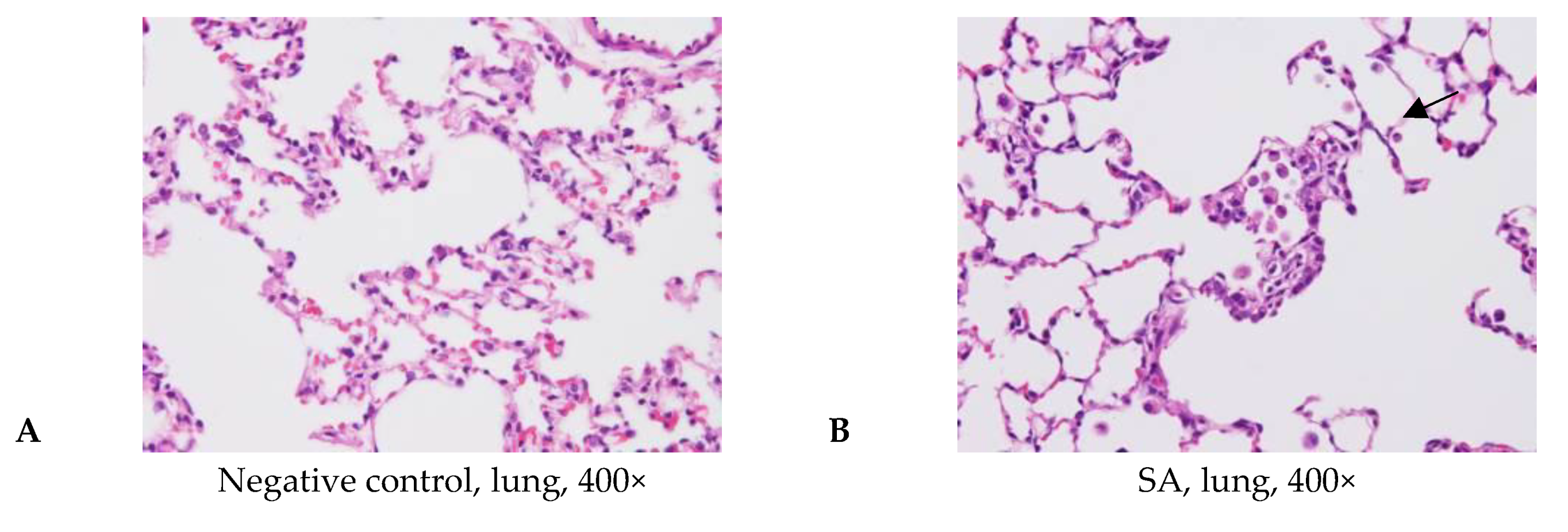
3.4. Acute Pulmonary Toxicity Test (1 D)
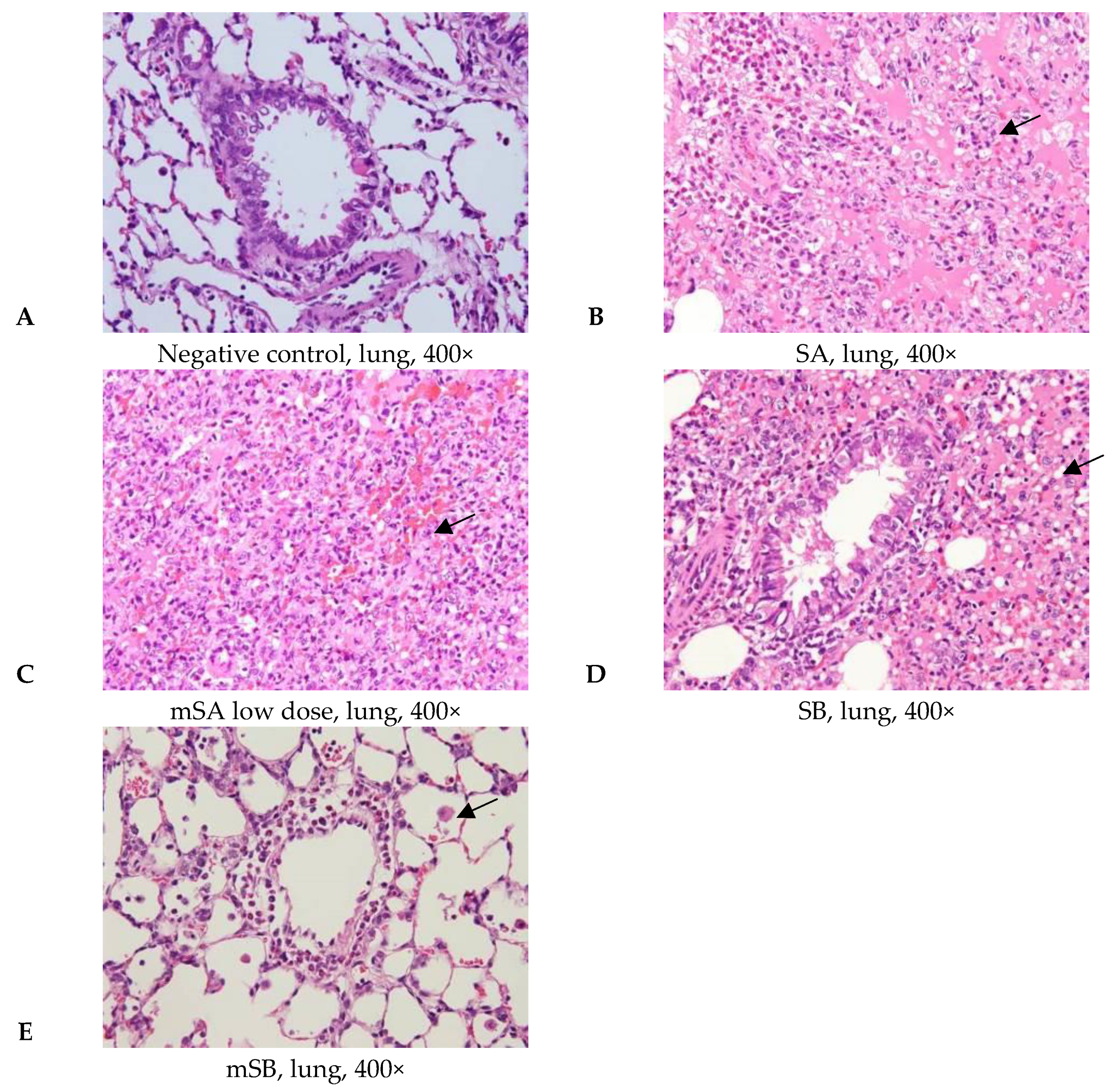
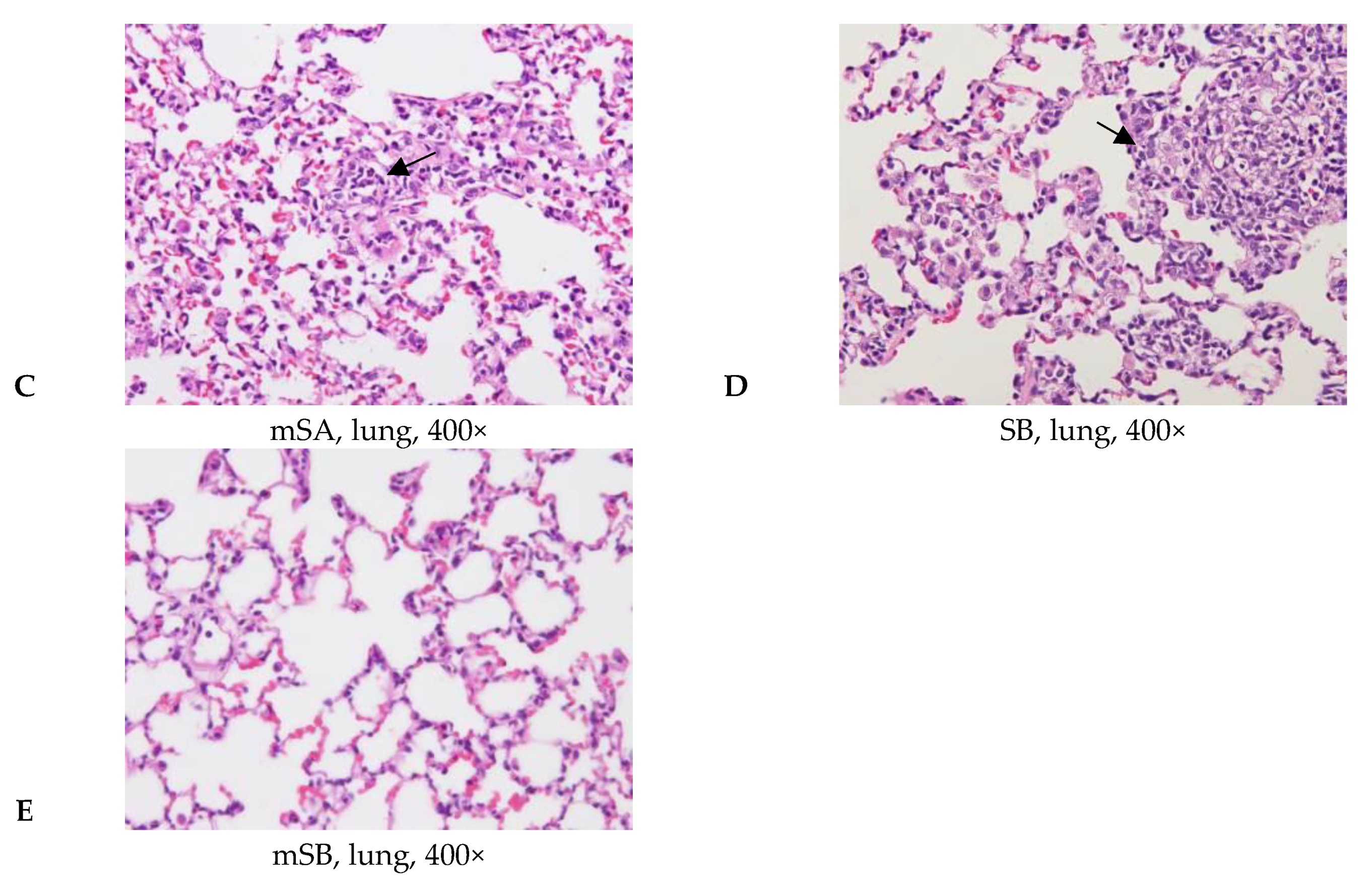
3.5. Acute Pulmonary Toxicity Test (14 D)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeevanandam, J.; Barhoum, A.; Chan, Y.S.; Dufresne, A.; Danquah, M. Review on nanoparticles and nanostructured materials: History, sources, toxicity and regulations. Beilstein. J. Nanotechnol. 2018, 9, 1050–1074. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Huang, Y.W.; Zhou, X.D.; Ma, Y. In vitro toxicity of silica nanoparticles in human lung cancer cells. Toxicol. Appl. Pharmacol. 2006, 217, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Yildirimer, L.; Thanh, N.T.K.; Loizidou, M.; Seifalian, A.M. Toxicology and clinical potential of nanoparticles. Nano Today 2011, 6, 585–607. [Google Scholar] [CrossRef] [Green Version]
- Warheit, D.B.; Brown, S.C. What is the impact of surface modifications and particle size on commercial titanium dioxide particle samples?—A review of in vivo pulmonary and oral toxicity studies-Revised 11-6-2018. Toxicol. Lett. 2019, 302, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhu, T.; Chen, C.; Liu, Y. Right or left: The role of nanoparticles in pulmonary disease. Int. J. Mol. Sci. 2014, 15, 17577–17600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberdorster, G. Lung particle overload: Implications for occupational exposures to particles. Regul. Toxicol. Pharmacol. 1995, 21, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Luanpitpong, S.; Castranova, V.; Tse, W.; Lu, Y.; Pongrakhananon, V.; Rojanasakul, Y. Carbon nanotubes induce malignant transformation and tumorigenesis of human lung epithelial cells. Nano. Lett. 2011, 11, 2796–2803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Downs, T.R.; Meredith, M.E.; Hu, T.; Kumar, S.; Sullivan, A.; Sarlo, K.; Reeder, B.; Lynch, M.; Wagner, M.; Mills, T.; et al. Silica nanoparticles administered at the maximum tolerated dose induce genotoxic effects through an inflammatory reaction while gold nanoparticles do not. Mutat. Res. 2012, 745, 38–50. [Google Scholar] [CrossRef]
- Yoshiura, Y.; Izumi, H.; Oyabu, T.; Hashiba, M.; Kambara, T.; Mizuguchi, Y.; Lee, B.W.; Okada, T.; Tomonaga, T.; Myojo, T. Pulmonary toxicity of well-dispersed titanium dioxide nanoparticles following intratracheal instillation. J. Nanopart. Res. 2015, 17, 241. [Google Scholar] [CrossRef] [Green Version]
