Abstract
A great deal of information related to rice cultivation has been published on the web. Conventionally, this information is studied by end-users to identify pests, and to prevent production losses from rice diseases. Despite its benefits, such information has not yet been encoded in a machine-processable form. This research closes the gap by modeling the knowledge-bases using ontologies and semantic technologies. Our modeled ontologies are externalized from existing reliable sources only, and offer axioms that describe abnormal appearances in rice diseases (and insects) and the corresponding controls. In addition, we developed an expert system called RiceMan, based on our ontologies, to support technical and non-technical users for diagnosing problems from observed abnormalities. We also introduce a composition procedure that aggregates users’ observation data with others for realizing spreadable diseases. This procedure, together with ontology reasoning, lies at the heart of our methodology. Finally, we evaluate our methodology practically with four groups of stakeholders in Thailand: senior agronomists, junior agronomists, agricultural students, and ontology specialists. Both ontologies and RiceMan are evaluated to verify their correctness, usefulness, and usability in various aspects. Our experimental results show that ontology reasoning is a promising approach for this domain problem.
1. Introduction
A critical challenge in rice cultivation is to handle rice diseases and paddy insects that affect the plants. Rice plants can be damaged by both abiotic factors (i.e., the environment and nutrient factors, such as drought and cold) and biotic factors (i.e., animals, bacteria, and viruses). When they are damaged, different kinds of observable abnormal characteristics can occur. In real-life situations, farmers generally handle these infections in manual and ad-hoc manners. For instance, they may either recall from their background knowledge or consult with experts (i.e., experienced farmers and agronomists). However, these practices are not efficient, since different experts may have diverse skills and familiarities. Indeed, knowledge should be refined and externalized to farmers so that they can recognize diseases from abnormal characteristics and pick-up treatments appropriately.
Information related to rice cultivation on diverse websites enables rice farmers to enhance their knowledge and adapt it to their circumstances. For instance, they can search and seek desirable information to identify why a disease occurs and how they should handle it. On the other hand, farmers need to consume vast and diverse sources to acquire the desired knowledge. Furthermore, such published information is usually not encoded in a machine-processable form, causing difficulties to expert systems for utilizing it properly. This problem motivated us to close the gap by constructing structured knowledge with well-defined semantics and develop an expert system for rice disease identification and control recommendation to support user decisions. At a high-level, our motivations amount to investigations of the design and modeling of rice disease ontologies and their adoption to practical applications. Therefore, our research questions (RQs) are set up as follows:
- RQ1: how could a knowledge base for rice disease and paddy insects be modeled from the available unstructured resources?
- RQ2: How could a knowledge base for rice disease be applied practically?
- RQ3: How can a designed expert system be evaluated to ensure its appropriateness, from the viewpoints of stakeholders?
This research is an extended study of our proceeding research, which developed the first preliminary version of the ontologies “RiceDO” and “TreatO”, covering 22 rice diseases and paddy insects, and introduced a semantic-based framework for supporting rice disease and control identification. In this paper, our novelties consist of two ontologies (RiceDO and TreatO version 2) which have been improved from RiceDO and TreatO to meet the accepted standards. The structures of RiceDO and TreatO version 2 are reorganized and are designed for retrieving possible rice pests and controls, respectively. Additionally, to practically utilize both ontologies, we extended and implemented a novel framework and expert system, called RiceMan, and then performed a thorough evaluation to ensure the correctness, usefulness, and usability. The contributions of this work are threefold:
- Existing ontologies were investigated, remodeled, revised, and evaluated, practically, from the viewpoints of four groups of stakeholders;
- An expert system, called RiceMan, was designed and developed for rice disease identification and control recommendation;
- Results of applying a expert system in the rice domain were evaluated and reported.
2. Related Works
2.1. Existing Ontologies and Vocabularies
Table 1 summarizes and compares existing ontologies and vocabularies in the agriculture domain, with respect to their main focus and covered knowledge.

Table 1.
Existing, relevant ontologies and vocabularies in the agriculture domain †.
AGROVOC [1] is a well-known vocabulary, covering many areas, such as food, nutrition, agriculture, fisheries, forestry, and the environment. Despite its huge size, the presented vocabulary is not applicable for rice disease identification and control recommendation due to the lack of specific rice plant axioms. Agriculture Activity Ontology (AAO) [2] is a part of the Common Agricultural Vocabulary (CAVOC). It was developed as a common vocabulary to support cooperation between different farm management systems. It defines farm activity expressions based on description logics, with eight kinds of essential attributes, e.g., purposes, actions, equipment, and crop names. The National Agricultural Library Thesaurus (NALT) (https://agclass.nal.usda.gov, accessed on 1 November 2021) is an agricultural vocabulary and glossary in English and Spanish. It covers 17 subject categories, such as food, animals, biological, farms, and insects. Phenotype and Trait Ontology (PATO) (https://github.com/pato-ontology/pato, accessed on 1 November 2021) [3] provides definitions of phenotypic qualities (properties and attributes) that can be used to define phenotypes across species’ domains, such as color (e.g., red, yellow), abnormal positions, temperatures (e.g., high, low), and symptoms (e.g., swelling) [4].
Planteome (https://planteome.org, accessed on 1 November 2021) [8] provides a suite of reference ontologies for plants, such as Plant Trait Ontology (TO) (https://browser.planteome.org/amigo, accessed on 1 November 2021) [5,6], accessed on 1 November 2021, Plant Ontology (PO) (https://browser.planteome.org/amigo, accessed on 1 November 2021) [7,8,9], and others. TO provides nine broad categories of plant traits, in which each category consists of measurable characteristics of plants. Planteome develops PO to model plant-related knowledge. Thus, PO describes anatomical structures and growth stages in the plant bodies (e.g., roots, stems, leaves, fruits, and seeds)
Several rice plant knowledge-bases were developed using the OWL, RDF, and OBO formats. For example, the Crop Ontology Rice Trait Ontology (CO_320) (https://cropontology.org/term/CO_320:ROOT, accessed on 1 November 2021) [10] is a part of Crop Ontology (CO) (https://cropontology.org/, accessed on 1 November 2021) [16,17], providing validated trait names as well as the terminology of phenotypic data of rice plants. Plant Disease Ontology (PDO) (https://github.com/Planteome/plant-disease-ontology, http://wiki.plantontology.org/index.php/Plant_Disease_Ontology_(PDO), accessed on 1 November 2021) [11] provides a list of maize, wheat, and rice diseases, structurally classified into three main categories, i.e., bacteria, fungi, and viruses. Plant Protection Ontology (PPOntology) (https://sites.google.com/site/ontoworks/ontologies, https://sites.google.com/site/ppontology/home, accessed on 1 November 2021) [12,13] classifies barley’s disorder into abiotic and biotic. The biotic disorder is also classified further into bacteria, fungi, and viruses. Rice Disease Ontology (RiceDO) and Treatment Ontology (TreatO) [14] initially model rice diseases and insects according to their abnormal characteristics of rice plants and defined relevant biological and chemical controls using ontological classes and relation expressions.
Regarding the engineering discipline of each ontology, CO_320 and PDO exploit annotations for modeling the pathogens and short descriptions of each disease. In contrast, PPOntology models disease names, appearances, and pathogens as individuals in the ontology. While these models can represent the domain knowledge, their choices cause difficulties in employing inference engines, such as reasoning for implicit diseases from a symptom expression. RiceDO and TreatO [14] address this issue by employing a different modeling approach using ontology class definitions. Despite their existence, they have not yet been evaluated and used empirically. These unaddressed points motivate us to justify and revise towards their practical implementation.
