Improving a Rapid Alignment Method of Tomography Projections by a Parallel Approach
Abstract
:Featured Application
Abstract
1. Introduction
1.1. Projection Misalignment Problem
1.2. Post-Acquisition Alignment
1.3. Proposed Solution
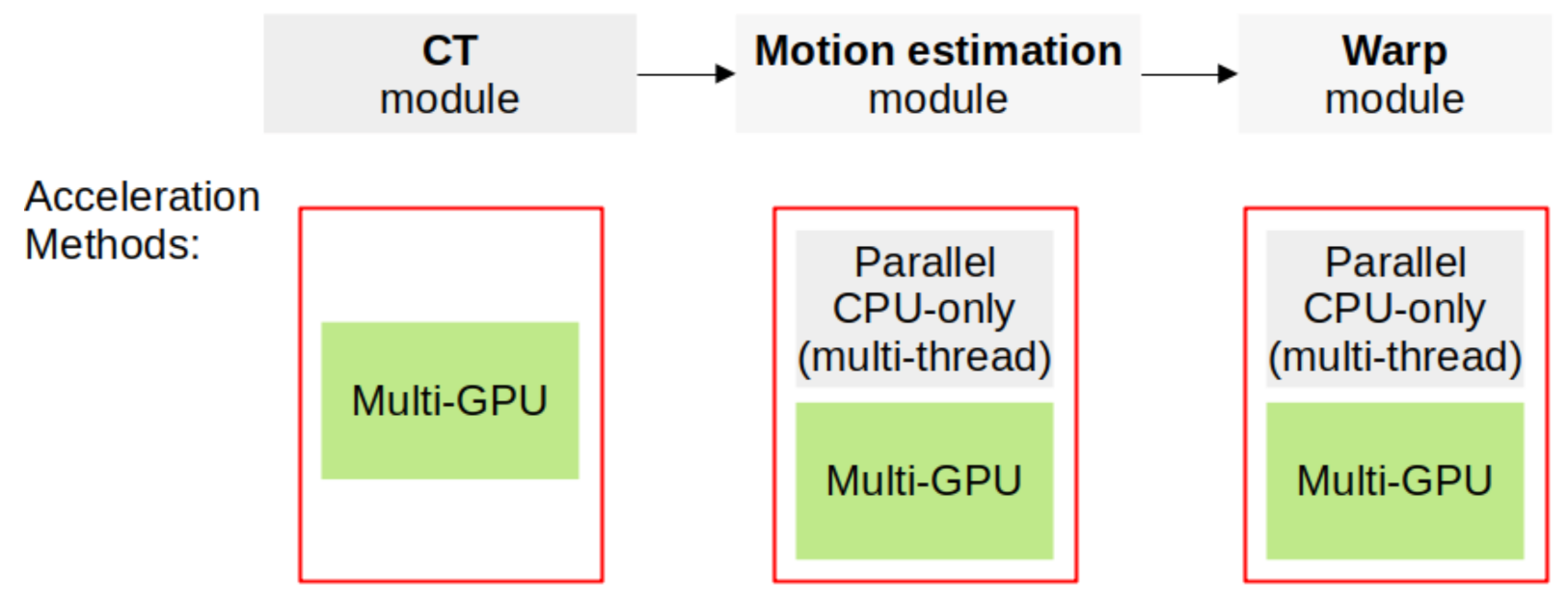
2. Computational Methods
2.1. Joint Reconstruction-Reprojection Method
2.2. Implementation Details
2.3. Tomography Module
2.4. Motion Estimation Module
2.5. Warp Module
3. Results and Discussion
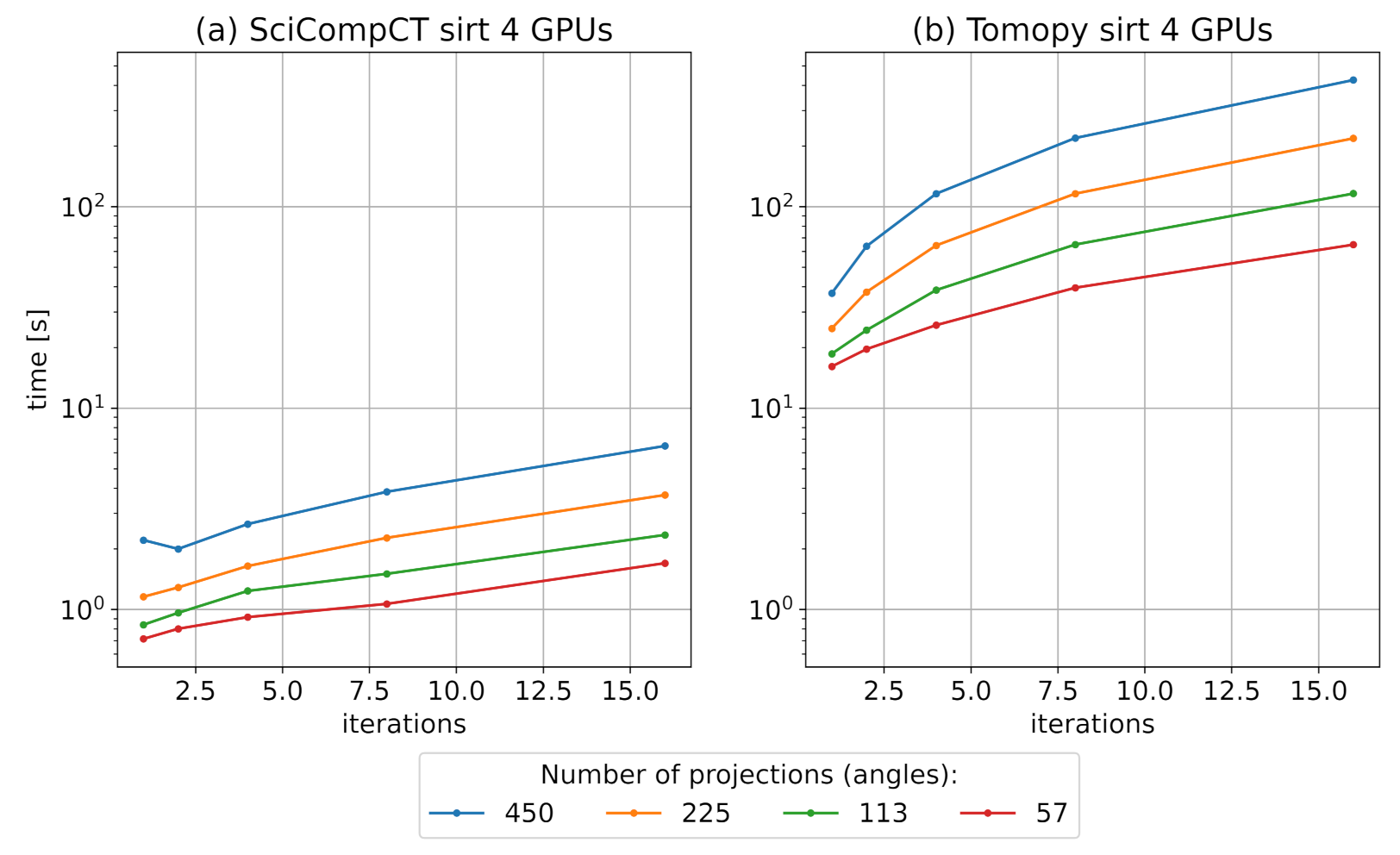
3.1. CT Module Benchmark
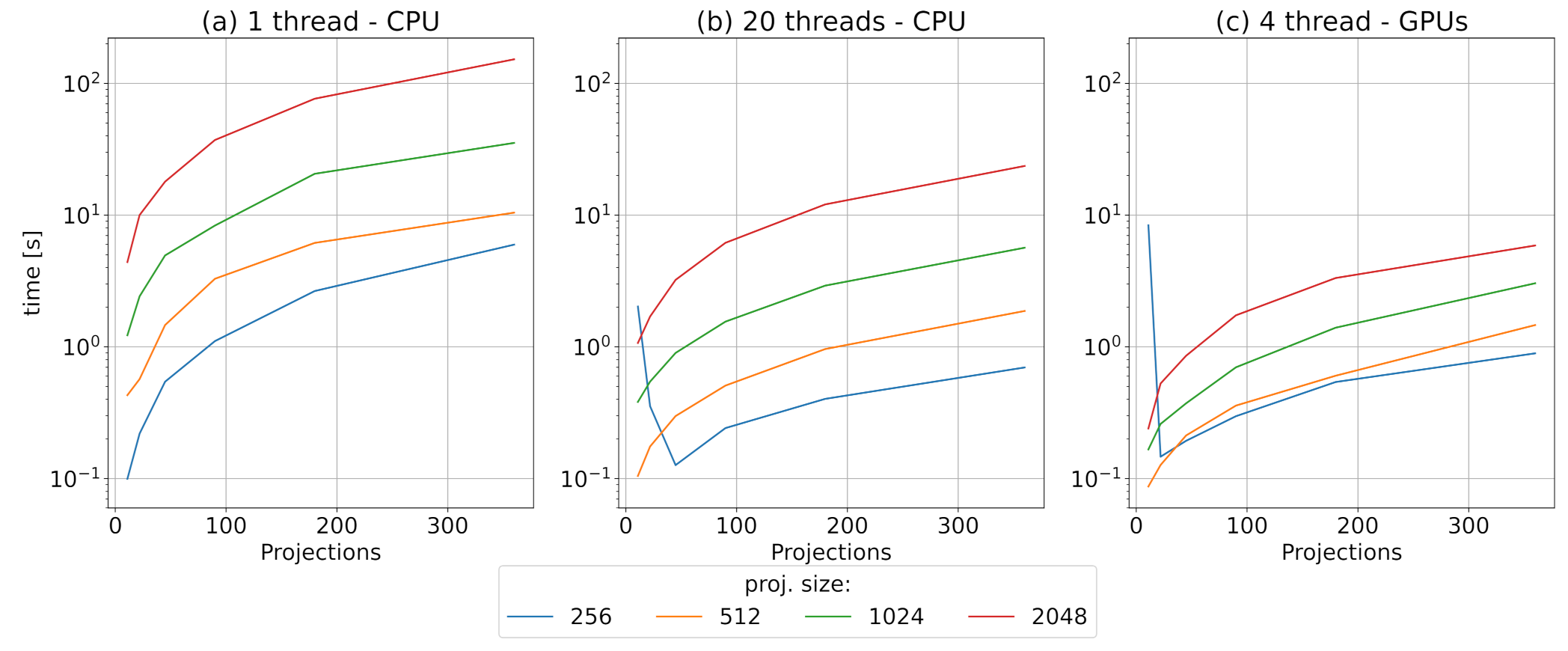
3.2. Motion Estimation Module Benchmark
3.3. Warp Module Benchmark
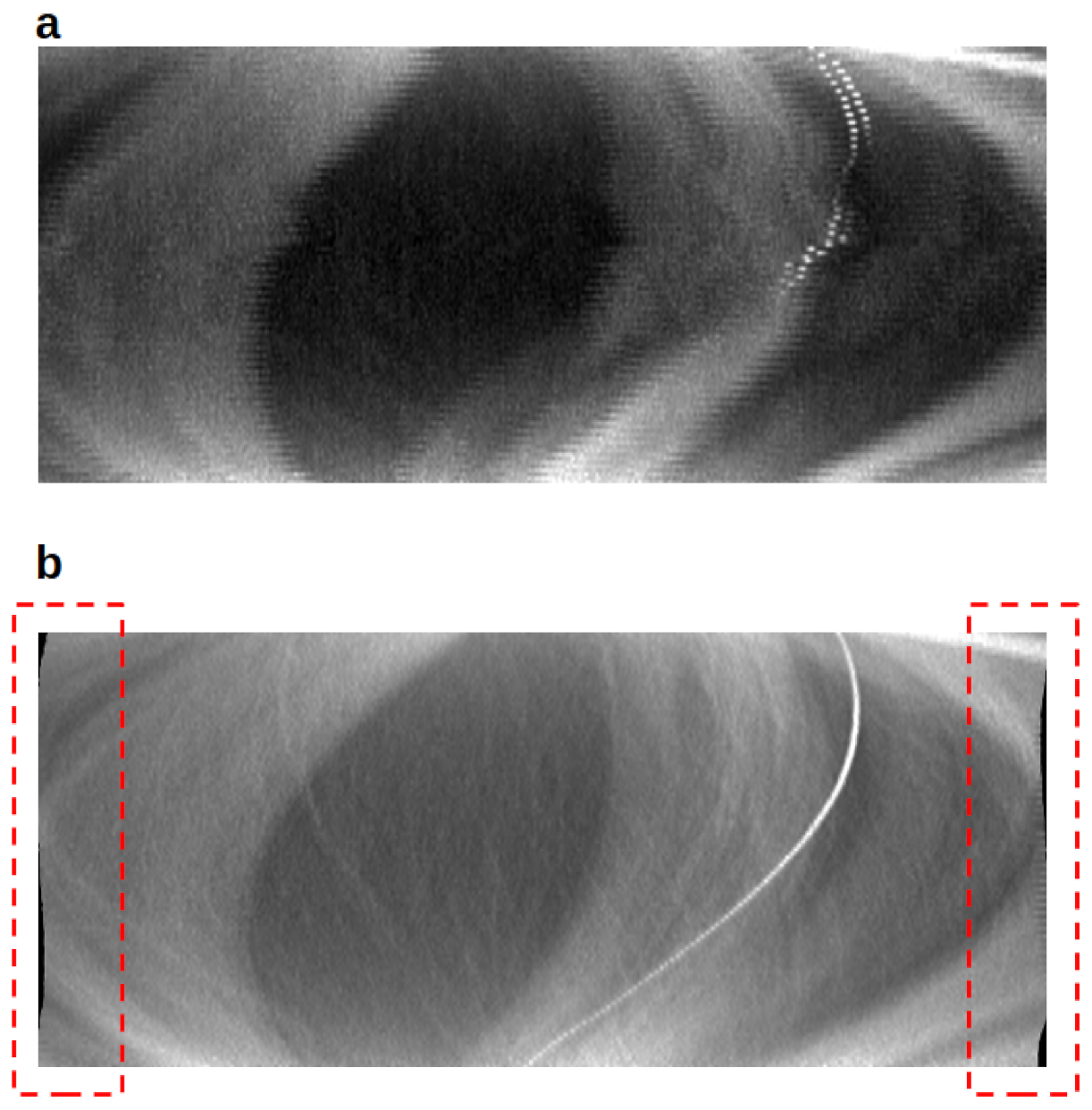
3.4. Entire Algorithm Test
3.5. Nanotomography Data
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| API | Application Program Interface |
| CCD | Charge Coupled Device |
| CPU | Central Processing Unit |
| CT | Computed Tomography |
| DFT | Discrete Fourier Transform |
| GPU | Graphics Processing Unit |
| HPC | High Performance Computing |
| MSE | Mean Square Error |
| RAM | Random Access Memory |
| SIRT | Simultaneous Iterative Reconstruction Technique |
| STN | Spatial Transformer Network |
References
- Pereiro, E.; Nicolás, J.; Ferrer, S.; Howells, M.R. A soft X-ray beamline for transmission X-ray microscopy at ALBA. J. Synchrotron Radiat. 2009, 16, 505–512. [Google Scholar] [CrossRef]