- Maser, E.; Schulz, M.; Sauer, U.G.; Wiemann, M.; Ma-Hock, L.; Wohlleben, W.; Hartwig, A.; Landsiedel, R. In vitro and in vivo genotoxicity investigations of differently sized amorphous SiO2 nanomaterials. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2015, 794, 57–74. [Google Scholar] [CrossRef]
- Manke, A.; Wang, L.; Rojanasakul, Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. BioMed Res. Int. 2013, 2013, 942916. [Google Scholar] [CrossRef] [Green Version]
- Fu, P.P.; Xia, Q.; Hwang, H.M.; Rat, P.C.; Yu, H. Mechanisms of nanotoxicity: Generation of reactive oxygen species. J. Food Drug Anal. 2014, 22, 64–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox. Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domej, W.; Oettl, K.; Renner, W. Oxidative stress and free radicals in COPD-implications and relevance for treatment. Int. J. Chronic Obstr. Pulm. Dis. 2014, 9, 1207–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, L.; Lukamowicz-Rajska, M.; Thomassen, L.C.; Kirschhock, C.E.; Leyns, L.; Lison, D.; Martens, J.A.; Elhajouji, A.; Kirsch-Volders, M. Co-assessment of cell cycle and micronucleus frequencies demonstrates the influence of serum on the in vitro genotoxic response to amorphous monodisperse silica nanoparticles of varying sizes. Nanotoxicology 2014, 8, 876–884. [Google Scholar] [CrossRef]
- Decan, N.; Wu, D.; Williams, A.; Bernatchez, S.; Johnston, M.; Hill, M.; Halappanavar, S. Characterization of in vitro genotoxic, cytotoxic and transcriptomic responses following exposures to amorphous silica of different sizes. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2016, 796, 8–22. [Google Scholar] [CrossRef]
- Guichard, Y.; Fontana, C.; Chavinier, E.; Terzetti, F.; Gaté, L.; Binet, S.; Darne, C. Cytotoxic and genotoxic evaluation of different synthetic amorphous silica nanomaterials in the V79 cell line. Toxicol. Ind. Health 2016, 32, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.; Dommershausen, N.; Schulz, M.; Landsiedel, R.; Reichardt, P.; Krause, B.-C.; Tentschert, J.; Luch, A. Genotoxicity testing of different surface-functionalized SiO2, ZrO2 and silver nanomaterials in 3D human bronchial models. Arch. Toxicol. 2017, 91, 3991–4007. [Google Scholar] [CrossRef]
- Park, M.V.; Verharen, H.W.; Zwart, E.; Hernandez, L.G.; van Benthem, J.; Elsaesser, A.; Barnes, C.; McKerr, G.; Howard, C.V.; Salvati, A.; et al. Genotoxicity evaluation of amorphous silica nanoparticles of different sizes using the micronucleus and the plasmid lacZ gene mutation assay. Nanotoxicology. 2011, 5, 168–181. [Google Scholar] [CrossRef]
