Docosahexaenoic Acid and Eicosapentaenoic Acid Intakes Modulate the Association of FADS2 Gene Polymorphism rs526126 with Plasma Free Docosahexaenoic Acid Levels in Overweight Children
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Anthropometric Measurement
2.3. Food Intake
2.4. Hematological and Biochemical Tests
2.5. Analysis of Genetic Variants
2.6. Fatty Acids Quantification
2.7. Statistical Analysis
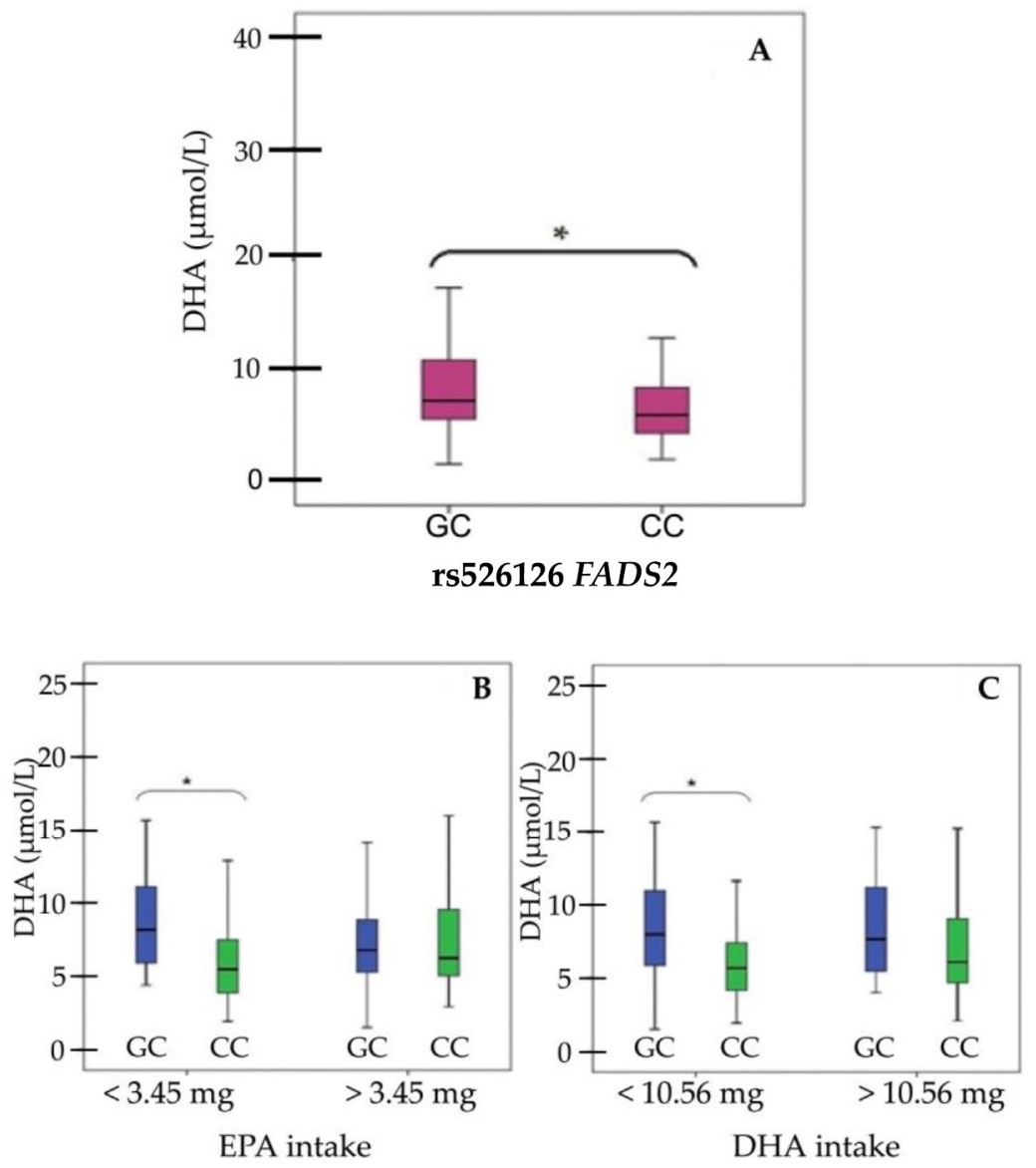
3. Results
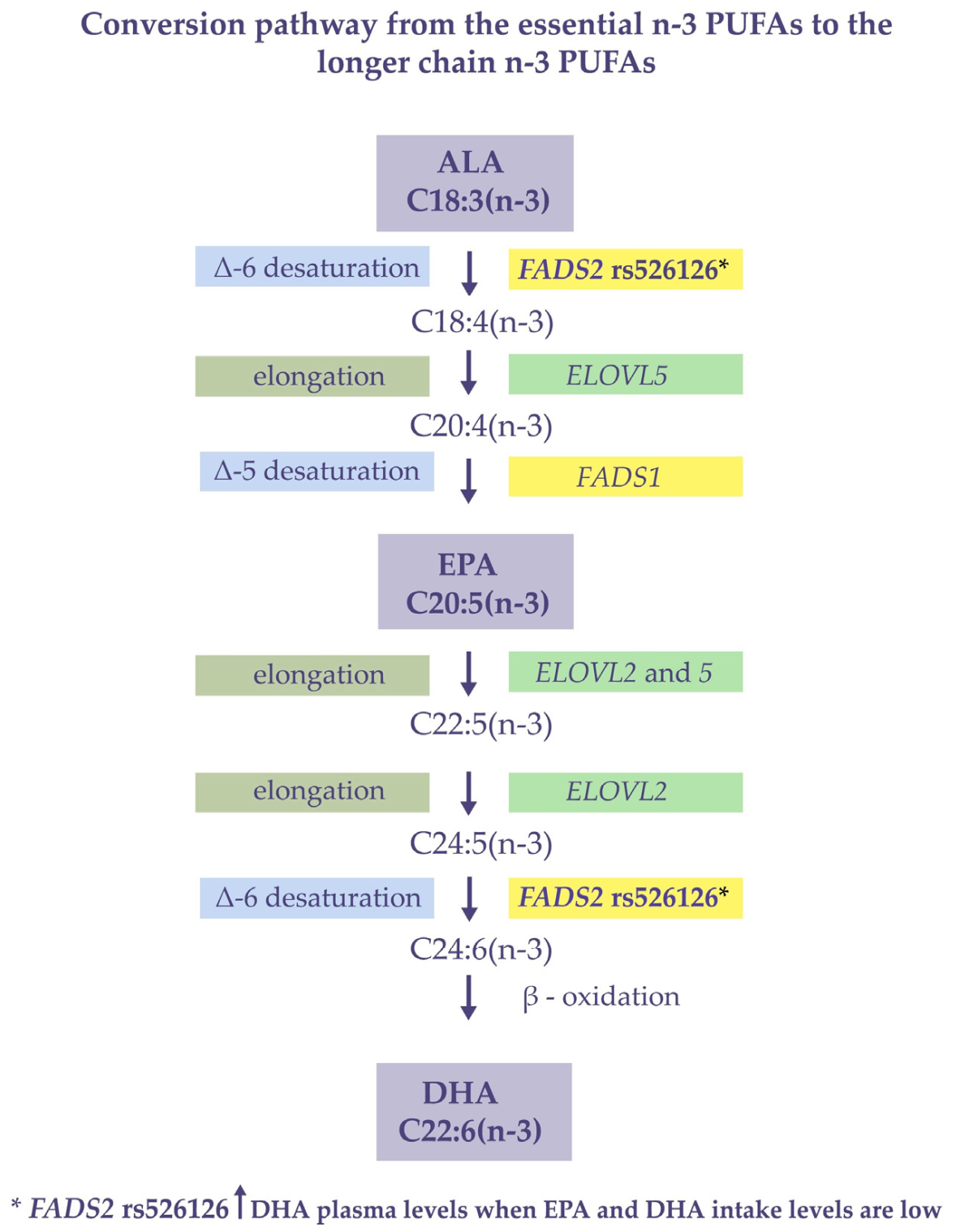
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Gene | Rs Chr Position GRCh37 | AA/AB/BB | AA% | AB% | BB% |
|---|---|---|---|---|---|
| FADS2 | rs2526678chr1161623793 | GG/GA/AA | 82.1 | 17.9 | 0.0 |
| rs526126chr1161624885 | GG/GC/CC | 3.1 | 26.3 | 70.6 |
References
- Chilton, F.H.; Murphy, R.C.; Wilson, B.A.; Sergeant, S.; Ainsworth, H.; Seeds, M.C.; Mathias, R.A. Diet-Gene Interactions and PUFA Metabolism: A Potential Contributor to Health Disparities and Human Diseases. Nutrients 2014, 6, 1993–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calcaterra, V.; Regalbuto, C.; Porri, D.; Pelizzo, G.; Mazzon, E.; Vinci, F.; Zuccotti, G.; Fabiano, V.; Cena, H. Inflammation in Obesity-Related Complications in Children: The Protective Effect of Diet and Its Potential Role as a Therapeutic Agent. Biomolecules 2020, 10, 1324. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chan, J.S.Y.; Ren, L.; Yan, J.H. Obesity Reduces Cognitive and Motor Functions across the Lifespan. Neural Plast. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Versini, M.; Jeandel, P.Y.; Rosenthal, E.; Shoenfeld, Y. Obesity in autoimmune diseases: Not a passive bystander. Autoimmun Rev. 2014, 13, 981–1000. [Google Scholar] [CrossRef]
- Kyrou, I.; Randeva, H.S.; Tsigos, C.; Kaltsas, G.; Weickert, M.O. Clinical Problems Caused by Obesity. In Endotext; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. Available online: https://www.ncbi.nlm.nih.gov/books/NBK278973 (accessed on 10 August 2021).