2.2. Farming Knowledge Systems and Expert Systems for Plant Disease Diagnosis
Table 2 presents existing online farming knowledge portals and plant disease diagnosis systems. PlantVillage (www.plantvillage.psu.edu/, accessed on 1 November 2021) [18] is an open-access database, providing knowledge of various crops, enabling the development of plant disease expert systems. Plantwise Knowledge Bank (www.plantwise.org/KnowledgeBank, accessed on 1 November 2021) [19,20] provides pest diagnostic tools and fact sheets on plants via a website and a mobile application. Evidence in [19] shows that the combination of online fact sheets and plant doctors, to advise farmers, is helpful in practice. Toepfer et al. [21] reports that 86% of trained plant doctors in China indeed provide comprehensive pest management recommendations, leading to sustainable farming. CROPROTECT (https://croprotect.com, accessed on 1 November 2021) [22] is a UK-based website that aims to provide remote farmers with access to knowledge about crops, pests, weeds, and disease management. RiceXpert (https://play.google.com/store/apps/details?id=com.icar.ricexpert&hl=en&gl=US, accessed on 1 November 2021) [23] is a mobile application for disseminating rice plant knowledge, e.g., rice diseases, paddy insects, non-insect pests, weeds, nutrients, as well as trendy news, a fertilizer calculator, and online advisory services from experts via text, images, and voice. Finally, Plantix [24,25] is a pest and disease diagnosis tool that employs machine learning algorithms to recommend a customized treatment corresponding to an identified disease based on an input image. It can support the knowledge of pests and diseases of more than 46 plants; it also provides a fertilization calculator.

Table 2.
A comparison of online farming knowledge portals and diagnosis systems.
Regarding image-based disease detection, ref. [31] reviews various modern feature extraction techniques for plant disease detection. In [32], the authors propose a C-GAN based deep learning approach to detect tomato diseases from leaf images. The residual-based CNN architecture is proposed in [33]. Still, there exist various expert systems for plant disease diagnosis. An android-based application in [34] is developed to identify fruit diseases from photos by using K-Means to segment each damaged area, using an artificial neuron network (ANN) to classify the diseases. A desktop application for diagnosing six papaya plant diseases [35] is developed as an IF-THEN rule-based system, using CLIPS and Delphi language. In [36], a fuzzy logic rule-based web application for diagnosing banana diseases and pests contains 37 symptoms for 5 pests and 4 diseases of banana plants. In [37], a web-based system for managing nine rice diseases and pests is introduced, covering 32 damages for a rice plant.
Several expert systems [14,38,39,40] utilize ontologies and semantic web technologies for crop planning and diagnosis of plant diseases. An expert system in [38] employs an ontology and rules for recommending rice varieties and theirs corresponding crop calendar based on geographic and temporal information. Its knowledge-base and rules cover rice varieties, disease and pest resistance, soil types, and ecosystems. For disease diagnosis purposes, ref. [39] presents phytopathology ontology covering plants, diseases, causes, a comprehensive set of symptoms, recommendations, etc. Its knowledge base is modeled as individuals (ABox) with rules developed based on SWRL. On the other hand, refs. [14,40] RiceDO and TreatO ontologies model rice diseases, abnormalities, and controls as TBox axioms using the description logic-based formalism. This paper aims to improve RiceDO and TreatO to meet the accepted standards and utilizes both ontologies for identifying rice diseases and controls with the developed expert system called RiceMan.
3. Ontology Development for RiceMan
RiceMan exploits OWL for modeling knowledge about rice diseases and controls. Hence, it enables the disease identification and control recommendation to be functioned by an ontology reasoner equipped within the framework.
3.1. Ontology Requirements
We carefully elaborate competency questions (CQs) to clarify the ontology usage’s scope. Ontologies do not merely store knowledge, but they also enable entailing implicitly valid knowledge to the users. To validate the entailment’s soundness, CQs are also used during the system evaluation tasks. The CQs used in this research are shown below:
- CQ1:
- What are the recognizable diseases when a rice plant has an appearance A?
- CQ2:
- What are the recognizable diseases when a rice plant has appearances and ⋯?
- CQ3:
- What are the possible appearances of a disease A?
- CQ4:
- What are the possible biological controls of a disease A?
- CQ5:
- What are the possible chemical controls of a disease A?
3.2. Ontology Design and Development
This subsection explains our two ontologies: RiceDO and TreatO, addressing CQ1–CQ3 and CQ4–CQ5, respectively.
3.2.1. RiceDO and TreatO: Original Design
RiceDO and TreatO, initially introduced in [14], model terminological knowledge with classes and properties related to traits and phenotype of various rice diseases in Thailand. Their main objectives are to support retrieval of potential diseases that might occur on a rice plant, given an expression of abnormal characteristics with a DL query language. Supported queries could be either in the form of a single appearance (such as a gray spot) or multiple ones (such as a gray spot with a yellow oval spot). The ontologies cover 22 rice diseases and paddy insects in Thailand and coincide with the aforementioned CQs. However, the modeled knowledge is verbose and impractical for the disease identification and control recommendation. To overcome these issues, we further investigate and redesign the ontologies. Thus, incorrect and improper design issues are identified, including (1) unused classes; (2) classes and properties with ungrammatical semantics; (3) classes with semantic overlapping; (4) inappropriateness of class-subclass relationships; and (5) lack of restrictions in domains/ranges of properties.
3.2.2. RiceDO v2: Revised Design
RiceDO v2 is redesigned to address the aforementioned issues and is enhanced to support interoperability with related ontologies. Three reliable knowledge sources are used to formulate axioms in RiceDO: (1) Rice Knowledge Bank Thai version (Thai-RKB) [41] maintained by the Rice Department, Thailand; (2) Rice Knowledge Bank (RKB) from IRRI [26]; and (3) Rice Ontology (CO_320) [10]. Here, we consider Thai-RKB as the primary knowledge source for modeling our ontology, while the other two are used in absence of the information in Thai-RKB.
We adopt the design principle of TBox-based knowledge representation from a well-known medical ontology called SNOMED CT (https://www.snomed.org/, accessed on 1 November 2021). Following this design principle, RiceDO v2 (http://purl.org/ricedo, accessed on 1 November 2021) [42] consists of nine top-level classes under the RiceDOTop and 11 object properties in the object property hierarchy, as shown in Figure 1a,b, respectively. RiceDO v2 uses the special-purpose property called abnormalityGroup to axiomatize a group of various characteristics that can occur together on a certain disease. We illustrate this in Figure 1c for the rice brown spot fungal disease, in which the property abnormalityGroup is used to group characteristics ‘light yellow halo spot on leaf during the tillering growth stage’; that is, abnormalityGroup some (SpotOnLeaf and (hasColor some LightYellow) and (hasGrowthStage some Tillering) and (hasShape some Halo)).