- Pfuhler, S.; Downs, T.R.; Allemang, A.J.; Shan, Y.; Crosby, M.E. Weak silica nanomaterial-induced genotoxicity can be explained by indirect DNA damage as shown by the OGG1-modified comet assay and genomic analysis. Mutagenesis 2017, 32, 5–12. [Google Scholar] [CrossRef]
- Gong, C.; Tao, G.; Yang, L.; Liu, J.; He, H.; Zhuang, Z. The role of reactive oxygen species in silicon dioxide nanoparticle-induced cytotoxicity and DNA damage in HaCaT cells. Mol. Biol. Rep. 2012, 39, 4915–4925. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, H.; Iijima, M. Surface modification and characterization for dispersion stability of inorganic nanometer-scaled particles in liquid media. Sci. Technol. Adv. Mater. 2010, 11, 044304. [Google Scholar] [CrossRef]
- Morris, A.S.; Adamcakova-Dodd, A.; Lehman, S.E.; Wongrakpanich, A.; Thorne, P.S.; Larsen, S.C.; Salem, A.K. Amine modification of nonporous silica nanoparticles reduces inflammatory response following intratracheal instillation in murine lungs. Toxicol. Lett. 2016, 241, 207–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ames, B.N.; McCann, J.; Yamasaki, E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat. Res. 1975, 31, 347–364. [Google Scholar] [CrossRef]
- Organization for Economic Co-operation and Development (OECD). In vitro mammalian chromosome aberration test. In OECD Guidelines for the Testing of Chemicals; Test Guideline No: 473; OECD: Paris, France, 1997; pp. 1–10. [Google Scholar]
- Shackelford, C.; Long, G.; Wolf, J.; Okerberg, C.; Herbert, R. Qualitative and quantitative analysis of nonneoplastic lesions in toxicology studies. Toxicol. Pathol. 2002, 30, 93–96. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, K.; Borm, P.J. The quartz hazard: A variable entity. Ann. Occup. Hyg. 1998, 42, 287–294. [Google Scholar] [CrossRef]
- Gibbs, A.R.; Attanoos, R.L. Environmental and Toxin Induced Lung Diseases. In Pulmonary Pathology; Zander, D.S., Farver, C.F., Eds.; Churchill Livingstone: London, UK, 2008; pp. 374–396. [Google Scholar]
- Rimal, B.; Greenberg, A.K.; Rom, W.N. Basic pathogenetic mechanisms in silicosis: Current understanding. Curr. Opin. Pulm. Med. 2005, 11, 169–173. [Google Scholar] [CrossRef]