- Baym, C.L.; Khan, N.A.; Monti, J.M.; Raine, L.B.; Drollette, E.S.; Moore, R.D.; Scudder, M.R.; Kramer, A.F.; Hillman, C.H.; Cohen, N.J. Dietary lipids are differentially associated with hippocampal-dependent relational memory in prepubescent children. Am. J. Clin. Nutr. 2014, 99, 1026–1033. [Google Scholar] [CrossRef]
- Koletzko, B.; Reischl, E.; Tanjung, C.; Gonzalez-Casanova, I.; Ramakrishnan, U.; Meldrum, S.; Simmer, K.; Heinrich, J.; Demmelmair, H. FADS1 and FADS2 Polymorphisms Modulate Fatty Acid Metabolism and Dietary Impact on Health. Ann. Rev. Nutr. 2019, 39, 21–44. [Google Scholar] [CrossRef]
- Figueiredo, P.S.; Inada, A.C.; Marcelino, G.; Lopes Cardozo, C.M.; de Cássia Freitas, K.; de Cássia Avellaneda Guimarães, R.; Pereira de Castro, A.; Aragão do Nascimento, V.; Aiko Hiane, P. Fatty Acids Consumption: The Role Metabolic Aspects Involved in Obesity and Its Associated Disorders. Nutrients 2017, 9, 1158. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, T.A.; Bremner, A.P.; Beilin, L.J.; Ambrosini, G.L.; Mori, T.A.; Huang, R.C.; Oddy, W.H. Polyunsaturated fatty acid intake and blood pressure in adolescents. J. Hum. Hypertens. 2012, 26, 178–187. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.M.; Ma, D.W. Are all n-3 polyunsaturated fatty acids created equal? Lipid Health Dis. 2009, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.; Grimble, R. Polyunsaturated fatty acids, inflammation and immunity. Eur. J. Clin. Nut. 2002, 56 (Suppl. 3), S14–S19. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A. An Increase in the Omega-6/Omega-3 Fatty Acid Ratio Increases the Risk for Obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef] [Green Version]
- Kawashima, H. Intake of arachidonic acid-containing lipids in adult humans: Dietary surveys and clinical trials. Lipids Health Dis. 2019, 18, 101. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.M.; Lee, H.; Kang, S.; Park, W.J. Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Bio-technological Advances. Nutrients 2016, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Mathias, R.A.; Pani, V.; Chilton, F.H. Genetic Variants in the FADS Gene: Implications for Dietary Recommendations for Fatty Acid Intake. Curr. Nutr. Rep. 2014, 3, 139–148. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Innis, S.M. Genetic Variants of the FADS1 FADS2 Gene Cluster Are Associated with Altered (n-6) and (n-3) Essential Fatty Acids in Plasma and Erythrocyte Phospholipids in Women during Pregnancy and in Breast Milk during Lactation. J. Nutr. 2008, 138, 2222–2228. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.C.; Chang, W.T.; Chang, H.Y.; Chung, H.F.; Chen, F.P.; Huang, Y.F.; Hsu, C.C.; Hwang, S.J. FADS Gene Polymorphisms, Fatty Acid Desaturase Activities, and HDL-C in Type 2 Diabetes. Int. J. Environ. Res. Public Health 2017, 14, 572. [Google Scholar] [CrossRef] [Green Version]
- Khodarahmi, M.; Nikniaz, L.; Abbasalizad Farhangi, M. The Interaction Between Fatty Acid Desaturase-2 (FADS2) rs174583 Genetic Variant and Dietary Quality Indices (DASH and MDS) Constructs Different Metabolic Phenotypes Among Obese Individuals. Front Nutr. 2021, 8, 669207. [Google Scholar] [CrossRef]
