Abstract
The asymptomatic nature of and lack of effective early-stage diagnostic tools in CKD, predisposes individuals to the risk of end-stage CKD and related complications. Whole blood microRNAs (miRNAs) have the potential for CKD risk screening. We evaluated the expression profile of six novel whole blood miRNAs as well as their ability to predict prevalent CKD in individuals with hypertension and/or diabetes. We included 911 individuals with hypertension and/or diabetes, of which 18.8% had prevalent CKD. The miRNA expression was analyzed using quantitative reverse transcription PCR (RT-PCR). Five of the six miRNAs, namely hsa-miR-novel-chr1_36178, hsa-miR-novel-chr2_55842, hsa-miR-novel-chr7_76196, hsa-miR-novel-chr5_67265, and hsa-miR-novel-chr13_13519, were significantly increased in people with CKD (all p < 0.028). Only the increased expression of hsa-miR-novel-chr2_55842 and hsa-miR-novel-chr7_76196 were independently associated with reduced estimated glomerular filtration rate (eGFR) (both p ≤ 0.038), while all the analyzed miRNAs were positively associated with prevalent CKD (all p ≤ 0.038). All the blood miRNAs were acceptable predictors of CKD (C-statistic > 0.7 for all), with similar predictive capacity (p = 0.202). However, hsa-miR-novel-chr13_13519 added to CKD prediction beyond conventional factors (p = 0.040). Novel whole blood miRNAs showed an acceptable discriminative power to predict prevalent CKD; thereby suggesting the potential use of these miRNAs, particularly hsa-miR-novel-chr13_13519, in clinical practice as a screening tool for CKD in high-risk individuals.
1. Introduction
The burden of chronic kidney disease (CKD) has increased substantially over the past three decades, progressing from the 36th ranked cause of death in 1990 to the 12th ranked cause of death worldwide in 2017 [1]. Globally, around 10% of the general adult population have CKD, with an estimated 16% for the African population [2].
It was suggested that the high prevalence of CKD observed in Africa is partly attributable to the high prevalence of hypertension (HTN) and increasing incidence of diabetes mellitus (DM) [2,3] as a result of urbanization, sedentary lifestyles, and longevity. DM, which is the leading cause of end-stage renal disease (ESRD), accounts for approximately 11% to 83.7% of CKD cases in Africa [4], depending on the method of diagnosis used for CKD. This is concerning as, according to the International Diabetes Federation (IDF), of the approximate 9.3% (463 million) of people with DM, 50% are unaware of their condition and the highest proportions of the undiagnosed DM population are found in the African region (59.7%) [5]. HTN, which is another independent modifiable risk factor of CKD development and progression to ESRD [6], is strongly associated with cardiovascular disease (CVD) and a leading cause of premature death [7]. Among the leading risk factors of CKD, HTN is the most prevalent, affecting approximately 31% (1.39 billion) of the adult population globally, with the highest proportions observed in the low- and middle-income countries [8]. A high prevalence of 57% was reported in the older adults of Africa [9].
The high incidences of CKD due to DM and HTN will result in significant social and economic ramifications in Africa in particular due to limited and inadequate health resources. Early identification of CKD will enable the early initiation of risk reducing therapies, which may subsequently prevent or delay progression to advanced CVDs, complications, or ESRD that require costly renal replacement therapy (RRT) [10]. However, due to its asymptomatic nature in the early stages, CKD is frequently diagnosed only during the advanced stages of the disease when prevention interventions are less likely to be effective. Therefore, screening individuals at high-risk of developing CKD, particularly those with HTN and DM, may prevent or halt the development or progression of CKD.
The expression profiles of microRNAs (miRNAs) in biofluids have been associated with diseases such as cancer [11], neurodegenerative disease [12], and CVDs [13], therefore suggesting the potential utility of these miRNAs as minimally invasive biomarkers for disease diagnosis, monitoring, and as therapeutic targets. As a family of small non-coding transcripts that regulate gene expression post-transcriptionally, miRNAs inhibit mRNA translation or trigger mRNA degradation, thereby preventing the synthesis of certain proteins [14]. They were shown to have important regulatory functions, such as proliferation, cell differentiation, development, apoptosis, and metabolism [15]. Therefore, dysregulation of miRNAs may result in impaired cellular function and disease development [16]. In addition to their intracellular function, studies showed that miRNAs are also secreted by cells into the blood and other body fluids, although the mechanism remains unclear [17]. Unlike RNA, miRNAs in blood and other body fluids are very stable, as they are released into circulation bound to proteins or encapsulated in microvesicles, thereby protecting them from degradation by ribonucleases [17,18].
Several studies showed that the expression of certain miRNAs were tissue specific and may be involved in the development, homeostasis, and physiology of the kidney [14,19]. Although it has been demonstrated that miRNAs exhibit functional dysregulation in various diseases, including CKD [19], HTN [20], DM [21], as well as HTN and DM-related kidney diseases [22,23], minimal evidence exists on the predictive ability of these miRNAs in relation to CKD, none of which is in sub-Saharan Africa. We previously identified novel whole blood miRNAs via deep sequencing, which were significantly dysregulated in HTN (hsa-miR-novel-chr1_36178 and hsa-miR-novel-chr15_18383) [24], pre-diabetes (hsa-miR-novel-chr2_55842) [25], and DM (hsa-miR-novel-chr7_76196, hsa-miR-novel-chr5_67265, and hsa-miR-novel-chr13_13519) (unpublished data). The current study aimed to: (1) evaluate the expression profile of these miRNAs in high-risk individuals (HTN and/or DM) with and without CKD; (2) determine the diagnostic ability of these six novel whole blood miRNAs to discriminate between individuals with CKD and those without; and (3) determine whether these miRNAs offer additional diagnostic advantage above and beyond conventional CKD risk factors.