- Ghiazza, M.; Polimeni, M.; Fenoglio, I.; Gazzano, E.; Ghigo, D.; Fubini, B. Does vitreous silica contradict the toxicity of the crystalline silica paradigm? Chem. Res. Toxicol. 2010, 23, 620–629. [Google Scholar] [CrossRef]
- Mortelmans, K.; Zeiger, E. The Ames Salmonella/microsome mutagenicity assay. Mutat. Res. 2000, 455, 29–60. [Google Scholar] [CrossRef]
- European Centre for Ecotoxicology and Toxicology of Chemicals (ECETOC). Synthetic Amorphous Silica; (CAS No. 7631-86-9); ECETOC: Brussels, Belgium, 2006; Volume 51. [Google Scholar]
- US Environmental Protection Agency. Screening-Level Hazard Characterization of Silane, Dichlorodimethyl-, Reaction Product with Silica; (CASRN 68611-44-9); US Environmental Protection Agency: Washington, DC, USA, 2011.
- Organization for Economic Cooperation and Development. SIDS Dossier on Synthetic Amorphous Silica and Silicates; Organization for Economic Cooperation and Development: Berlin, Germany, 2004. [Google Scholar]
- Li, P.R.; Wei, J.C.; Chiu, Y.F.; Su, H.L.; Peng, F.C.; Lin, J.J. Evaluation on cytotoxicity and genotoxicity of the exfoliated silicate nanoclay. ACS Appl. Mater. Interfaces 2010, 2, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Mateuca, R.; Lombaert, N.; Aka, P.V.; Decordier, I.; Kirsch-Volders, M. Chromosomal changes: Induction, detection methods and applicability in human biomonitoring. Biochimie 2006, 88, 1515–1531. [Google Scholar] [CrossRef]
- Albertini, R.J.; Anderson, D.; Douglas, G.R.; Hagmar, L.; Hemminki, K.; Merlo, F.; Natarajan, A.T.; Norppa, H.; Shuker, D.E.; Tice, R. IPCS guidelines for the monitoring of genotoxic effects of carcinogens in humans. Mutat. Res. Rev. Mutat. Res. 2000, 463, 111–172. [Google Scholar] [CrossRef]
- Hagmar, L.; Stromberg, U.; Bonassi, S.; Hansteen, I.L.; Knudsen, L.E.; Lindholm, C.; Norppa, H. Impact of types of lymphocyte chromosomal aberrations on human cancer risk: Results from Nordic and Italian cohorts. Cancer. Res. 2004, 64, 2258–2263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, J.Y.; Kim, H.L.; Lee, J.Y.; Ju, Y.H.; Kim, J.S.; Kang, S.H.; Kim, Y.R.; Lee, J.K.; Jeong, J.; Kim, M.K. Undetectable levels of genotoxicity of SiO2 nanoparticles in in vitro and in vivo tests. Int. J. Nanomedicine. 2014, 9 (Suppl. S2), 173–181. [Google Scholar] [PubMed] [Green Version]
- Krishna, G.; Hayashi, M. In vivo rodent micronucleus assay: Protocol, conduct and data interpretation. Mutat. Res. 2000, 455, 155–166. [Google Scholar] [CrossRef]
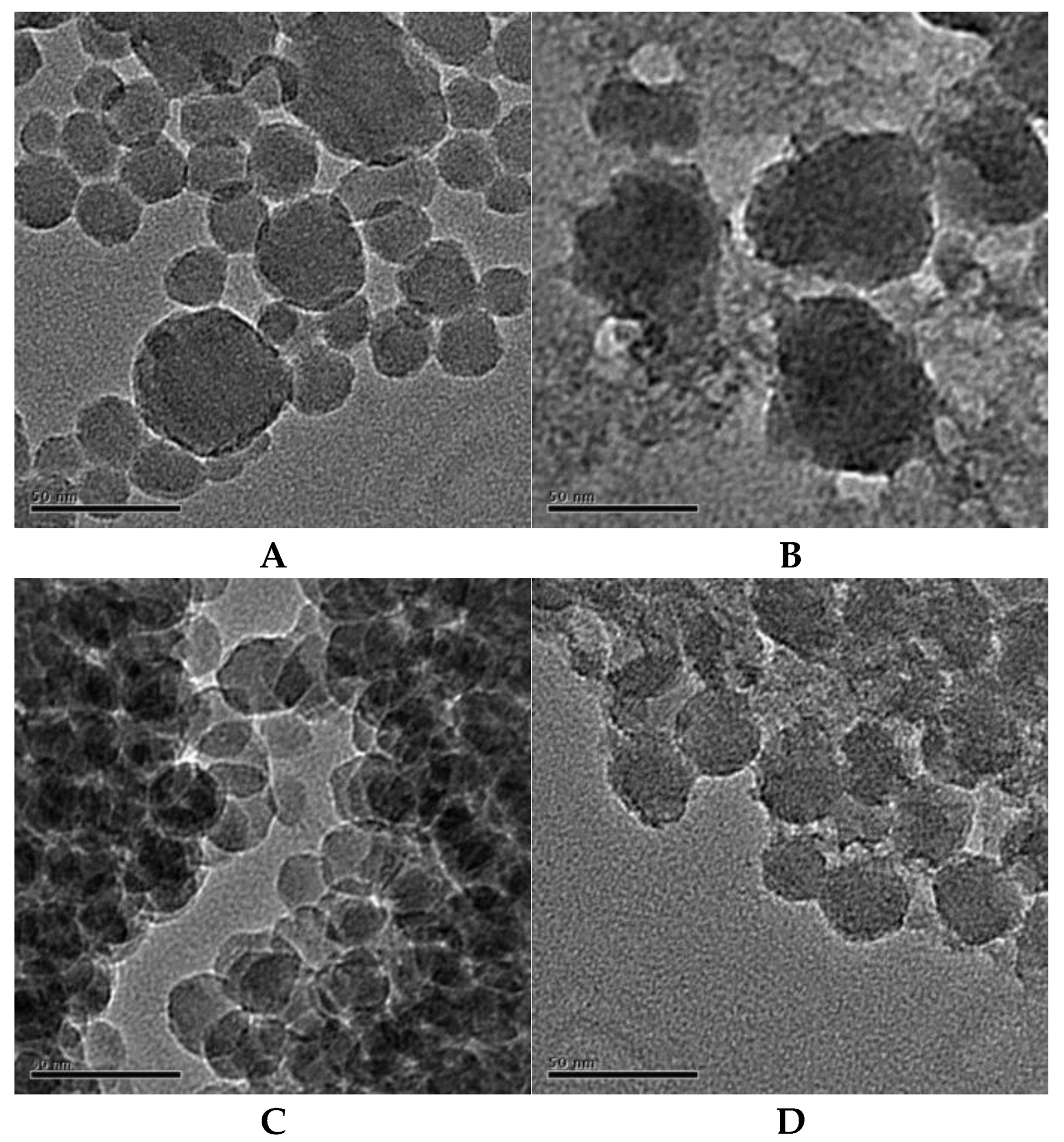