- Sorrentino, A.; Nicolás, J.; Valcárcel, R.; Chichón, F.J.; Rosanes, M.; Avila, J.; Tkachuk, A.; Irwin, J.; Ferrer, S.; Pereiro, E. MISTRAL: A transmission soft X-ray microscopy beamline for cryo nano-tomography of biological samples and magnetic domains imaging. J. Synchrotron Radiat. 2015, 22, 1112–1117. [Google Scholar] [CrossRef]
- Carrascosa, J.L.; Chichón, F.J.; Pereiro, E.; Rodríguez, M.J.; Fernández, J.J.; Esteban, M.; Heim, S.; Guttmann, P.; Schneider, G. Cryo-X-ray tomography of vaccinia virus membranes and inner compartments. J. Struct. Biol. 2009, 168, 234–239. [Google Scholar] [CrossRef]
- Arhatari, B.D.; Stevenson, A.W.; Abbey, B.; Nesterets, Y.I.; Maksimenko, A.; Hall, C.J.; Thompson, D.; Mayo, S.C.; Fiala, T.; Quiney, H.M.; et al. X-ray phase-contrast computed tomography for soft tissue imaging at the imaging and medical beamline (IMBL) of the australian synchrotron. Appl. Sci. 2021, 11, 4120. [Google Scholar] [CrossRef]
- Bertero, M.; Lantéri, H.; Zanni, L. Iterative image reconstruction: A point of view. Math. Methods Biomed. Imaging Intensity Modul. Radiat. Ther. (IMRT) 2008, 7, 37–63. [Google Scholar]