2. Materials and Methods
2.1. Study Design and Procedures
The current study, which is based on data collected between 2014 and 2016, forms part of the ongoing Vascular and Metabolic Health (VMH) study, an extension of the Cape Town Bellville South study, previously described in detail [26]. Ethical clearance for the VMH study was granted by the research ethics committees of the Cape Peninsula University of Technology (CPUT) and Stellenbosch University (NHREC: REC—230, 408–014, and N14/01/003, respectively). For the current study, ethical clearance was granted by the Faculty of Health and Wellness Sciences Research Ethics Committee of the CPUT (CPUT/HW-REC 2020/H11). The participants gave signed and written consent after they were informed about their rights and the procedures were fully explained in the language of their choice. Research was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki).
A total of 1989 individuals of mixed-ancestry were recruited between 2014 and 2016 as part of the VMH Study and of these 911 individuals had HTN and DM, with age range between 20 and 91, a median age of 56 years and were thus selected for the present analysis (18.8% with CKD and 22 % males). Of the total sample 6.4%, 3.6%, 7.1%, 1.2% and 0.4% of the individuals were in CKD stages 1–5 respectively.
The detailed data collection procedures, using standardized questionnaires and physical examination, were explained elsewhere [25]. Briefly, clinical measurements, including body weight, height, hip circumference (HC), waist circumference (WC), and blood pressure (BP), were taken by trained personnel via standardized methods. Body mass index (BMI) was calculated as weight in kilograms divided by the square of height in meters (kg/m2). Participants were classified as underweight (BMI < 18.5 kg/m2), normal weight (BMI 18.5–4.9 kg/m2), overweight (BMI 25.0–29.99 kg/m2), and obese (BMI ≥ 30.0 kg/m2). BP measurements were performed according to WHO guidelines [27], using an automatic digital BP monitor (Omron M6 Comfort-preformed Cuff Blood Pressure Monitor, Omron), with the participants sitting quietly in a relaxed position for at least 5 min. Two BP readings, including systolic BP (SBP) and diastolic BP (DBP), at three-minute intervals, were taken and the lowest SBP and the corresponding DBP were chosen as the participant’s BP. Pulse pressure (PP) was estimated by subtracting the DBP from the SBP. HTN was defined as SBP ≥ 140 mmHg and/or DBP ≥ 90 mmHg or self-reported ongoing use of antihypertensive medications.
All biochemical analysis was conducted by an ISO accredited pathology practice (PathCare Laboratory, Cape Town, South Africa). All participants, excluding those who self-reported diabetes mellitus (confirmed by either participant medical card record or use of diabetic medication), underwent a 75 g oral glucose tolerance test (OGTT) after an overnight fast, according to WHO guidelines [28]. DM was defined as a fasting blood glucose (FBG) of ≥7.0 mmol/L and/or a 2 h postprandial glucose (Glucose 2 HR) of ≥11.1 mmol/L, or ongoing use of glucose control medications. Plasma glucose concentrations were measured using the enzymatic hexokinase method (Beckman AU, Beckman Coulter, Brea, CA, USA), glycated hemoglobin (HbA1c) was measured using high performance liquid chromatography (HPLC) (Biorad Variant Turbo, BioRad, Hercules, CA, USA), urine albumin levels were measured using the colorimetric (bromocresol purple) method (Beckman AU, Beckman Coulter, Brea, CA, USA), and serum and urinary creatinine was measured by the modified Jaffe kinetic method (Beckman AU, Beckman Coulter, Brea, CA, USA). For serum samples, fasting and 2-h blood samples were collected in a plain tube (with no clotting factors) and this was centrifuged at 2500 RPM for 15 min using a Beckman General Purpose centrifuge (Beckman Coulter Inc., CA, USA) to obtain serum and for urine samples, the urine was collected in specimen containers supplied by the laboratory and all samples were transported daily in an ice-box for processing at pathology practice (PathCare Laboratory, Cape Town, South Africa). The level of kidney function was measured using estimated glomerular filtration rate (eGFR) which was calculated using the 4-variable Modification of Diet in Renal Disease (MDRD) (eGFR = 175 × (SCr)−1.154 × (age)−0.203 × 0.742 [if female]) [29], and Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equations (eGFR = 141 × min(SCr/κ, 1)α × max(SCr /κ, 1)−1.209 × 0.993Age × 1.018 [if female], where SCr was defined as serum creatinine, κ was 0.7 (females) or 0.9 (males), α was −0.329 (females) or −0.411 (males), min indicated the minimum of SCr/κ or 1 and max indicated the maximum of SCr/κ or 1. The correction factor for African American ethnicity was not included as recommended for our study population [30] Only the MDRD data are shown, as similar results were obtained for both equations. CKD was defined as an eGFR of <90 mL/min/1.73 m2 and/or albumin-to-creatinine ratio (ACR) >3 mg/mmol as recommended by the current Kidney Disease: Improving Global Outcomes (KDIGO) guidelines [29]. Participants were classified a “current smoker”, if serum cotinine levels were >15 ng/mL [31] and a “drinker” if they self-reported the consumption of alcohol.