| Sample | Principal Component | Solid Content (%) | Diameter (nm) | pH |
|---|---|---|---|---|
| Sample A | Colloidal SiO2 | 41.13 | 55.16 | 7~9 |
| Modified sample A | Sample A TEOS a H3FSi | 3.77 | 82.29 | 2~4 |
| Sample B | Colloidal SiO2 | 35.8 | 22 | 7~9 |
| Modified sample B | Sample B TEOS a H3FSi | 3.77 | 86.64 | 2~4 |
| Group Code | Substance | Dose | Animal Number | |
|---|---|---|---|---|
| (mg/kg) | (mL/kg) | |||
| NC 1 | R.D.W. | - | 0.5, i.t. | 6 |
| SA | 80× diluted SA | 2.58 | 0.5, i.t. | 6 |
| mSAH | 80× diluted mSA | 0.24 | 0.5, i.t. | 5 |
| mSAL | 320× diluted mSA | 0.06 | 0.5, i.t. | 3 |
| SB | 80× diluted SB | 2.24 | 0.5, i.t. | 6 |
| mSB | 80× diluted mSB | 0.24 | 0.5, i.t. | 6 |
| Group Code | Substance | Dose | Animal Number | |
|---|---|---|---|---|
| (mg/kg) | (mL/kg) | |||
| NC 1 | R.D.W. | - | 0.5, i.t. | 5 |
| PC | Cyclophosphamide | 20 | 10, i.p. | 4 |
| SA | 320× diluted SA | 0.645 | 0.5, i.t. | 5 |
| mSA | 320× diluted mSA | 0.06 | 0.5, i.t. | 5 |
| SB | 80× diluted SB | 2.24 | 0.5, i.t. | 5 |
| mSB | 80× diluted mSB | 0.24 | 0.5, i.t. | 5 |
| Sample | SA | mSA | SB | mSB | |
| Number of Revertants (Colony, CFU/Plate) | |||||
| NC 1 | 22.7 ± 2.1 2 | 28.0 ± 5.1 | 25.0 ± 3.7 | 25.0 ± 3.7 | |
| 4NQO (positive control) | 213.7 ± 5.7 * | 586.3 ± 75.9 * | 468.3 ± 7.0 * | 468.3 ± 7.0 * | |
| TA98/without S9 activation (mg/plate) | 0.0295 | - | 31.3 ± 0.9 | - | 20.7 ± 1.2 |
| 0.0589 | 22.0 ± 1.4 | 26.3 ± 5.4 | 25.0 ± 6.4 | 22.7 ± 3.1 | |
| 0.1178 | 20.3 ± 0.5 | 23.7 ± 1.7 | 23.0 ± 2.9 | 21.7 ± 2.5 | |
| 0.2356 | 23.0 ± 1.4 | 27.3 ± 1.2 | 21.0 ± 0.8 | 21.0 ± 2.2 | |
| 0.4713 | 23.7 ± 2.6 | 37.7 ± 3.7 | 25.3 ± 0.9 | 22.7 ± 1.2 | |
| 0.9425 | 22.7 ± 1.9 | - | 20.3 ± 0.5 | - | |
| Sample | SA | mSA | SB | mSB | |
| Number of Revertants (Colony, CFU/Plate) | |||||
| NC | 27.3 ± 2.1 | 27.3 ± 2.1 | 34.0 ± 3.7 | 34.0 ± 3.7 | |
| 2-AA (positive control) | 2590.7 ± 138.9 * | 2590.7 ± 138.9 * | 1829.3 ± 236.1 * | 1829.3 ± 236.1 * | |
| TA98/with S9 activation (mg/plate) | 0.0295 | - | 26.7 ± 2.5 | - | 34.7 ± 4.6 |
| 0.0589 | 28.7 ± 1.7 | 26.3 ± 3.1 | 33.3 ± 2.6 | 35.7 ± 0.9 | |
| 0.1178 | 31.3 ± 2.5 | 26.7 ± 3.4 | 34.0 ± 3.7 | 29.3 ± 1.2 | |
| 0.2356 | 27.7 ± 2.9 | 28.7 ± 3.4 | 35.0 ± 2.2 | 32.0 ± 2.2 | |
| 0.4713 | 26.0 ± 0.8 | 26.0 ± 3.7 | 31.7 ± 2.6 | 31.3 ± 4.2 | |
| 0.9425 | 27.7 ± 2.1 | - | 29.3 ± 2.6 | - | |
| Sample | SA | mSA | SB | mSB | |
| Number of Revertants (Colony, CFU/Plate) | |||||