- Grejda, R.; Marsh, E.; Vallance, R. Techniques for calibrating spindles with nanometer error motion. Precis. Eng. 2005, 29, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Gürsoy, D.; De Carlo, F.; Xiao, X.; Jacobsen, C. TomoPy: A framework for the analysis of synchrotron tomographic data. J. Synchrotron Radiat. 2014, 21, 1188–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; De Carlo, F.; Phatak, C.; Gürsoy, D. A convolutional neural network approach to calibrating the rotation axis for X-ray computed tomography. J. Synchrotron Radiat. 2017, 24, 469–475. [Google Scholar] [CrossRef] [Green Version]
- Longo, R.; Arfelli, F.; Bonazza, D.; Bottigli, U.; Brombal, L.; Contillo, A.; Cova, M.A.; Delogu, P.; Di Lillo, F.; Di Trapani, V.; et al. Advancements towards the implementation of clinical phase-contrast breast computed tomography at Elettra. J. Synchrotron Radiat. 2019, 26, 1343–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Xia, S.; Wei, C.; Mao, Y.; Larsson, D.; Xiao, X.; Pianetta, P.; Yu, Y.S.; Liu, Y. Automatic projection image registration for nanoscale X-ray tomographic reconstruction. J. Synchrotron Radiat. 2018, 25, 1819–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jonge, M.D.; Kingston, A.M.; Afshar, N.; Garrevoet, J.; Kirkham, R.; Ruben, G.; Myers, G.R.; Latham, S.J.; Howard, D.L.; Paterson, D.J.; et al. Spiral scanning X-ray fluorescence computed tomography. Opt. Express 2017, 25, 23424–23436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gürsoy, D.; Hong, Y.P.; He, K.; Hujsak, K.; Yoo, S.; Chen, S.; Li, Y.; Ge, M.; Miller, L.M.; Chu, Y.S.; et