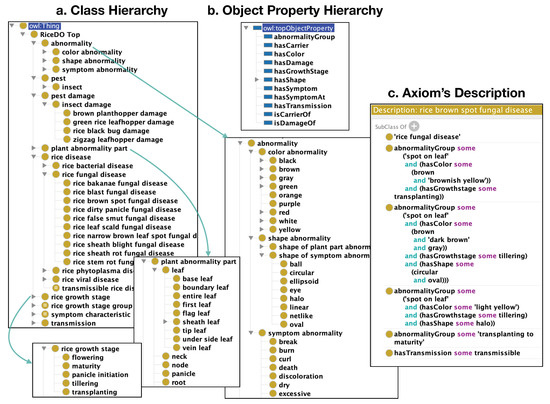
Figure 1.
The structure of RiceDO ontology is divided into three parts: (a) the class hierarchy of RiceDO; (b) the object properties of RiceDO; and (c) an example of axioms representing abnormalities of the rice brown spot fungal disease.
Another radical change in RiceDO v2 is to redefine Insect as a kind of Pest instead of RiceDisease as in the first version. In addition, the class PestDamage is redefined to characterize damages caused by pests. Furthermore, the class PlantDisease subsumes 18 diseases, which are categorized under four classes, namely PlantBacterialDisease, PlantFungalDisease, PlantPhytoplasmaDisease, and PlantViralDisease.
Moreover, RiceDO v2 complies with good modeling practices of W3C. Regarding the interoperability, RiceDO v2 reuses terms and handles potential mappings with existing ontologies: PDO, PO, and PATO. Rice disease names are imported from PDO. Besides, terms of plant parts and growth stages are mapped to existing equivalent classes in PO by using owl:equivalentClass. Similarly, terms of colors and shapes are mapped to the existing ones in PATO. Mapping terms to PO and PATO are used instead of importing in order to maintain the meaning and simplicity of RiceDO v2’s class hierarchy.
3.2.3. TreatO V2: Revised Design
In [43], four approaches for handling rice diseases are discussed: following the recommended physical practices, using host resistance, applying biological controls, and applying chemical controls. Coincided with CQs, the original TreatO focuses on modeling the biological and chemical control knowledge of each disease and paddy insects based on three knowledge sources: RKB’s fact sheets [26] and plant disease management knowledge [43,44].
The revised design of TreatO v2 contains corrected classes and properties to resolve ungrammatical semantics on the ‘is-a’ relation. In addition, it classifies control agents into bacteria, fungi, and viruses. Moreover, it has restrictions on all properties, and includes the class RiceDisease from RiceDO v2. Figure 2 shows the new TreatO v2 (http://purl.org/treato, acsessed on 1 November 2021) [45] with an example of a class description of the chemical control agent to its related disease.
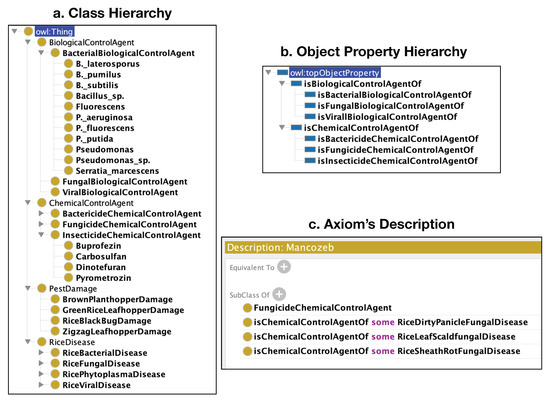
Figure 2.
The structure of TreatO ontology is divided into three parts: (a) the class hierarchy of TreatO; (b) the object properties of TreatO; and (c) an example of axioms representing Mancozeb (a fungicide chemical control agent for various rice diseases).
4. RiceMan Application Development
This section designs and develops the RiceMan application corresponding to the previously defined CQs and the elicited requirements to support user decisions.
4.1. System Requirements
Figure 3 depicts the use case diagram of RiceMan, covering three groups of target users: farmers, agronomists, and scholars. Firstly, farmers can use RiceMan to understand possible causes and suitable treatments for their plants. However, according to the discussion with agronomists, many traditional farmers in Thailand prefer to consult with agronomists. Thus, we consider agronomists who are responsible for supporting farmers to seek suitable treatments as the second target group. They need to understand the relationship between abnormalities and diseases and the relationship between diseases and treatments quite well. Without intelligent tools, they need to memorize all complex relationships to support farmers. Although the domain knowledge covers 18 diseases, its search space is complex since any disease can be indicated from various combinations of abnormalities: 28 colors, 11 shapes, 29 symptoms, 24 plant parts, and 5 growth stages. While RiceDO’s disease axioms do not contain all combinations, a particular disease could contain up to 15 combinations. Hence, RiceMan advances agronomists’ working methods by offering intelligent functionalities about diseases and treatments. RiceMan provides a more reliable approach than utilizing solely offline documentations. Lastly, scholars use RiceMan to support their activities. Since RiceMan identifies diseases based on observations and suggests treatments based on the identified diseases, then students and researchers can utilize this functionality to simulate specific scenarios for their study and research. The reasoning capability equipped with RiceMan is a perfect tool for this purpose. Important use cases of the system are explained as follows:
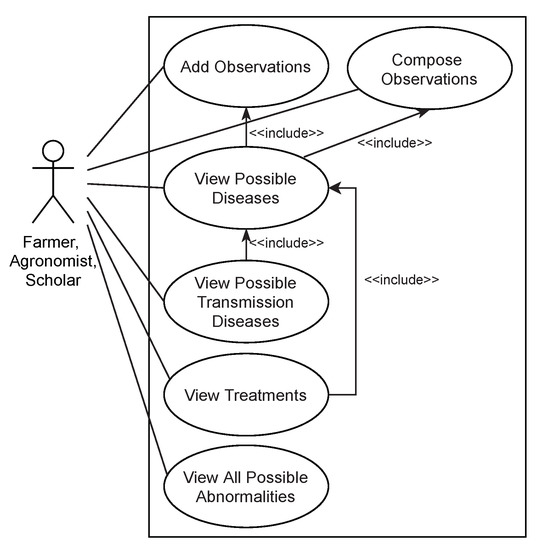
Figure 3.
The RiceMan use case diagram representing six use cases of the proposed RiceMan system and three actors (i.e., farmer, agronomist, and scholar).
- Add observations: upon the finding of abnormal appearances on a rice plant, a user can use RiceMan to create observation data digitally that represents the observed abnormal appearances.
- Find possible diseases: to find possible diseases, both explicit and implicit diseases corresponding to an observation, RiceMan employs logical reasoning to consider the logical relationships defined in RiceDO for the identification.
- Compose observations (with respect to a distance and a duration): concerning the fact that some rice diseases and paddy insects are spreadable to nearby areas, a user may choose to include observations made within the specific distance and duration in his/her consideration of the disease diagnosis. This functionality is called the observation composition. It calculates the distance between different latitudes and longitudes and considers the duration in the unit of days.
- Find possible transmittable diseases: once users consider composing observations, the identified results can be classified into (1) possible diseases with respect to distance and duration; and (2) possible transmittable diseases, with respect to distance and duration.
- Find possible controls: RiceMan considers possible controls for a disease based on the knowledge defined in TreatO by an employment of logical reasoning.
- Find possible abnormalities of a given disease: RiceMan can return all possible abnormal appearances on a rice plant if it is infected by a particular disease. This enables users to compare their observations with the retrieved results and make a better decision.