| NC 1 | 194.7 ± 2.1 2 | 194.7 ± 2.1 | 143.0 ± 6.7 | 187.7 ± 7.8 | |
| Sodium azide (positive control) | 1090.3 ± 52.1 * | 1090.3 ± 52.1 * | 1819.0 ± 196.5 * | 1665.3 ± 32.3 * | |
| TA100/without S9 activation (mg/plate) | 0.0295 | 195.0 ± 10.7 | 188.3 ± 10.7 | - | - |
| 0.0589 | 188.0 ± 11.6 | 201.7 ± 15.5 | - | 182.3 ± 6.2 | |
| 0.1178 | 208.0 ± 11.4 | 195.7 ± 4.7 | 153.0 ± 7.8 | 192.3 ± 9.2 | |
| 0.2356 | 199.3 ± 7.0 | 184.7 ± 6.2 | 162.7 ± 10.6 | 189.0 ± 4.5 | |
| 0.4713 | 214.0 ± 13.4 | 204.7 ± 3.7 * | 225.0 ± 3.6 * | 172.7 ± 8.3 | |
| 0.9425 | - | - | 251.3 ± 13.5 * | 172.3 ± 2.1 | |
| 1.885 | - | - | 354.0 ± 5.9 * | - | |
| Sample | SA | mSA | SB | mSB | |
| Number of Revertants (Colony, CFU/Plate) | |||||
| NC | 145.0 ± 10.6 | 145.0 ± 10.6 | 185.0 ± 8.3 | 185.0 ± 8.3 | |
| 2-AA (positive control) | 2449.3 ± 216.6 * | 2449.3 ± 216.6 * | 3288.3 ± 181.5 * | 3288.3 ± 181.5 * | |
| TA100/with S9 activation (mg/plate) | 0.0295 | 135.3 ± 9.4 | 128.3 ± 6.9 | - | - |
| 0.0589 | 134.7 ± 7.7 | 130.0 ± 6.4 | - | 192.7 ± 9.3 | |
| 0.1178 | 128.7 ± 2.1 | 137.7 ± 10.3 | 168.7 ± 9.0 | 175.0 ± 5.7 | |
| 0.2356 | 136.7 ± 8.7 | 171.7 ± 9.5 | 192.3 ± 9.0 | 178.0 ± 7.5 | |
| 0.4713 | 129.0 ± 5.1 | 150.3 ± 8.7 | 182.3 ± 6.9 | 177.7 ± 5.2 | |
| 0.9425 | - | - | 193.0 ± 4.2 | 180.7 ± 7.1 | |
| 1.885 | - | - | 205.3 ± 9.0 | - | |
| Group | Dosage (mg/mL) | Frequency of Aberration (%) | Chromatic Type | Chromosome Type | |||
|---|---|---|---|---|---|---|---|
| Deletion | Intrachange | Interchange | Ring | Gap | |||
| NC 1 | 0 | 2.7 ± 1.5 | 2.7 ± 1.5 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Mitomycin C | 2.5 | 18.0 ± 2.6 * | 13.3 ± 6.1 * | 0.0 ± 0.0 | 3.3 ± 0.6 | 0.7 ± 0.6 | 5.0 ± 3.5 * |
| SA | 1.387 | 2.3 ± 1.5 | 1.7 ± 0.6 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.7 ± 1.2 |
| mSA | 0.778 | 3.0 ± 2.0 | 2.7 ± 2.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.6 |
| SB | 0.353 | 9.7 ± 1.5 *,a | 6.0 ± 2.0 a | 0.0 ± 0.0 | 0.7 ± 1.2 | 0.0 ± 0.0 | 4.0 ± 1.0 *,a |
| mSB | 1.569 | 2.7 ± 1.5 a | 2.0 ± 1.0 a | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.7 ± 1.2 a |
| Group | Dosage (mg/mL) | Frequency of Aberration (%) | Chromatic Type | Chromosome Type | |||
|---|---|---|---|---|---|---|---|
| Deletion | Intrachange | Interchange | Ring | Gap | |||
| NC 1 | 0 | 2.7 ± 2.1 | 1.3 ± 1.2 | 0.0 ± 0.0 | 0.3 ± 0.6 | 0.0 ± 0.0 | 1.0 ± 1.0 |
| Cyclophophamide | 25 | 12.7 ± 3.1 * | 13.7 ± 4.7 * | 0.0 ± 0.0 | 0.3 ± 0.6 | 0.0 ± 0.0 | 1.7 ± 0.6 |