2.2. MicroRNA Analysis
The detailed methodology of whole blood miRNA extraction and analysis was described previously [25]. Briefly, whole blood samples were collected in Tempus RNA tubes and stored at −20 degrees for circulating miRNA extraction and analysis. The samples were sent to Arraystar Inc (Rockville, USA) for small RNA library construction, deep sequencing, and data processing. The identified miRNAs that showed dysregulation between the groups were selected for validation, in a larger study sample, by quantitative reverse transcription polymerase chain reaction (RT-qPCR) to assess the reproducibility of the results. Following isolation, using the MagMAXTM for Stabilized Blood Tubes RNA Isolation Kit (Life Technologies, Waltham, MA, USA), total RNA was reverse transcribed into complementary DNA (cDNA) using the TaqManTM Advanced cDNA Synthesis Kit, and in accordance with the manufacturer’s specifications (Applied Biosystems, Waltham, MA, USA, 2015). Following reverse transcription, the quantification of miRNA expression was then performed using the TaqManTM miRNA assay protocol, as per the manufacturer’s instructions, with a Quantum Studio 7 (Life Technologies, Carlsbad, CA, USA). The delta Ct (2−ΔCt) method was used to assess the miRNA expression level in each sample by normalization to endogenous control (miR-16-5p) expression levels. The relative miRNA expression between samples was calculated using the delta-delta Ct (2−ΔΔCT) method [32].
2.3. Statistical Analysis
The Shapiro-Wilk W test was used to check the data distribution. Due to the non-Gaussian distribution of most variables, the general participant characteristics were presented as median (25–75th percentiles) or count and percentages. Wilcoxon rank-sum tests (continuous variables) and chi-square tests (categorical variables) were used for comparisons between individuals with CKD and those without CKD. Robust linear regression models, unadjusted and adjusted for age, gender, smoking status, drinking status, HTN, and DM status were used to assess the association between whole blood miRNAs and eGFR. The models used were as follows: Model 1: Crude; Model 2: Model 1 + age + gender; Model 3: Model 2 + smoking status + drinking status; Model 4: Model 3 + HTN status + DM status. Logistic regression models with a similar level of adjustment were used to analyze the ability of whole blood miRNAs to predict prevalent CKD. The area under the receiver operating characteristics (ROC) curve (AUC) was used to determine the discriminatory power of each miRNA (hsa-miR-novel-chr1_36178, hsa-miR-novel-chr15_18383, hsa-miR-novel-chr2_55842, hsa-miR-novel-chr7_76196, hsa-miR-novel-chr5_67265, and hsa-miR-novel-chr13_13519), alone and as a collective, and alongside conventional risk factors to distinguish participants with CKD from those without. Statistical significance was defined as a p-value of <0.05.
3. Results
3.1. General Characteristics of the Study Population
A total of 1989 individuals of mixed-ancestry were recruited, between 2014 and 2016, as part of the VMH study. Of these, 911 individuals had HTN and DM, and were thus selected for the present analysis (18.8% with CKD and 22 % males). The clinical characteristics of the study participants by CKD status are summarized in Table 1. Of the total sample 6.4%, 3.6%, 7.1%, 1.2%, and 0.4% of the individuals were in CKD stages 1–5, respectively. Individuals with CKD were significantly older (63 vs. 55 years, p < 0.0001), had a larger waist circumference (98.5 vs. 94.4 cm, p = 0.023), higher fasting plasma glucose (5.3 vs. 5.2, p = 0.0026), 2 h glucose (7.1 vs. 6.4, p = 0.0047), HbA1c (6.2 vs. 5.9, p < 0.0001), 2 h insulin (51.5 vs. 41.8, p = 0.032), SBP levels (149 vs. 146, 0.047), and a higher proportion of DM (45 vs. 27%, p < 0.001) compared to those without CKD. Conversely, participants without CKD had a higher proportion of smokers (47.7 vs. 31.2%) and alcohol consumers (28.5 vs. 7.6%) than those with CKD (p < 0.0001 for both).

Table 1.
General characteristics of the study participants at risk of developing CKD, categorized by CKD status.
3.2. Relative Expression Levels of Whole Blood miRNAs
The relative expression levels of the six novel whole blood miRNAs are shown in Figure 1A–F. The expression levels of whole blood miRNAs (hsa-miR-novel-chr1_36178, hsa-miR-novel-chr2_55842, hsa-miR-novel-chr7_76196, hsa-miR-novel-chr5_67265, and hsa-miR-novel-chr13_13519) were significantly higher in individuals with CKD as compared to those without CKD (all p < 0.028), whereas the expression of hsa-miR-novel-chr15_18383 showed no differences between the two groups (p = 0.197).
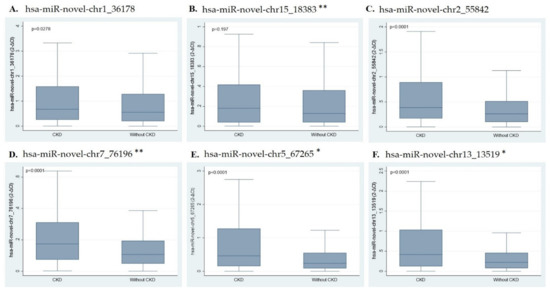
Figure 1.