4.2. System Design
Figure 4 depicts the RiceMan’s architectural design, based on a client-server architecture, and implemented by Java, OWL API (http://semanticweb.org/wiki/OWL_API.html, accessed on 1 November 2021) [46], and MySQL (for storing observation data). The main reasoning technique used in RiceMan is subsumption [47], which enables to logically infer if a class expression subsumes one another or not. Thus, to enable the search of recognizable diseases and potential controls with subsumption reasoning, we convert each observation object as a class expression. Once the potential diseases are indicated, we also employ the subsumption reasoner to suggest corresponding controls.
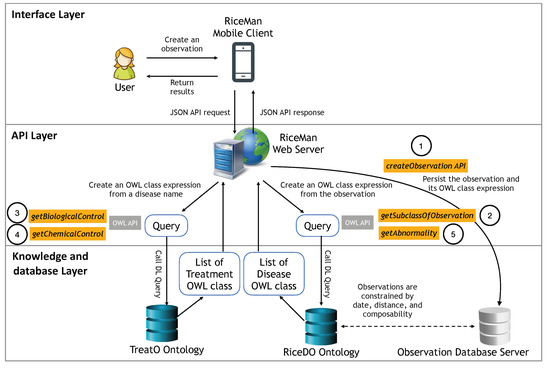
Figure 4.
The RiceMan system architecture classified into three layers: (1) interface layer which indicates the client side’s environment; (2) API layer with five main APIs; and (3) knowledge and database layer, which contains our two ontologies and their environment.
The web server exposes five main APIs. First, createObservation provides a service to store observations into the database. Second, getSubclassOfObservation returns a list of disease names, which are sub-categories of an observation. This API can also be parameterized for the composition with other observations if a distance and a duration are satisfied. Internally, when this API is called, a corresponding OWL class expression will be determined and be subsequently used by the OWL reasoner to find their subclasses from the RiceDO ontology. The third and fourth APIs are getBiologicalTreatmentsOfDisease and getChemicalTreatmentsOfDisease, respectively. As indicated by their names, they return a list of (biological and chemical) treatments corresponding to a given disease. Lastly, API getAbnormalities returns a group of abnormality for a specified disease.
4.3. Query Construction and User Interface
Since RiceDO and TreatO ontologies are developed by the OWL language, then the appropriate query language is thus the DL query expressed in the Manchester OWL syntax. DL query uses a class expression as an input to logically determine implicit facts inside an ontology. This research uses this competency of DL query to search for certain classes using subsumption reasoning. Note that we employ subsumption in DL query because our prime objective is to classify whether an input observation expression belongs to any disease classes or not. For this purpose, a user’s input is needed to transform into a DL query before it can be used to query for a certain pest. Therefore, we develop two procedures for creating DL queries from a user’s input by complying with the design of our ontologies.
The first procedure is for converting an observation object in the database into an OWL class expression based on the following three steps: (1) we nest a set of symptoms, places, shapes, growth stages, transmission, and colors using existential quantification via properties hasSymptom, hasSymptomAt, hasShape, hasGrowthStage, hasTransmission, and hasColor, respectively; (2) we conjunct these nested expressions; and (3) we nest all using existential quantification via property abnormalityGroup. Figure 5a illustrates a constructed OWL class expression of an input observation object using this procedure.
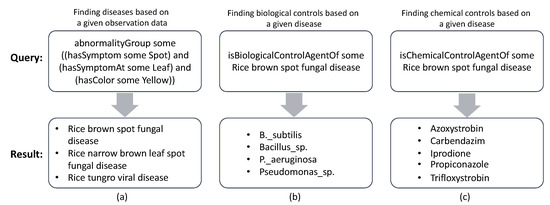
Figure 5.
DL queries and results of finding possible diseases and controls for a disease: (a) a DL query of finding the corresponding rice diseases given an observation, (b) a DL query of finding biological controls for the rice brown spot fungal disease, and (c) a DL query of finding chemical controls for the rice brown spot fungal disease.
The second procedure is for treatment recommendation. That is, it nests a given disease name via property isBiologicalControlAgentOf (or isChemicalControlAgentOf depending upon a user’s request) using existential quantification. Figure 5b,c illustrates an application of this procedure to recommend controls for a recognizable disease.
Figure 6 shows a sequence of RiceMan’s user interfaces that display the results of our queries as follows. From Steps 1–3, the user inputs the observed abnormalities as a brown oval spot on a leaf. Step 4 allows the user to define thresholds for a distance and a duration. In Step 5, RiceMan returns the identified disease(s). In Step 6, the user can click on the disease identified to get recommended treatments. Steps 7–8 show the groups of all possible abnormalities of a disease followed by the suggested controls.
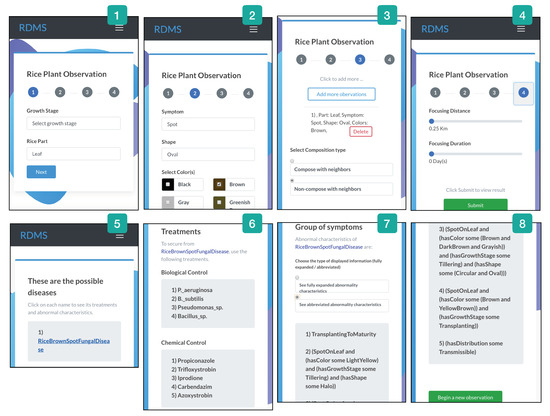
Figure 6.
Sequential steps of using the RiceMan’s user interfaces from providing observation data (a user’s input) to receiving the results.
4.4. Observation Data Composition
RiceMan implements the observation composition mechanism to identify diseases from multiple observations. If observations are located nearby each other, occur coincidentally, and are composable, then they are composed and are collectively treated as a single observation. Figure 7 illustrates an example in which each observation and occurs in different geolocations and dates as described in Table 3.
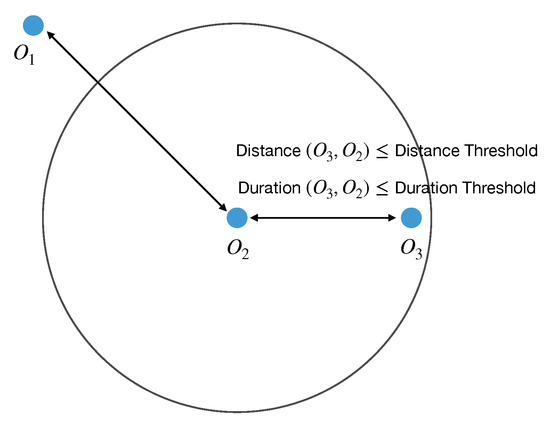
Figure 7.
An example of practical observations and in the study where falls outside the distance and duration threshold of and falls inside the thresholds of .

Table 3.
Details of observations and .
Here, suppose that is collected first; after that, and are collected. RiceMan users can compose past observations with the current observation for disease identification, which is implemented as follows:
- 1
- Past observations that satisfy three conditions are retrieved: a distance threshold, a date threshold, and a composability boolean, from the database;
- 2
- An OWL class expression is created from a database object, representing a current observation manifested with certain characteristics and intersects it with the retrieved past OWL class expressions;
- 3
- The new observation is used for the subsequent disease identification based on the subsumption reasoning.