| SA | 1.387 | 3.3 ± 0.6 | 2.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.6 | 0.3 ± 0.6 | 0.7 ± 0.6 |
| mSA | 0.778 | 4.7 ± 2.1 | 2.3 ± 2.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.7 ± 0.6 | 2.0 ± 1.7 |
| SB | 0.353 | 2.3 ± 0.6 | 2.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| mSB | 1.569 | 3.3 ± 1.5 | 3.0 ± 2.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.6 |
| Group/ Intervals | RETs/NCEs 1 (‰) | Mn-RETs/RETs (‰) |
|---|---|---|
| 48 h | ||
| NC 2 | 37.2 ± 7.4 3 | 8.1 ± 2.8 |
| PC | 6.2 ± 1.9 * | 62.1 ± 36.5 * |
| SA | 38.9 ± 7.0 | 7.5 ± 4.4 |
| mSA | 35.7 ± 13.2 | 6.9 ± 4.5 |
| SB | 25.7 ± 3.0 * | 12.4 ± 10.9 |
| mSB | 28.1 ± 5.0 | 7.1 ± 4.6 |
| 72 h | ||
| NC | 46.3 ± 8.2 | 4.6 ± 1.5 |
| PC | 19.2 ± 4.9 * | 8.0 ± 2.7 * |
| SA | 44.9 ± 6.6 | 4.2 ± 1.4 |
| mSA | 38.4 ± 12.1 | 8.0 ± 4.1 |
| SB | 34.9 ± 3.1 * | 8.8 ± 3.5 *,a |
| mSB | 33.6 ± 4.6 * | 4.7 ± 1.3 a |
| Group | Animal No. | Clinical Sign | Hours After Treatment | Mortality (%) 3 | ||
|---|---|---|---|---|---|---|
| 1 | 12 | 24 | ||||
| NC 1 | 6 | Normal | 0 2 | 0 | 0 | 0 |
| SA | 6 | Normal | 0 | 0 | 0 | 0 |
| mSAL | 5 | Normal | 0 | 0 | 0 | 0 |
| SB | 6 | Normal | 0 | 0 | 0 | 0 |
| mSB | 6 | Normal | 0 | 0 | 0 | 0 |
| mSAH | 3 | Dyspnea | 0 | 1 | 1 | 66.6 |
| Pathological Incidence | ||||||
| Organ | Lesions | Group | ||||
| NC 1 | SA | mSAL | SB | mSB | ||
| Lung | ||||||
| Edema, extensive | - | 4/6 | 2/5 | 3/6 | - | |
| Hemorrhage, focal | - | - | 3/5 | 3/6 | 1/6 | |
| Inflammation, alveolar, focal | - | 6/6 | 4/5 | 6/6 | 3/6 | |
| Necrosis, alveolar wall, focal | - | 5/6 | 2/5 | 4/6 | - | |
| Pathological Scores | ||||||
| Organ | Lesions | Group | ||||
| NC | SA | mSAL | SB | mSB | ||
| Lung | ||||||
| Edema, extensive | 0.0 ± 0.0 2,3 | 1.8 ± 1.5 * | 1.0 ± 1.4 | 2.0 ± 2.2 *,b | 0.0 ± 0.0 b | |
| Hemorrhage, focal | 0.0 ± 0.0 | 0.0 ± 0.0 a | 1.2 ± 1.1*,a | 1.0 ± 1.1 * | 0.2 ± 0.4 | |
| Inflammation, alveolar, focal | 0.0 ± 0.0 | 2.7 ± 0.8 * | 2.0 ± 1.6 * | 3.5 ± 1.2 *,b | 0.7 ± 0.8 b | |
| Necrosis, alveolar wall, focal | 0.0 ± 0.0 | 2.2 ± 1.2 * | 1.0 ± 1.4 | 2.0 ± 1.7 *,b | 0.0 ± 0.0 b | |
| Total score 4 | 0.0 ± 0.0 | 6.7 ± 3.2 * | 5.2 ± 5.2 * | 8.5 ± 5.5 *,b | 0.8 ± 1.2 b | |
| Group | WBC 2 (105) | E (105) | N (105) | L (105) | M (105) |
|---|---|---|---|---|---|
| NC 1 | 6.1 ± 2.9 3 | 0.0 ± 0.0 | 0.1 ± 0.1 | 0.3 ± 0.2 | 5.7 ± 2.7 |
| SA | 23.7 ± 12.2 *,a | 0.4 ± 1.0 | 10.1 ± 8.1 * | 1.1 ± 0.7 * | 12.2 ± 3.9 * |