- Nakayama, K.; Bayasgalan, T.; Tazoe, F.; Yanagisawa, Y.; Gotoh, T.; Yamanaka, K.; Ogawa, A.; Munkhtulga, L.; Chimedregze, U.; Kagawa, Y.; et al. A single nucleotide polymorphism in the FADS1/FADS2 gene is associated with plasma lipid profiles in two genetically similar Asian ethnic groups with distinctive differences in lifestyle. Hum. Genet. 2010, 127, 685–690. [Google Scholar] [CrossRef]
- Khamlaoui, W.; Mehri, S.; Hammami, S.; Hammouda, S.; Chraeif, I.; Elosua, R.; Hammami, M. Association Between Genetic Variants in FADS1-FADS2 and ELOVL2 and Obesity, Lipid Traits, and Fatty Acids in Tunisian Population. Clin. Appl. Thromb. Hemost. 2020, 26. [Google Scholar] [CrossRef]
- Maguolo, A.; Zusi, C.; Giontella, A.; Miraglia Del Giudice, E.; Tagetti, A.; Fava, C.; Morandi, A.; Maffeis, C. Influence of genetic variants in FADS2 and ELOVL2 genes on BMI and PUFAs homeostasis in children and adolescents with obesity. Int. J. Obes. 2021, 45, 56–65. [Google Scholar] [CrossRef]
- Serafim, V.; Tiugan, D.A.; Andreescu, N.; Mihailescu, A.; Paul, C.; Velea, I.; Puiu, M.; Niculescu, M.D. Development and Validation of a LC–MS/MS-Based Assay for Quantification of Free and Total Omega 3 and 6 Fatty Acids from Human Plasma. Molecules 2019, 24, 360. [Google Scholar] [CrossRef] [Green Version]
- Bokor, S.; Dumont, J.; Spinneker, A.; Gonzalez-Gross, M.; Nova, E.; Widhalm, K.; Moschonis, G.; Stehle, P.; Amouyel, P.; De Henauw, S.; et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J. Lipid. Res. 2010, 51, 2325–2333. [Google Scholar] [CrossRef] [Green Version]
- Park, H.G.; Park, W.J.; Kothapalli, K.S.D.; Brenna, J.T. The fatty acid desaturase 2 (FADS2) gene product catalyzes Δ4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015, 29, 3911–3919. [Google Scholar] [CrossRef] [Green Version]
- Park, H.G.; Kothapalli, K.S.D.; Park, W.J.; DeAllie, C.; Liu, L.; Liang, A.; Lawrence, P.; Brenna, J.T. Palmitic acid (16:0) competes with omega-6 linoleic and omega-3 α-linolenic acids for FADS2 mediated Δ6-desaturation. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2016, 1861, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Roke, K.; Ralston, J.C.; Abdelmagid, S.; Nielsen, D.E.; Badawi, A.; El-Sohemy, A.; Ma, D.V.L.; Mutch, D.M. Variation in the FADS1/2 gene cluster alters plasma n-6 PUFA and is weakly associated with hsCRP levels in healthy young adults. Prostaglandins Leukot. Essent. Fat. Acids 2013, 89, 257–263. [Google Scholar] [CrossRef]
- Risé, P.; Eligini, S.; Ghezzi, S.; Colli, S.; Galli, C. Fatty acid composition of plasma, blood cells and whole blood: Relevance for the assessment of the fatty acid status in humans. Prostaglandins Leukot. Essent. Fat. Acids 2007, 76, 363–369. [Google Scholar] [CrossRef]
- Ouellet, M.; Emond, V.; Chen, C.T.; Julien, C.; Bourasset, F.; Oddo, S.; LaFerla, F.; Bazinet, R.P.; Calon, F. Diffusion of docosahexaenoic and eicosapentaenoic acids through the blood–brain barrier: An in situ cerebral perfusion study. Neurochem. Int. 2009, 55, 476–482. [Google Scholar] [CrossRef]