al. Rapid alignment of nanotomography data using joint iterative reconstruction and reprojection. Sci. Rep. 2017, 7, 11818. [Google Scholar] [CrossRef] [Green Version]
- van Aarle, W.; Palenstijn, W.J.; Cant, J.; Janssens, E.; Bleichrodt, F.; Dabravolski, A.; De Beenhouwer, J.; Joost Batenburg, K.; Sijbers, J. Fast and flexible X-ray tomography using the ASTRA toolbox. Opt. Express 2016, 24, 25129. [Google Scholar] [CrossRef]
- Han, R.; Wan, X.; Wang, Z.; Hao, Y.; Zhang, J.; Chen, Y.; Gao, X.; Liu, Z.; Ren, F.; Sun, F.; et al. AuTom: A novel automatic platform for electron tomography reconstruction. J. Struct. Biol. 2017, 199, 196–208. [Google Scholar] [CrossRef] [Green Version]
- Dehaeze, T.; Collette, C.; Magnin-Mattenet, M. Sample Stabilization for Tomography Experiments in Presence of Large Plant Uncertainty. In Proceedings of the 10th Mechanical Engineering Design of Synchrotron Radiation Equipment and Instrumentation, Paris, France, 25–29 June 2018. [Google Scholar]
- Odstrčil, M.; Holler, M.; Raabe, J.; Guizar-Sicairos, M. Alignment methods for nanotomography with deep subpixel accuracy. Opt. Express 2019, 27, 36637–36652. [Google Scholar] [CrossRef] [Green Version]