Box and whisker plots showing miRNA expression. (A) hsa-miR-novel-chr1_36178, (B) hsa-miR-novel-chr15_18383 **, (C) hsa-miR-novel-chr2_55842, (D) hsa-miR-novel-chr7_76196 **, (E) hsa-miR-novel-chr5_67265 *, and (F) hsa-miR-novel-chr13_13519 *. * Represent miRNAs factored by 10 and ** represent miRNAs factored by 100 as the values were very low. Abbreviations: CKD (chronic kidney disease); miR (microRNA).
3.3. Relationship between Whole Blood miRNAs, eGFR, and Prevalent CKD
In the robust linear regression models (Table 2), the increased expression of blood miRNAs (hsa-miR-novel-chr2_55842 and hsa-miR-novel-chr7_76196) were significantly associated with reduced eGFR, independent of age, gender, smoking status, drinking status, DM, and HTN status (Models 1–5, p < 0.038 for all). Blood miRNAs hsa-miR-novel-chr5_67265 and hsa-miR-novel-chr13_13519 showed significant association with lower eGFR levels, independent of age and gender (Models 1–2, all p ≤ 0.05), but not after further adjustment for smoking status, alcohol consumption, DM, and HTN status (Models 3–4, p < 0.062). The miRNAs hsa-miR-novel-chr1_36178 and hsa-miR-novel-chr15_18383 showed no association with eGFR (p ≥ 0.311 for all). Table 3 presents the odds ratios (ORs) with 95% confidence intervals (CIs) from logistic regression analysis of whole blood miRNAs for the prediction of CKD. All the whole blood miRNAs were positively associated with CKD in people with HTN and/or DM, even after adjustment for a range of confounders (p ≤ 0.038, for all).

Table 2.
Robust linear regression models for the association between whole blood miRNAs and eGFR.

Table 3.
Logistic regression analysis of blood miRNAs for prediction of CKD.
3.4. Diagnostic Value of Whole Blood miRNAs to Predict CKD
Figure 2 presents the ROC curves for the discriminatory ability of whole blood miRNAs to identify people with CKD. All the whole blood miRNAs had acceptable discriminatory power for prevalent CKD in people with HPT and/or DM (all AUC > 0.7), however AUC comparison showed no significant difference between miRNAs in predicting prevalent CKD (p = 0.202). Moreover, when comparing the standard model (including only age, gender, and smoking, drinking, HTN, and DM status) with the models containing the miRNAs, Figure 3 only the model with hsa-miR-novel-chr13_13519 was significantly different to the standard model (p = 0.0397).
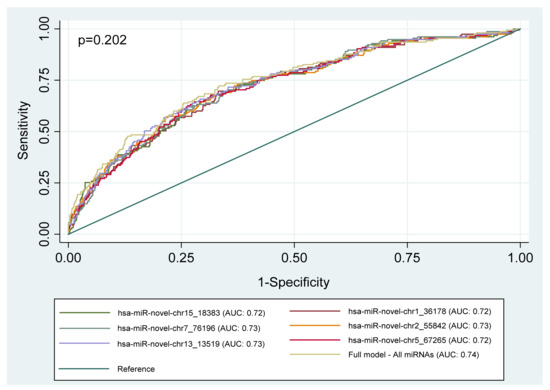
Figure 2.
Area under the receiver operating characteristics (ROC) curves (AUC), illustrating the diagnostic ability of six novel whole blood miRNAs to discriminate between individuals with CKD and those without in a group of high-risk individuals. Models: miRNA + age + gender + smoking status + drinking status + DM status + HTN status. Full model includes all six miRNAs. Abbreviation: miRNA (microRNA).
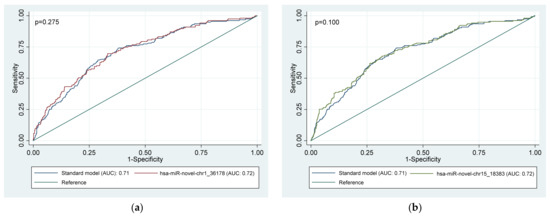
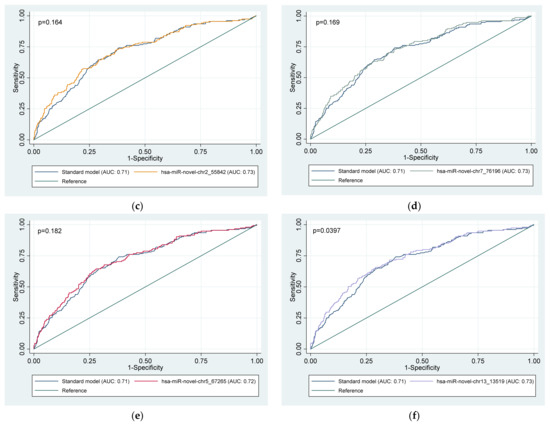
Figure 3.
Area under the receiver operator characteristic (ROC) curves (AUC), illustrating the diagnostic advantage of six (a–f) novel whole blood miRNAs to predict CKD in a group of high-risk individuals above that of conventional risk factors. Standard model: age + gender + smoking status + drinking status + diabetes status + hypertension status. Model for each miRNA separately: miRNA + age + gender + smoking status + drinking status + diabetes status + hypertension status. miRNA, microRNA.