- Chen, C.T.; Kitson, A.P.; Hopperton, K.E.; Domenichiello, A.F.; Trépanier, M.O.; Lin, L.E.; Ermini, L.; Post, M.; Thies, F.; Bazinet, R.P. Plasma non-esterified docosahexaenoic acid is the major pool supplying the brain. Sci. Rep. 2015, 5, 15791. [Google Scholar] [CrossRef] [Green Version]
- Lassandro, C.; Banderali, G.; Radaelli, G.; Borghi, E.; Moretti, F.; Verduci, E. Docosahexaenoic Acid Levels in Blood and Metabolic Syndrome in Obese Children: Is There a Link? Int. J. Mol. Sci. 2015, 16, 19989–20000. [Google Scholar] [CrossRef] [Green Version]
- Duvall, M.G.; Levy, B.D. DHA- and EPA-derived resolvins, protectins, and maresins in airway inflammation. Eur. J. Pharmacol. 2016, 785, 144–155. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.D.; Bailes, J. Neuroprotection for the Warrior: Dietary Supplementation With Omega-3 Fatty Acids. Mil. Med. 2011, 176, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Damsgaard, C.T.; Eidner, M.B.; Stark, K.D.; Hjorth, M.F.; Sjödin, A.; Andersen, M.R.; Andersen, R.; Tetens, I.; Astrup, A.; Michaelsen, K.F.; et al. Eicosapentaenoic acid and docosahexaenoic acid in whole blood are differentially and sex-specifically associated with cardiometabolic risk markers in 8-11-year-old danish children. PLoS ONE. 2014, 9, e109368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serafim, V.; Chirita-Emandi, A.; Andreescu, N.; Tiugan, D.A.; Tutac, P.; Paul, C.; Velea, I.; Mihailescu, A.; Șerban, C.L.; Zimbru, C.G.; et al. Single nucleotide polymorphisms in PEMT and MTHFR genes are associated with omega 3 and 6 fatty acid levels in the red blood cells of children with obesity. Nutrients 2019, 11, 2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Variables | All n = 196 | Females n = 101 | Males n = 95 | p Value * (Males vs. Females) | |||
|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | Median | IQR | ||
| Anthropometric data | |||||||
| Age (years) | 12 | 5 | 12.00 | 5.0 | 12.00 | 4.75 | 0.787 |
| zBMI | 3.13 | 1.20 | 2.83 | 1.21 | 3.44 | 1.37 | 0.001 * |
| Biochemical analysis | |||||||
| TC (mg/dL) | 174.00 | 54.00 | 166.00 | 52.00 | 182.50 | 55.50 | 0.109 |
| TG (mg/dL) | 129.00 | 86.50 | 125.00 | 88.00 | 133.00 | 99.75 | 0.237 |
| HDL (mg/dL) | 46.00 | 18.00 | 43.00 | 19.00 | 48.00 | 6.75 | 0.085 |
| AST (U/L) | 29.00 | 14.00 | 29.00 | 14.00 | 29.50 | 15.50 | 0.127 |
| ALT (U/L) | 32.00 | 15.00 | 31.00 | 16.00 | 33.50 | 14.75 | 0.167 |
| CRP (mg/dL) | 4.70 | 7.90 | 4.80 | 8.70 | 4.75 | 7.72 | 0.315 |
| Homocysteine (µmoL/L) | 14.78 | 8.78 | 14.16 | 8.08 | 15.33 | 9.28 | 0.398 |
| Measurements of free PUFA in plasma | |||||||
| ALA (µmol/L) | 3.72 | 4.27 | 3.24 | 3.95 | 4.09 | 4.54 | 0.145 |
| EPA (µmol/L) | 0.23 | 0.14 | 0.2 | 0.13 | 0.26 | 0.14 | 0.008 * |
| DHA (µmol/L) | 6.26 | 4.82 | 5.88 | 4.01 | 6.76 | 6.09 | 0.217 |
| LA (µmol/L) | 137.2 | 122.12 | 126.39 | 105.35 | 159.08 | 124.62 | 0.215 |
| ARA (µmol/L) | 7.15 | 4.44 | 6.8 | 4.05 | 7.78 | 4.82 | 0.025 * |
| Selected daily nutrient intakes evaluated in 24 h dietary recalls | |||||||