| mSAL | 39.0 ± 10.8 *,a | 0.6 ± 0.7 | 20.4 ± 7.9 * | 2.0 ± 1.3 * | 16.1 ± 10.0 * |
| SB | 33.5 ± 19.2 *,b | 0.4 ± 0.4 * | 15.6 ± 14.0 *,b | 2.2 ± 1.4 *,b | 15.3 ± 6.5 * |
| mSB | 10.7 ± 10.7 b | 0.1 ± 0.2 | 0.8 ± 1.5 b | 0.5 ± 0.5 b | 9.4 ± 8.7 |
| Pathological Incidence | ||||||
| Organ | Lesions | Group | ||||
| NC 1 | SA | mSA | SB | mSB | ||
| Lung | ||||||
| Aggregate of macrophage, alveolar multifocal | - | 4/5 2 | 5/5 | 5/5 | 0/5 | |
| Granulomatous inflammation, alveolar, focal | - | - | - | 5/5 | 0/5 | |
| Pathological Scores | ||||||
| Organ | Lesions | Group | ||||
| NC | SA | mSA | SB | mSB | ||
| Lung, left lobe | ||||||
| Aggregate of macrophage, alveolar multifocal | 0.0 ± 0.0 3,4 | 2.2 ± 1.5 * | 1.6 ± 0.5 * | 2.2 ± 0.4 *,a | 0.0 ± 0.0 a | |
| Granulomatous inflammation, alveolar, focal | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.6 ± 0.5 *,a | 0.0 ± 0.0 a | |
| Total score 5 | 0.0 ± 0.0 | 2.2 ± 1.5 * | 1.6 ± 0.5 * | 3.8 ± 0.8 *,a | 0.0 ± 0.0 a | |
| Group | M (105) | N (105) | L (105) | E (105) |
|---|---|---|---|---|
| NC 1 | 3.41 ± 0.88 2 | 0.03 ± 0.01 | 0.07 ± 0.05 | 0.00 ± 0.00 |
| SA | 3.94 ± 0.49 | 0.18 ± 0.16 | 0.16 ± 0.08 | 0.01 ± 0.03 |
| mSA | 7.28 ± 4.11 | 0.03 ± 0.03 | 0.19 ± 0.13 | 0.00 ± 0.00 |
| SB | 5.09 ± 2.97 | 0.29 ± 0.24 * | 0.39 ± 0.41 | 0.00 ± 0.00 |
| mSB | 3.97 ± 2.31 | 0.23 ± 0.34 | 0.16 ± 0.07 | 0.00 ± 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-T.; Lue, P.-Y.; Chen, P.-W.; Chueh, P.-J.; Tsai, F.-J.; Liao, J.-W. Comparison of Genotoxicity and Pulmonary Toxicity Study of Modified SiO2 Nanomaterials. Appl. Sci. 2021, 11, 11990. https://doi.org/10.3390/app112411990
Chen Y-T, Lue P-Y, Chen P-W, Chueh P-J, Tsai F-J, Liao J-W. Comparison of Genotoxicity and Pulmonary Toxicity Study of Modified SiO2 Nanomaterials. Applied Sciences. 2021; 11(24):11990. https://doi.org/10.3390/app112411990
Chicago/Turabian StyleChen, Yng-Tay, Po-Yi Lue, Po-Wei Chen, Pin-Ju Chueh, Fuu-Jen Tsai, and Jiunn-Wang Liao. 2021. "Comparison of Genotoxicity and Pulmonary Toxicity Study of Modified SiO2 Nanomaterials" Applied Sciences 11, no. 24: 11990. https://doi.org/10.3390/app112411990
APA StyleChen, Y.-T., Lue, P.-Y., Chen, P.-W., Chueh, P.-J., Tsai, F.-J., & Liao, J.-W. (2021). Comparison of Genotoxicity and Pulmonary Toxicity Study of Modified SiO2 Nanomaterials. Applied Sciences, 11(24), 11990. https://doi.org/10.3390/app112411990