4. Discussion
The key findings of this study are that all six novel whole blood miRNAs were significantly associated with prevalent CKD, independent of conventional risk factors. The expression profile of miRNAs (hsa-miR-novel-chr1_36178, hsa-miR-novel-chr2_55842, hsa-miR-novel-chr7_76196, hsa-miR-novel-chr5_67265, and hsa-miR-novel-chr13_13519) were significantly higher in CKD individuals as compared to those with normal kidney function. While all the miRNAs had acceptable and comparable discriminatory power for prevalent CKD, only hsa-miR-novel-chr13_13519 added to predictions beyond conventional risk factors. The findings of the current investigation suggest that these novel whole blood miRNAs have a potential to contribute to CKD risk screening in people with HTN and/or DM.
Various studies have reported on the dysregulated patterns of miRNAs in CKD in individuals with HTN and DM. Kato et al. were among the first to demonstrate the role of miRNAs in the development of diabetic kidney disease (DKD) [21,33]. They showed that the expression of miR-192 was significantly elevated in the glomeruli of a diabetic mouse model [21] and, in a further study, showed that miR-192 induced the transforming growth factor beta 1 (TGF-𝛽1) signaling, accelerating the progression of DKD [33]. Another study explored the expression profile and clinical significance of plasma miRNAs in Chinese individuals with DKD and found that the expression profiles of miR-150-5p, miR-155-5p, miR-30e, miR-320e, and miR-3196 were significantly decreased during the early stages of DKD [23]. A study performed in Egyptians with type 2 DM showed that levels of miR-451 were elevated and reduced in plasma and urine samples, respectively, in different stages of DKD [34]. Studies by Liu et al. demonstrated that downregulation of miR-214-3p may be associated with the development of chronic kidney injury in HTN, and showed that upregulation of this miRNA may offer a protective role in the kidneys [35,36]. A study by Lu et al. showed that serum- and urine-derived miR-103a-3p were significantly higher in individuals with hypertensive nephropathy and hypertensive mice infused with angiotensin II hormone compared to normal controls. The authors found that this hormone-induced kidney injury via the activation of the SNRK /NF-κB/p65 signaling pathway and was positively associated with increased levels of miR-103a-3p [37]. Berillo et al. recently examined the expression profile of let-7g-5p and miR-191-5p in platelet-poor plasma and found that the decreased levels of these miRNAs were independently associated with CKD among individuals with HTN. This, therefore, suggested that let-7g-5p and miR-191-5p may be involved in the pathophysiology of CKD and may serve as potential biomarkers for disease diagnosis [38].
In the current study, we observed that increased expression of hsa-miR-novel-chr2_55842 and hsa-miR-novel-chr7_76196 were independently associated with reduced eGFR, similar to other studies that reported a positive association with eGFR, albeit with other miRNAs [23,38]. However, the association between hsa-miR-novel-chr5_67265 and hsa-miR-novel-chr13_13519 and eGFR was influenced by smoking. Previous studies, like the one by Yokoyama et al., likewise found that exposure to cigarette smoke mediated the regulation of certain miRNAs. In their case, miR-155 and miR-21 were upregulated and the expression of miR-126-3p was downregulated [39]. Moreover, the current study showed that all six novel miRNAs were positively associated with prevalent CKD in individuals with HTN and/or DM, independent of conventional risk factors, like age and gender. Furthermore, all six novel miRNAs had an acceptable ability to predict CKD (AUC > 0.7). However, the prediction model containing hsa-miR-novel-chr13_13519 offered an additional advantage in predicting CKD, above that of conventional risk factors (age, gender. smoking status, drink status, DM, and HPT status). These findings demonstrate that, although the studied miRNAs are all acceptable predictors of CKD, only hsa-miR-novel-chr13_13519 seems to offer an additional advantage.
Contrary to our study, previous studies, which explored the predictive value of miRNAs analyzed for CKD in individuals with HTN or DM, quantified their expression profile in plasma [23,34], platelet poor plasma [38], serum [37], and urine [34,37], while in the current study we used whole blood. As reviewed by Witwer in 2015 [40], the establishment of an accurate and reliable circulating miRNA biomarker for disease has proven to be quite challenging as it may be affected by pre-analytical factors, such as the starting material of biofluid, processing, and the type of normalization miRNA used. Although platelet-poor plasma is not widely biased by coagulation due to a lack of platelets [41], similarly to plasma and serum, the concentration of total miRNA is reduced after extraction [42] and may be affected by pre-analytical processes, such as sample handling and bias due hemolysis [13,41]. Urine samples also present with some shortcomings, in particular with regard to the use of normalization control, with some studies showing that the inclusion of a normalization control might reduce the predictive value of urine miRNAs, whereas the exclusion of it might affect the accuracy of the results [43]. In the current study, we analyzed miRNA expression in whole blood samples, comprised of multiple different blood cell types with their own specific miRNA expression profile, which might have contributed to the profile of miRNAs observed. Individuals with CKD generally have lower levels of red blood cell (RBC) count, due to anemia, compared to their control counterparts [44]. It was reported that RBC-derived miRNAs constitute the majority of miRNAs expressed in whole blood [45]. However, we found high levels of miRNA expression in CKD individuals supposedly with low RBC count, therefore it is likely that our novel miRNAs are not highly expressed in RBCs and our results were not affected. Furthermore, Keller et al. performed a statistical evaluation on the effect of different blood cell counts on a miRNA expression profile for different human diseases to test for disease-specific alterations. The authors found that different blood cell counts only partly affected the profile of miRNAs and that they did not significantly affect the feasibility of associating a miRNA profile and human disease, therefore supporting the use of whole blood for miRNA profile analysis as the basis for the detection of disease [46]. Moreover, whole blood has a high concentration of total miRNA after extraction and is not affected by pre-sample analysis and cell lysis [47].