| Kilocalories | 1201.59 | 494.72 | 1201.49 | 440.11 | 1205.77 | 580.32 | 0.889 |
| Protein (g) | 66.60 | 22.11 | 66.63 | 23.00 | 66.48 | 23.80 | 0.832 |
| Lipids (g) | 44.59 | 25.08 | 44.17 | 22.99 | 45.67 | 28.42 | 0.781 |
| Carbohydrates (g) | 135.93 | 58.15 | 138.52 | 51.27 | 129.93 | 61.29 | 0.266 |
| Water (g) | 21.96 | 829.49 | 2146.43 | 749.58 | 2280.80 | 912.78 | 0.207 |
| Cholesterol (mg) | 260.54 | 157.94 | 255.68 | 129.83 | 274.79 | 158.47 | 0.853 |
| ALA (mg) | 77.16 | 217.21 | 57.29 | 196.85 | 87 | 272.44 | 0.280 |
| DHA (mg) | 10.63 | 24 | 10.16 | 19.47 | 10.95 | 24.74 | 0.547 |
| EPA (mg) | 3.4 | 5.99 | 3.09 | 5.81 | 4.00 | 6.20 | 0.971 |
| LA (mg) | 943.08 | 2306.07 | 953.40 | 2072.57 | 943.08 | 2334.76 | 0.527 |
| EPA Plasma | DHA Plasma | ALA Intake | EPA Intake | DHA Intake | Age | ||
|---|---|---|---|---|---|---|---|
| ALA plasma | rho | 0.519 ** | 0.523 ** | 0.093 | 0.159 * | 0.069 | −0.237 ** |
| p-value | <0.001 | <0.001 | 0.227 | 0.037 | 0.37 | 0.001 | |
| EPA plasma | rho | 0.682 ** | 0.108 | 0.13 | 0.056 | −0.217 ** | |
| p-value | <0.001 | 0.158 | 0.089 | 0.469 | 0.002 | ||
| DHA plasma | rho | −0.003 | 0.182 * | 0.159 * | −0.195 ** | ||
| p-value | 0.973 | 0.017 | 0.038 | 0.007 | |||
| ALA intake | rho | 0.195 ** | 0.117 | −0.074 | |||
| p-value | 0.01 | 0.126 | 0.337 | ||||
| EPA intake | rho | 0.769 ** | −0.122 | ||||
| p-value | <0.001 | 0.109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mihailescu, A.; Serafim, V.; Paul, C.; Andreescu, N.; Tiugan, D.-A.; Tutac, P.; Velea, I.; Zimbru, C.G.; Serban, C.L.; Ion, A.I.; et al. Docosahexaenoic Acid and Eicosapentaenoic Acid Intakes Modulate the Association of FADS2 Gene Polymorphism rs526126 with Plasma Free Docosahexaenoic Acid Levels in Overweight Children. Appl. Sci. 2021, 11, 9845. https://doi.org/10.3390/app11219845
Mihailescu A, Serafim V, Paul C, Andreescu N, Tiugan D-A, Tutac P, Velea I, Zimbru CG, Serban CL, Ion AI, et al. Docosahexaenoic Acid and Eicosapentaenoic Acid Intakes Modulate the Association of FADS2 Gene Polymorphism rs526126 with Plasma Free Docosahexaenoic Acid Levels in Overweight Children. Applied Sciences. 2021; 11(21):9845. https://doi.org/10.3390/app11219845
Chicago/Turabian StyleMihailescu, Alexandra, Vlad Serafim, Corina Paul, Nicoleta Andreescu, Diana-Andreea Tiugan, Paul Tutac, Iulian Velea, Cristian G. Zimbru, Costela Lacrimioara Serban, Adina Iuliana Ion, and et al. 2021. "Docosahexaenoic Acid and Eicosapentaenoic Acid Intakes Modulate the Association of FADS2 Gene Polymorphism rs526126 with Plasma Free Docosahexaenoic Acid Levels in Overweight Children" Applied Sciences 11, no. 21: 9845. https://doi.org/10.3390/app11219845
APA StyleMihailescu, A., Serafim, V., Paul, C., Andreescu, N., Tiugan, D.-A., Tutac, P., Velea, I., Zimbru, C. G., Serban, C. L., Ion, A. I., David, V. L., Ionescu, A., Puiu, M., & Niculescu, M. D. (2021). Docosahexaenoic Acid and Eicosapentaenoic Acid Intakes Modulate the Association of FADS2 Gene Polymorphism rs526126 with Plasma Free Docosahexaenoic Acid Levels in Overweight Children. Applied Sciences, 11(21), 9845. https://doi.org/10.3390/app11219845