- De Andrade, V.; Nikitin, V.; Wojcik, M.; Deriy, A.; Bean, S.; Shu, D.; Mooney, T.; Peterson, K.; Kc, P.; Li, K.; et al. Fast X-ray Nanotomography with Sub-10 nm Resolution as a Powerful Imaging Tool for Nanotechnology and Energy Storage Applications. Adv. Mater. 2021, 33, 2008653. [Google Scholar] [CrossRef]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Mastronarde, D.N.; Held, S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 2017, 197, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Mastronarde, D.N. Dual-Axis Tomography: An Approach with Alignment Methods That Preserve Resolution. J. Struct. Biol. 1997, 120, 343–352. [Google Scholar] [CrossRef]
- Brun, F.; Pacilè, S.; Accardo, A.; Kourousias, G.; Dreossi, D.; Mancini, L.; Tromba, G.; Pugliese, R. Enhanced and Flexible Software Tools for X-ray Computed Tomography at the Italian Synchrotron Radiation Facility Elettra. Fundam. Inform. 2015, 141, 233–243. [Google Scholar] [CrossRef]
- Brun, F.; Massimi, L.; Fratini, M.; Dreossi, D.; Billé, F.; Accardo, A.; Pugliese, R.; Cedola, A. SYRMEP Tomo Project: A graphical user interface for customizing CT reconstruction workflows. Adv. Struct. Chem. Imaging 2017, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Nickell, S.; Förster, F.; Linaroudis, A.; Net, W.D.; Beck, F.; Hegerl, R.; Baumeister, W.; Plitzko, J.M. TOM software toolbox: Acquisition and analysis for electron tomography. J. Struct. Biol. 2005, 149, 227–234. [Google Scholar] [CrossRef]