The current study had some limitations, which need to be taken into consideration when interpreting the findings. The cross-sectional design of the study limited the exploration of a causal relationship between the whole blood miRNAs and CKD, therefore future longitudinal analysis is recommended to elucidate the causal relationship between studied miRNAs and the pathogenesis of CKD in individuals with HTN and/or DM. Although the present study had a large study sample, we had a small number of individuals with CKD in various stages of the disease, thereby not allowing for the exploration of miRNA profiles over the various stages of CKD. CKD was diagnosed based on eGFR estimated from a single time point creatinine measurement, which is not ideal. However, we included levels of ACR, which are important for interpretation when eGFR levels are above 60 mL/min/1.73 m2 as recommended by the KDIGO guidelines [29]. HTN was diagnosed with BP measurements taken on one visit, although the International Society of Hypertension recommends that diagnosis should be made based on measurements taken at two or more visits separated by a period of one week [48]. Moreover, we did not exclude individuals on antihypertensive/antidiabetic medication and take into consideration the duration of the disease, therefore we cannot exclude the possibility that these might have influenced the findings of this study. However, we adjusted for some of the common risk factors associated with CKD, as well as miRNAs, thereby eliminating their confounding bias. Moreover, the fact that we explored the differentially expressed miRNAs that were identified in our population and evaluated their potential role as screening tools for CKD, especially in high-risk individuals with HTN and or DM considering their reported high incidences, serves as a strength for this study. Our findings form a basis to the potential pathophysiological importance of whole blood miRNAs in CKD.
5. Conclusions
Taken together, the findings of the present study demonstrate that hsa-miR-novel-chr13_13519 that was positively associated with CKD prevalent, may be used as a potential screening tool for CKD risk screening particularly in people with HTN and DM for whom early initiation of treatment may prevent the onset or halt the progression of CKD. However, these miRNAs are still novel, and they have only been reported in our study population. Future studies are, therefore, necessary to validate our findings in a large study sample and, most importantly, explore the origin of these miRNAs, elucidate their physiological roles, and target genes and pathways.
Author Contributions
Conceptualization, R.T.E., A.P.K. and T.E.M.; Data curation, D.M.M., C.J.W. and S.F.G.D.; Formal analysis, D.D.M., C.G. and S.F.G.D.; Funding acquisition, R.T.E., A.P.K. and T.E.M.; Investigation, D.D.M., D.M.M. and C.J.W.; Methodology, D.D.M., D.M.M. and C.J.W.; Project administration, R.T.E., A.P.K. and T.E.M.; Resources, T.E.M.; Software, C.G.; Supervision, C.G. and T.E.M.; Validation, C.G.; Visualization, D.D.M.; Writing—original draft, D.D.M.; Writing—review and editing, D.D.M., C.G., D.M.M., C.J.W., R.T.E., A.P.K. and T.E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by T.E. Matsha, Cape Peninsula University of Technology, via funding from the South African Medical Research Council (MRC), with funds from the National Treasury under its Economic Competitiveness and Support Package [MRC-RFA-UFSP-01-2013/VMH Study].
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the CPUT Faculty of Health and Wellness Sciences Research Ethics Committee, reference number (CPUT/HW-REC 2020/H11).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The dataset used for this study is available from the corresponding author upon reasonable request.
Acknowledgments
The authors would like to acknowledge the population of Bellville South (Ward 009) community for participation in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bikbov, B.B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Adebayo, O.M.; Afarideh, M.; Agarwal, S.K.; Agudelo-Botero, M.; et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef]
- Kaze, A.D.; Ilori, T.; Jaar, B.G.; Echouffo-Tcheugui, J.B. Burden of chronic kidney disease on the African continent: A systematic review and meta-analysis. BMC Nephrol. 2018, 19, 125. [Google Scholar] [CrossRef]
- George, J.A.; Brandenburg, J.T.; Fabian, J.; Crowther, N.J.; Agongo, G.; Alberts, M.; Ali, S.; Asiki, G.; Boua, P.R.; Gómez-Olivé, F.X.; et al. Kidney damage and associated risk factors in rural and urban sub-Saharan Africa (AWI-Gen): A cross-sectional population study. Lancet Glob. Health 2019, 7, 1632–1643. [Google Scholar] [CrossRef]
- Noubiap, J.J.N. Diabetic nephropathy in Africa: A systematic review. World J. Diabetes 2015, 6, 759. [Google Scholar] [CrossRef]
- Internation Diabetes Foundation. IDF Diabetes Atlas, 9th ed.; Internation Diabetes Foundation: Brussels, Belgium, 2019; Available online: https://diabetesatlas.org/en/ (accessed on 10 August 2021).