The observation composition mechanism helps RiceMan users delve into specific results when a single observation captures partial or broad abnormalities. Nevertheless, this mechanism may yield the same results as considering a single observation. That is when there are no observations that satisfy the three conditions. For instance, using the composition function with yields the same results as without using this function, i.e., it returns the rice brown spot fungal disease. This is because there are no nearby observations with . On the other hand, RiceMan yields the rice blast fungal disease, the rice sheath rot fungal disease, and the rice brown spot fungal disease for when the composition function is disabled (cf. the top part of Figure 8). In contrast, it yields the rice brown spot fungal disease for when this composition functionality is opted in (cf. the bottom part of Figure 8). When a single observation contains broad abnormal characteristics, diverse results can be obtained. Hence, this composition mechanism helps users narrow the disease identification from many possible disease candidates.
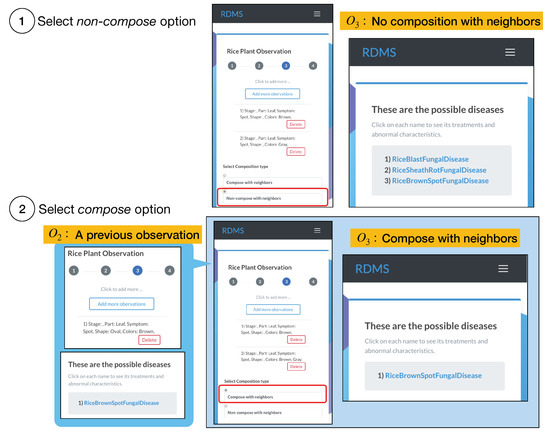
Figure 8.
The usage of our composition function: (1) selecting the non-composition option and (2) selecting the composition option.
5. RiceMan: Ontology and System Evaluation
Figure 9 illustrates our evaluation design to validate important aspects of the developed ontologies and the RiceMan system. The evaluation comprises three important dimensions: RiceDO and TreatO ontology validation, ontology usage, and RiceMan application usability. The following subsections then elaborate each evaluation dimension in more details.
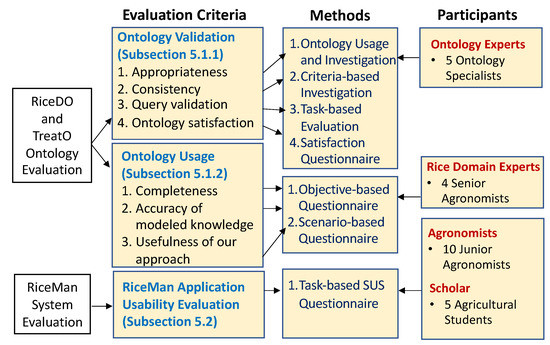
Figure 9.
Our evaluation design for both ontologies and RiceMan (the evaluation criteria, the methods used by each evaluation and the specific participants are shown).
5.1. Ontology Evaluation
5.1.1. Ontology Validation
We carried out this part of the evaluation with five ontology experts, who were ontology engineer experts from NECTEC (National Electronics and Computer Technology Center) and AIT (Asian Institute of Technology) with experiences in modeling and applying ontologies in their work. Four important criteria were considered:
- 1.
- Appropriateness: we concentrated on verifying whether each defined class, axiom, and the object property was appropriately modeled based on the domain knowledge and practical usage from rice plant experts’ perspectives. Here, the participants investigated both ontologies by using Protégé and provided scores for the defined eight sub-criteria, shown in Table 4. That is, RiceDO and TreatO were modeled with 95.2% and 89.4% appropriateness, respectively.
 Table 4. Verifying the appropriateness of RiceDO and TreatO ontologies.
Table 4. Verifying the appropriateness of RiceDO and TreatO ontologies. - 2.
- Consistency: we performed consistency checking in RiceDO and TreatO according to the six sub-criteria proposed in [48]: (1) no duplicated classes; (2) no class cycles in the class hierarchy; (3) no duplicated properties; (4) no property cycles; (5) no invalid ranges; and (6) no invalid domains. As a result, the participants found a few mistakes in some object properties’ domains, and we had corrected them. Finally, all participants agreed that the revised ontologies contain no inconsistency according to the mentioned criteria.
- 3.
- Query validation: to ensure the practicability of applying the ontologies with respect to the defined CQs, we carried out a task-based evaluation, in which we presented for each task the prepared queries corresponding to the defined CQs and their results. The participants verified whether the results were correct, and the queries were written efficiently. Table 5 shows that every query was formulated correctly and efficiently except for Query#4 (i.e., 92%) due to its semantics’ incorrectness. Note that we skipped the evaluation of CQ 3 (i.e., finding the abnormalities of any disease) because it was natural to retrieve axioms of abnormalities modeled in RiceDO. Besides, we validated that our shortened axioms usage enabled us to write concise queries. Though this aspect was not part of CQs, it was helpful to realize the effectiveness towards writing practical queries.
 Table 5. Query validation.
Table 5. Query validation. - 4.
- Ontology satisfaction: the participants were given a questionnaire to analyze their ontology satisfaction usage. In summary, the participants were satisfied with RiceDO in terms of identifying rice diseases based on either single or multiple abnormalities as well as its convenience in querying by means of the shortened axioms (score 100%). For TreatO, the participants were satisfied with its query facilities for searching of controls (score 88–92%). Moreover, the participants agreed that both ontologies could improve work efficiency (score 100%) and they were willing to use them (score 96%). Overall, it shows the ontologies gain high satisfaction with an average score of 95.5%. For more details, see Table A1 in the Appendix A.
5.1.2. Ontology Usage
This evaluation was conducted to verify that our ontologies are valuable and can effectively facilitate our users with respect to the three important criteria: completeness, accuracy, and usefulness. Two groups of senior agronomists: two plant pathologists and two entomologists, from DRRD (The Division of Rice Research and Development under the Rice Department, Ministry of Agriculture and Cooperatives, Thailand), with 8-year experience on average, were involved.
- 1.
- Evaluating the completeness of diseases and paddy insects: we evaluated the completeness in terms of the number of diseases and insects provided in RiceDO (cf. Table 6). Two levels of completeness checking were executed. Firstly, we compared the modeled knowledge with the Rice Knowledge Bank maintained by the Rice Department of Thailand (Thai-RKB) [41]. Secondly, we verified the completeness based on domain experts’ knowledge to ensure that it was applicable in practice. RiceDO covers most of the rice diseases in Thailand; however, it covers relatively fewer insects. Though the number of insects’ coverage seems insufficient, two of them frequently occur in rice fields, and three of them are carriers for some rice diseases (such as gall dwarf disease and rice tungro disease).
 Table 6. RiceDO’s completeness (number of diseases and paddy insects).
Table 6. RiceDO’s completeness (number of diseases and paddy insects). - 2.
- Evaluating the accuracy of modeled knowledge: two aspects were considered: (1) the correctness of the modeled axioms; and (2) the appropriateness of our vocabulary. Precisely, the participants were asked to consider if the modeled axioms were correct or not. Similar steps were carried out to verify the appropriateness of the vocabulary. Table 7 shows the results of this evaluation in percentage. Note that 13 (out of 18) rice diseases, frequently found in Thailand rice fields, and four paddy insects were selected for this validation. For the error analysis, the correctness of rice disease axioms was lower than those of other classes due to two main reasons. The first reason was the inconsistency between knowledge sources and domain experts. Some participants voted the axioms as incorrectly defined because the axioms did not coincide with their experiences, although they were modeled properly according to the referenced knowledge sources (IRRI and Thai-RKB). Concerning the appropriateness of the used vocabulary, some participants argued that the terms referenced from the knowledge sources were not suitable. We found that the vocabulary’s appropriateness relies on a proper term selection and a proper English–Thai translation.