- Anderson, A.H.; Yang, W.; Townsend, R.R.; Pan, Q.; Chertow, G.M.; Kusek, J.W.; Charleston, J.; He, J.; Kallem, R.; Lash, J.P.; et al. Time-updated systolic blood pressure and the progression of chronic kidney disease: A cohort study. Ann. Intern. Med. 2015, 162, 258–265. [Google Scholar] [CrossRef]
- Mills, K.T.; Stefanescu, A.; He, J. The global epidemiology of hypertension. Nat. Rev. Nephrol. 2020, 16, 223–237. [Google Scholar] [CrossRef]
- Mills, K.T.; Bundy, J.D.; Kelly, T.N.; Reed, J.E.; Kearney, P.M.; Reynolds, K.; Chen, J.; He, J. Global disparities of hypertension prevalence and control. Circulation 2016, 134, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Bosu, W.K.; Reilly, S.T.; Aheto, J.M.K.; Zucchelli, E. Hypertension in older adults in Africa: A systematic review and meta-analysis. PLoS ONE 2019, 14, e0214934. [Google Scholar] [CrossRef]
- US Renal Data System. USRDS 2013 Annual Data Report. In Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States; National Institutes of Health, National Institute of Diabetes and Digestiveand Kidney Diseases: Bethesda, MD, USA, 2013; Volume 2. [Google Scholar]
- Yuan, H.L.; Wang, T.; Zhang, K.H. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. OncoTargets Ther. 2018, 11, 3891. [Google Scholar] [CrossRef]
- Danborg, P.B.; Simonsen, A.H.; Waldemar, G.; Heegaard, N.H.H. The potential of microRNAs as biofluid markers of neurodegenerative diseases—A systematic review. Biomarkers 2014, 19, 259–268. [Google Scholar] [CrossRef]
- Felekkis, K.; Papaneophytou, C. Challenges in using circulating micro-RNAs as biomarkers for cardiovascular diseases. Int. J. Mol. Sci. 2020, 21, 561. [Google Scholar] [CrossRef]
- Bhatt, K.; Mi, Q.S.; Dong, Z. MicroRNAs in kidneys: Biogenesis, regulation, and pathophysiological roles. Am. J. Physiol. Ren. Physiol. 2011, 300, 602–610. [Google Scholar] [CrossRef]
- Brodersen, P.; Voinnet, O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009, 10, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, M. Differential expression of microRNAs in different disease states. Circ. Res. 2012, 110, 638–650. [Google Scholar] [CrossRef]
- Keller, S.; Ridinger, J.; Rupp, A.K.; Janssen, J.W.G.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, K.; Karolina, D.S.; Sepramaniam, S.; Armugam, A.; Wintour, E.M.; Bertram, J.F.; Jeyaseelan, K. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012, 81, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.J.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef]
- Kato, M.; Zhang, J.; Wang, M.; Lanting, L.; Yuan, H.; Rossi, J.J.; Natarajan, R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-β-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 2007, 104, 3432–3437. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kwan, B.C.H.; Lai, F.M.M.; Choi, P.C.L.; Chow, K.M.; Li, P.K.T.; Szeto, C.C. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am. J. Hypertens. 2010, 23, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, G.; Liang, Y.; Zhou, X. Expression profiling and clinical significance of plasma microRNAs in diabetic nephropathy. J. Diabetes. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Matshazi, D.M.; Weale, C.J.; Erasmus, R.T.; Kengne, A.P.; Davids, S.F.; Raghubeer, S.; Davison, G.M.; Matsha, T.E. Two novel microRNAs and their association with absolute blood pressure parameters in an urban South African community. Mol. Biol. Rep. 2021, 48, 2553–2560. [Google Scholar] [CrossRef] [PubMed]
- Matsha, T.E.; Kengne, A.P.; Hector, S.; Mbu, D.L.; Yako, Y.Y.; Erasmus, R.T. MicroRNA profiling and their pathways in South African individuals with prediabetes and newly diagnosed type 2 diabetes mellitus. Oncotarget 2018, 9, 30485–30498. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Erasmus, R.T.; Soita, D.J.; Hassan, M.S.; Blanco-Blanco, E.; Vergotine, Z.; Kengne, A.P.; Matsha, T.E. High prevalence of diabetes mellitus and metabolic syndrome in a South African coloured population: Baseline data of a study in Bellville, Cape Town. S. Afr. Med. J. 2012, 102, 841–844. [Google Scholar] [CrossRef]
- Chalmers, J.O.H.N.; MacMahon, S.; Mancia, G.; Whitworth, J.; Beilin, L.; Hansson, L.; Neal, B.; Rodgers, A.; Mhurchu, N.; Clark, T. World Health Organization-International Society of Hypertension Guidelines for the management of hypertension. Guidelines sub-committee of the World Health Organization. Clin. Exp. Hypertens. 1999, 21, 1009–1060. [Google Scholar] [PubMed]
- Alberti, K.G.M.M.; Zimmet, P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus. Provisional report of a WHO consultation. Diabet Med. 1998, 15, 539–553. [Google Scholar] [CrossRef]