 Table 7. Accuracy of the modeled knowledge.
Table 7. Accuracy of the modeled knowledge. - 3.
- Evaluating the usefulness of inferred knowledge: to evaluate the practical applicability of the ontologies, we asked the participants to review the prepared inferred knowledge based on six scenarios. Specifically, scenarios #1–#2 identify rice diseases from single and multiple abnormality groups (usefulness score: 48% and 70%); Scenario #3 queries for all abnormality groups of disease (76%); Scenarios #4–#5 query for a disease’s biological and chemical control agents (42% and 48%); Scenario #6 focuses on the disease classification into possible diseases and warning diseases with respect to nearby and recent observation data (90%) (cf. Table A2). From the results, Scenarios #2, #3, and #6 obtained prominently high scores, indicating that the proposed functions help the participants reduce the searching times compared to the manual search. The participants agreed that the multiple observation data and the composition mechanism, and ontology reasoning are promising for those scenarios.
5.2. RiceMan Application Usability Evaluation
Based on the proposed RiceMan system architectural design, the application was developed with the UI in both English and Thai languages. The usability evaluation was then conducted with two groups of participants: senior-year agricultural students and junior agronomists. The first group of participants were five senior-year undergraduate students (Chulalongkorn University, School of Agriculture Resources) who were knowledgeable in rice diseases and paddy insects from their degree program’s coursework, but did not have direct experience in real-world practice. The students used RiceMan via their mobile phones to complete four provided tasks. For tasks #1 and #2, the participants were asked to identify rice diseases from a given single observation and multiple observations, respectively. For tasks #3 and #4, they were asked to identify biological and chemical controls from the returned diseases. After their completion, the participants provided their opinions by answering ten questions based on System Usability Scale (SUS) [49] (cf. Table A3 in the Appendix A for more information). The average SUS score among the students was 70.5. Since it was above 68, then RiceMan was acceptable from the agricultural students’ viewpoints. The participants were interested in applying ontology reasoning techniques to identify diseases and treatments. The feedback shows that the application can support their study and improve the search in many real-life use cases. Since they had experience in farm work, they believed that the application can benefit the farmers, especially those interested in using new technologies to support their works. However, most users were facing difficulties with using the application due to some graphical interfaces.
The second group of participants were ten junior agronomists from Ayutthaya Rice Research Center, who worked on transferring technologies to farmers, focused on rice variety research and development, and advised farmers. Similarly, they performed the same four tasks on their mobile phones, and then answered a SUS questionnaire. The average SUS score obtained was 64.3, close to the standard average of SUS.
Although the score was less than the standard average, the participants agreed that the application could significantly help them identify diseases and treatments, compared with searching through manuals and websites. It shows that RiceMan is applicable for use from the participants’ viewpoints since it offers helpful and sufficient functionalities for real-life situations even though the current version seems challenging for farmers to use. We summarize our lessons learned as follows:
UI/UX design:
- 1.
- The design of the observation data input form should be improved to make it more human-friendly for use and to support various kinds of abnormality appearances.
- 2.
- Since RiceMan is built as a responsive web application, its input form varies among iOS and Android devices. Here, the participants who used an iOS device faced some difficulty as the input form was a picker instead of a drop-down box.
- 3.
- The treatment names shown on RiceMan should be italicized for easy-looking.
Displayed information:
- 1.
- The participants preferred to have photos of plant parts to support the observation data input forms and photos of diseases and insects on the result page.
- 2.
- The control names should be translated into Thai and provided with the product/brand names.
- 3.
- Some agronomists were specialists in insects. Therefore, they preferred RiceMan to contain more knowledge of paddy insects.
6. Conclusions
This study proposes the design and development of RiceMan, an ontology-based expert system for rice disease identification and control recommendation. As part of our development, we make further progress on improving the two ontologies for rice disease in Thailand: RiceDO and TreatO. Together with the composition mechanism, these two ontologies lie at the heart of RiceMan’s functionalities. RiceMan facilitates users to identify diseases with respect to RiceDO’s TBox axioms and suggests controls with respect to TreatO’s TBox axioms. Though both ontologies are initially built for RiceMan, they can be potentially extended for other expert systems in the agriculture domain with some revisions to comply with the adapted problems.
To ensure its usability and practicability, we evaluate the developed ontologies and the expert system in comprehensive viewpoints with four user groups: (1) ontology specialists; (2) senior agronomists; (3) junior agronomists; and (4) scholars/agricultural students. From the ontology specialists’ perspective, RiceDO and TreatO are modeled soundly, appropriately, and consistently; hence, they can support the users to write efficient queries. From the senior agronomists’ perspectives, the knowledge related to rice diseases is modeled almost completely, but the knowledge regarding paddy insects may be insufficient. The correctness of the modeled axioms is acceptable, but requires some improvement in terms of the vocabulary. Regarding the usability evaluation, the participants found themselves pleasant with the Thai version, even though they faced some difficulties in the observation data input with the current user interface. These results demonstrate that RiceMan is a promising approach to the rice disease identification and control recommendation problem.
There are several future directions. Firstly, RiceDO and TreatO can be enhanced with modeling of axioms from various aspects of information and reliable knowledge sources. Secondly, we aim at collaborating more closely with agronomists at multiple stages of ontology development to clean up some inconsistency that may happen between the adopted knowledge sources and domain experts’ tacit knowledge. Besides, we are interested in applying ontology learning techniques to (semi-)automatically model and enhance the constructed knowledge base. Finally, considering potential applications of our ontologies with machine learning for disease identification, we plan to investigate and study the development of latent representation from RiceDO and TreatO. Inspired by [50], each class and property can be trained based on self-supervised learning to find relevant vector representations from the ontologies. In [50], ontologies from BioPortal (https://bioportal.bioontology.org/, accessed on 1 November 2021) are used to develop semantic word embedding for applications of monitoring diabetes and blood pressure (BP) patients. In the near future, we plan to attempt similar directions based on our ontologies for monitoring diseases from a multi-modal context.
Author Contributions
Conceptualization, W.J. and C.A.; methodology, W.J.; software, T.R.; validation, W.J., C.A., T.R. and F.A.; formal analysis, T.R.; investigation, W.J.; resources, W.J.; data curation, W.J.; writing—original draft preparation, W.J. and T.R.; writing—review and editing, W.J., C.A., T.R. and F.A.; visualization, W.J.; supervision, C.A. and F.A.; funding acquisition, F.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Institute of Informatics, Japan.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to acknowledge the support in the evaluation processes from the following three institutes: (1) the Division of Rice Research and Development (DRRD) of the Rice Department, Ministry of Agriculture and Cooperatives, Thailand; (2) Phra Nakhon Si Ayutthaya Rice Research Center, especially Kritkamol Paothong; and (3) School of Agriculture Resources, Chulalongkorn University.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Ontology satisfaction from the ontology specialists’ perspective.
Table A1.
Ontology satisfaction from the ontology specialists’ perspective.
| # | Ontology Usage Satisfaction | Average (%) |
|---|---|---|
| 1 | Searching for possible rice diseases, which are related to a single abnormality, can be facilitated by queries on RiceDO. | 100 |
| 2 | Searching for possible rice diseases, which are related to many abnormalities, can be facilitated by queries on RiceDO. | 100 |
| 3 | Using the defined subclasses of SymptomCharacteristic (shortened axioms) is more convenient than writing the full definitions in queries. | 100 |
| 4 | Searching for possible treatments (Bio. and Chem.) for a disease can be facilitated by queries on TreatO. | 88 |
| 5 | Searching for possible treatments (Bio. and Chem.) for many diseases can be facilitated by queries on TreatO. | 92 |
| 6 | RiceDO and TreatO ontologies can be extended to cover other factors e.g. image of symptoms, image of shapes, and detailed treatment, etc. | 88 |
| 7 | Assume that you are not a domain expert (Rice Plant), you feel that querying for diseases and treatments can save your time than finding from a website or a book. | 100 |
| 8 | If you want to identify rice diseases and treatments, you would like to use both ontologies. | 96 |
| Overall ontology usage satisfaction score | 95.5% |

Table A2.
Usefulness of inferred knowledge in six scenarios, evaluated by senior agronomists.
Table A2.
Usefulness of inferred knowledge in six scenarios, evaluated by senior agronomists.
| # | Scenarios | Usefulness Avg. (%) |
|---|---|---|
| 1 | Searching for possible diseases from a single abnormality group. | 48 |
| 2 | Searching for possible diseases from multiple abnormality groups. | 70 |
| 3 | Searching for all abnormality groups of a disease. | 76 |
| 4 | Searching for biological controls of a disease. | 42 |
| 5 | Searching for chemical controls of a disease. | 48 |
| 6 | The classification of diseases into possible diseases and warning diseases with respect to considering nearby observations. | 90 |
| Overall ontology usefulness score | 62.3% |

Table A3.
The usability testing of RiceMan based on the System Usability Scale (SUS), evaluated by junior agronomists and students.
Table A3.
The usability testing of RiceMan based on the System Usability Scale (SUS), evaluated by junior agronomists and students.
| # | System Usability Scale Items | Students Average Score | Junior Agronomists Average Score |
|---|---|---|---|
| 1 | I think that I would like to use this system frequently. | 4 | 4 |
| 2 | I found the system unnecessarily complex. | 2.2 | 2.7 |
| 3 | I thought the system was easy to use. | 3.2 | 3.9 |
| 4 | I think that I would need the support of a technical person to be able to use this system. | 2.2 | 3.3 |
| 5 | I found the various functions in this system were well integrated. | 3.6 | 3.5 |
| 6 | I thought there was too much inconsistency in this system. | 1.8 | 1.8 |
| 7 | I would imagine that most people would learn to use this system very quickly. | 4 | 3.9 |
| 8 | I found the system very cumbersome to use. | 1.4 | 2 |
| 9 | I felt very confident using the system. | 3.8 | 3.5 |
| 10 | I needed to learn a lot of things before I could get going with this system. | 2.8 | 3.3 |
| SUS Score | 70.5 | 64.3 |
References
- Caracciolo, C.; Stellato, A.; Morshed, A.; Johannsen, G.; Rajbhandari, S.; Jaques, Y.; Keizer, J. The AGROVOC linked dataset. Semant. Web 2013, 4, 341–348. [Google Scholar] [CrossRef] [Green Version]
- Joo, S.; Koide, S.; Takeda, H.; Horyu, D.; Takezaki, A.; Yoshida, T. Agriculture Activity Ontology: An Ontology for Core Vocabulary of Agriculture Activity. In International Semantic Web Conference (Posters & Demos); CEUR-WS.org: Aachen, Germany, 2016; Volume 33. [Google Scholar]
- Gkoutos, G.V.; Green, E.C.; Mallon, A.M.; Hancock, J.M.; Davidson, D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005, 6, 1–10. [Google Scholar]
- Gkoutos, G. Phenotype And Trait Ontology. Available online: https://www.obofoundry.org/ontology/pato.html (accessed on 20 February 2021).
- Jaiswal, P.; Ware, D.; Ni, J.; Chang, K.; Zhao, W.; Schmidt, S.; Pan, X.; Clark, K.; Teytelman, L.; Cartinhour, S.; et al. Gramene: Development and integration of trait and gene ontologies for rice. Comp. Funct. Genom. 2002, 3, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, E.; Cooper, L.; Shrestha, R.; Menda, N.; Nelson, R.T.; Matteis, L.; Skofic, M.; Bastow, R.; Jaiswal, P.; Mueller, L.; et al. Towards a reference plant trait ontology for modeling knowledge of plant traits and phenotypes. In International Conference on Knowledge Engineering and Ontology Development; SciTePress: Setúbal, Portugal, 2012; Volume 2, pp. 220–225. [Google Scholar]
- Cooper, L.; Walls, R.L.; Elser, J.; Gandolfo, M.A.; Stevenson, D.W.; Smith, B.; Preece, J.; Athreya, B.; Mungall, C.J.; Rensing, S.; et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013, 54, e1. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L.; Meier, A.; Laporte, M.A.; Elser, J.L.; Mungall, C.; Sinn, B.T.; Cavaliere, D.; Carbon, S.; Dunn, N.A.; Smith, B.; et al. The Planteome database: An integrated resource for reference ontologies, plant genomics and phenomics. Nucleic Acids Res. 2018, 46, D1168–D1180. [Google Scholar] [CrossRef] [Green Version]
- Walls, R.L.; Cooper, L.; Elser, J.; Gandolfo, M.A.; Mungall, C.J.; Smith, B.; Stevenson, D.W.; Jaiswal, P. The plant ontology facilitates comparisons of plant development stages across species. Front. Plant Sci. 2019, 10, 631. [Google Scholar] [CrossRef] [PubMed]
- Pietragalla, J.; Valette, L.; Shrestha, R.; Laporte, M.A.; Hazekamp, T.; Arnaud, E. Guidelines for Creating Crop-Specific Ontologies to Annotate Phenotypic Data, Version 2.0; Alliance Bioversity International-CIAT: Rome, Italy, 2020. [Google Scholar]
- Planteome.org. Repository for the Plant Disease Ontology. 2016. Available online: https://github.com/Planteome/plant-disease-ontology (accessed on 12 February 2021).
- Ammar, H. Ontology for Plant Protection. 2009. Available online: https://sites.google.com/site/ppontology/home (accessed on 12 February 2021).
- Ontology Based Research Group at IITM. Ontologies & RDF Datasets. Available online: https://sites.google.com/site/ontoworks/ontologies (accessed on 12 February 2021).