- Levin, A.; Stevens, P.E.; Bilous, R.W.; Coresh, J.; De Francisco, A.L.; De Jong, P.E.; Griffith, K.E.; Hemmelgarn, B.R.; Iseki, K.; Lamb, E.J.; et al. Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Suppl. 2013, 3, 1–150. [Google Scholar]
- Holness, J.L.; Bezuidenhout, K.; Davids, M.R.; Warwick, J.M. Validation of equations to estimate glomerular filtration rate in South Africans of mixed ancestry. S. Afr. Med. J. 2020, 110, 229–234. [Google Scholar] [CrossRef]
- Pirkle, J.L.; Flegal, K.M.; Bernert, J.T.; Brody, D.J.; Etzel, R.A.; Maurer, K.R. Exposure of the US population to environmental tobacco smoke: The Third National Health and Nutrition Examination Survey, 1988 to 1991. JAMA 1996, 275, 1233–1240. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 24, 402–408. [Google Scholar] [CrossRef]
- Kato, M.; Arce, L.; Wang, M.; Putta, S.; Lanting, L.; Natarajan, R. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011, 80, 358–368. [Google Scholar] [CrossRef]
- Abdelsalam, M.; Wahab, A.M.; El Sayed, Z.M.; Motawea, M. MicroRNA-451 as an Early Predictor of Chronic Kidney Disease in Diabetic Nephropathy. Int. J. Nephrol. 2020, 2020, 8075376. [Google Scholar] [CrossRef]
- Liu, Y.; Usa, K.; Wang, F.; Liu, P.; Geurts, A.; Li, J.; Williams, A.M.; Regner, K.R.; Kong, Y.; Liu, H.; et al. MicroRNA-214-3p in the kidney contributes to the development of hypertension. J. Am. Soc. Nephrol. 2018, 29, 2518–2528. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, D.; Wang, F.; Liu, J.; Huang, B.; Baker, M.A.; Yin, J.; Wu, R.; Liu, X.; Regner, K.R.; et al. Endogenous miR-204 protects the kidney against chronic injury in hypertension and diabetes. Am. Soc. Nephrol. 2020, 31, 1539–1554. [Google Scholar] [CrossRef]
- Lu, Q.; Ma, Z.; Ding, Y.; Bedarida, T.; Chen, L.; Xie, Z.; Song, P.; Zou, M.H. Circulating miR-103a-3p contributes to angiotensin II-induced renal inflammation and fibrosis via a SNRK/NF-κB/p65 regulatory axis. Nat. Commun. 2019, 10, 2145. [Google Scholar] [CrossRef]
- Berillo, O.; Huo, K.G.; Fraulob-Aquino, J.C.; Richer, C.; Briet, M.; Boutouyrie, P.; Lipman, M.L.; Sinnett, D.; Paradis, P.; Schiffrin, E.L. Circulating let-7g-5p and miR-191-5p are independent predictors of chronic kidney disease in hypertensive patients. Am. J. Hypertens. 2020, 33, 505–513. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Mise, N.; Suzuki, Y.; Tada-Oikawa, S.; Izuoka, K.; Zhang, L.; Zong, C.; Takai, A.; Yamada, Y.; Ichihara, S. MicroRNAs as potential mediators for cigarette smoking induced atherosclerosis. Int. J. Mol. Sci. 2018, 19, 1097. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rius, A.; Martinez-Perez, A.; López, S.; Sabater-Lleal, M.; Souto, J.C.; Soria, J.M. Expression of microRNAs in human platelet-poor plasma: Analysis of the factors affecting their expression and association with proximal genetic variants. Epigenetics 2020, 15, 1396–1406. [Google Scholar] [CrossRef]
- Sunderland, N.; Skroblin, P.; Barwari, T.; Huntley, R.P.; Lu, R.; Joshi, A.; Lovering, R.C.; Mayr, M. MicroRNA biomarkers and platelet reactivity: The clot thickens. Circ. Res. 2017, 120, 418–435. [Google Scholar] [CrossRef]
- Cochetti, G.; Cari, L.; Nocentini, G.; Maulà, V.; Suvieri, C.; Cagnani, R.; De Vermandois, J.A.R.; Mearini, E. Detection of urinary miRNAs for diagnosis of clear cell renal cell carcinoma. Sci. Rep. 2020, 10, 21290. [Google Scholar] [CrossRef] [PubMed]
- George, C.; Matsha, T.E.; Erasmus, R.T.; Kengne, A.P. Haematological profile of chronic kidney disease in a mixed-ancestry South African population: A cross-sectional study. BMJ Open 2018, 8, e025694. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yu, Y.; Niu, B.; Wang, D. Red Blood cells as potential repositories of microRNAs in the circulatory system. Front. Genet. 2020, 11, 442. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Bauer, A.; ElSharawy, A.; Haas, J.; Backes, C.; Wendschlag, A.; Giese, N.; Tjaden, C.; Ott, K.; et al. Toward the blood-borne miRNome of human diseases. Nat. Methods 2011, 8, 841–843. [Google Scholar] [CrossRef] [PubMed]
- Grasedieck, S.; Sorrentino, A.; Langer, C.; Buske, C.; Döhner, H.; Mertens, D.; Kuchenbauer, F. Circulating microRNAs in hematological diseases: Principles, challenges, and perspectives. Blood 2013, 121, 4977–4984. [Google Scholar] [CrossRef]
- Unger, T.; Borghi, C.; Charchar, F.; Khan, N.A.; Poulter, N.R.; Prabhakaran, D.; Ramirez, A.; Schlaich, M.; Stergiou, G.S.; Tomaszewski, M.; et al. 2020 International Society of Hypertension global hypertension practice guidelines. Hypertension 2020, 75, 1334–1357. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).