- Jearanaiwongkul, W.; Anutariya, C.; Andrès, F. A Semantic-Based Framework for Rice Plant Disease Management-Identification, Early Warning, and Treatment Recommendation Using Multiple Observations. New Gener. Comput. 2019, 37, 499–523. [Google Scholar] [CrossRef]
- Ruangrajitpakorn, T.; Kongkachandra, R.; Songmuang, P.; Supnithi, T. Automatic Ontology Development from Semi-structured Data in Web-Portal: Towards Ontology of Thai Rice Knowledge. In Joint International Semantic Technology Conference; Springer: Berlin/Heidelberg, Germany, 2018; pp. 262–276. [Google Scholar]
- Shrestha, R.; Arnaud, E.; Mauleon, R.; Senger, M.; Davenport, G.F.; Hancock, D.; Morrison, N.; Bruskiewich, R.; McLaren, G. Multifunctional crop trait ontology for breeders’ data: Field book, annotation, data discovery and semantic enrichment of the literature. AoB Plants 2010, 2010, plq008. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.; Matteis, L.; Skofic, M.; Portugal, A.; McLaren, G.; Hyman, G.; Arnaud, E. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Physiol. 2012, 3, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Cameron, K.H.; Somachandra, K.; Curry, C.N.; Jenner, W.H.; Hobbs, S.L. Delivering actionable plant health knowledge to smallholder farmers through the plantwise program. J. Agric. Food Inf. 2016, 17, 212–229. [Google Scholar] [CrossRef] [Green Version]
- Leach, M.C.; Hobbs, S.L. Plantwise knowledge bank: Delivering plant health information to developing country users. Learn. Publ. 2013, 26, 180–185. [Google Scholar]
- Toepfer, S.; Zhang, T.; Wang, B.; Qiao, Y.; Peng, H.; Luo, H.; Wan, X.; Gu, R.; Zhang, Y.; Ji, H.; et al. Sustainable Pest Management through Improved Advice in Agricultural Extension. Sustainability 2020, 12, 6767. [Google Scholar] [CrossRef]
- Bruce, T.J. The CROPROTECT project and wider opportunities to improve farm productivity through web-based knowledge exchange. Food Energy Secur. 2016, 5, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Nayak, A.; Mohapatra, S.; Lal, B.; Tripathi, R.; Shahid, M.; Gautam, P.; Patnaik, S.; Patil, N.; Raghu, S.; Yadav, M.; et al. “riceXpert” One-Step Revolution in Agriculture. 2017. Available online: https://play.google.com/store/apps/details?id=com.icar.ricexpert&hl=en&gl=US (accessed on 4 November 2021).
- GmbH, P. Plantix Library: Pests and Diseases. 2015. Available online: https://plantix.net/en/library/plant-diseases (accessed on 15 February 2021).
- GmbH, P. Plantix Preview—Your Crop Doctor. 2016. Available online: https://play.google.com/store/apps/details?id=com.peat.GartenBank.preview (accessed on 15 February 2021).
- International Rice Research Institute. The Fact Sheets of International Rice Research Institute. Available online: http://www.knowledgebank.irri.org/training/fact-sheets (accessed on 15 February 2021).
- International Rice Research Institute (IRRI). Rice Doctor Diagnostic Tool. Available online: http://www.knowledgebank.irri.org/decision-tools/rice-doctor/diagnostic-tool (accessed on 15 February 2021).
- International Rice Research Institute (IRRI). Rice Doctor Application. 2017. Available online: https://play.google.com/store/apps/details?id=com.lucidcentral.mobile.ricedoctor (accessed on 4 November 2021).
- Studionoframe. Agrobase-Weed, Disease, Insect. 2018. Available online: https://play.google.com/store/apps/details?id=lt.farmis.apps.farmiscatalog (accessed on 4 November 2021).
- Rice Department of Thailand. Smart Rice Farm v.2. 2018. Available online: https://apkgk.com/rice.ictc (accessed on 4 November 2021).
- Vishnoi, V.K.; Kumar, K.; Kumar, B. Plant disease detection using computational intelligence and image processing. J. Plant Dis. Prot. 2021, 128, 19–53. [Google Scholar] [CrossRef]
- Abbas, A.; Jain, S.; Gour, M.; Vankudothu, S. Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput. Electron. Agric. 2021, 187, 106279. [Google Scholar] [CrossRef]
- Shah, D.; Trivedi, V.; Sheth, V.; Shah, A.; Chauhan, U. ResTS: Residual deep interpretable architecture for plant disease detection. Inf. Process. Agric. 2021. [Google Scholar] [CrossRef]
- Gajanan, D.E.; Shankar, G.G.; Keshav, G.V. Android Based Plant Disease Identification System Using Feature Extraction Technique. Int. Res. J. Eng. Technol. 2018, 5, 861–864. [Google Scholar]
- Abu-Saqer, M.M.; Abu-Naser, S.S. Developing an Expert System for Papaya Plant Disease Diagnosis. Int. J. Acad. Eng. Res. 2019, 3, 14–21. [Google Scholar]
- Budiyanto, G.; Ipnuwati, S.; Al Gifari, S.A.; Huda, M.; Jalal, B.; Maseleno, A.; Hananto, A. Web based expert system for diagnosing disease pest on banana plant. Int. J. Eng. Technol. 2018, 7, 4715–4721. [Google Scholar]
- Adi, K.; Isnanto, R.R. Rice crop management expert system with forwarding chaining method and certainty factor. J. Phys. Conf. Ser. 2020, 1524, 012037. [Google Scholar]
- Kawtrakul, A.; Amorntarant, R.; Chanlekha, H. Development of an expert system for personalized crop planning. In Proceedings of the 7th International Conference on Management of Computational and Collective Intelligence in Digital EcoSystems, Caraguatatuba, Brazil, 25–29 October 2015; pp. 250–257. [Google Scholar]
- Lagos-Ortiz, K.; Medina-Moreira, J.; Paredes-Valverde, M.A.; Espinoza-Morán, W.; Valencia-García, R. An ontology-based decision support system for the diagnosis of plant diseases. J. Inf. Technol. Res. 2017, 10, 42–55. [Google Scholar] [CrossRef]
- Jearanaiwongkul, W.; Anutariya, C.; Andres, F. An ontology-based approach to plant disease identification system. In Proceedings of the 10th International Conference on Advances in Information Technology, Bangkok, Thailand, 10–13 December 2018; pp. 1–8. [Google Scholar]
- Rice Department of Thailand. Rice Knowledge Bank, Thailand. Available online: http://www.ricethailand.go.th/rkb3/Disease.htm (accessed on 15 February 2021).
- Jearanaiwongkul, W.; Anutariya, C.; Racharak, T.; Andres, F. RiceDO Version 2: Rice Disease Ontology; IEEE Dataport: Piscataway, NJ, USA, 2021. [Google Scholar] [CrossRef]
- Thind, T.S. Diseases of Field Crops and Their Management; Daya Publishing House: New Delhi, India, 2005. [Google Scholar]
- Gnanamanickam, S.S. Biological Control of Rice Diseases; Springer Science & Business Media: Berlin, Germany, 2009; Volume 8. [Google Scholar]
- Jearanaiwongkul, W.; Anutariya, C.; Racharak, T.; Andres, F. TreatO Version 2: Rice Disease Control Ontology; IEEE Dataport: Piscataway, NJ, USA, 2021. [Google Scholar] [CrossRef]
- Horridge, M.; Bechhofer, S. The owl api: A java api for owl ontologies. Semant. Web 2011, 2, 11–21. [Google Scholar] [CrossRef]
- Horrocks, I.; Tobies, S. Reasoning with Axioms: Theory and Pratice. arXiv 2000, arXiv:0005012. [Google Scholar]
- Mendel-Gleason, G.; Feeney, K.; Brennan, R. Ontology Consistency and Instance Checking for Real World Linked Data. In Linked Data Quality, Proceedings of the ESWC 2015; Trinity College Dublin: Dublin, Ireland, 2015; Volume 1376, p. 1376. [Google Scholar]
- Brooke, J. SUS—A quick and dirty usability scale. Usability Eval. Ind. 1996, 189, 4–7. [Google Scholar]
- Ali, F.; El-Sappagh, S.; Islam, S.R.; Ali, A.; Attique, M.; Imran, M.; Kwak, K.S. An intelligent healthcare monitoring framework using wearable sensors and social networking data. Future Gener. Comput. Syst. 2021, 114, 23–43. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).