- Heymann, J.B.; Belnap, D.M. Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007, 157, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Winkler, H.; Taylor, K.A. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 2006, 106, 240–254. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Keszthelyi, B.; Branlund, E.; Lyle, J.M.; Braunfeld, M.B.; Sedat, J.W.; Agard, D.A. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J. Struct. Biol. 2007, 157, 138–147. [Google Scholar] [CrossRef]
- Pyle, E.; Zanetti, G. Current data processing strategies for cryo-electron tomography and subtomogram averaging. Biochem. J. 2021, 478, 1827–1845. [Google Scholar] [CrossRef]
- Guarnieri, G.; Fontani, M.; Guzzi, F.; Carrato, S.; Jerian, M. Perspective registration and multi-frame super-resolution of license plates in surveillance videos. Forensic Sci. Int. Digit. Investig. 2021, 36, 301087. [Google Scholar] [CrossRef]
- Cop, M.; Dengler, J. A multi-resolution approach to the 3D reconstruction of a 50S ribosome from an EM-tilt series solving the alignment problem without gold particles. In Proceedings of the International Conference on Pattern Recognition, Atlantic City, NJ, USA, 16–21 June 1990; Volume 1, pp. 733–737. [Google Scholar]
- Latham, S.J.; Kingston, A.M.; Recur, B.; Myers, G.R.; Sheppard, A.P. Multi-resolution radiograph alignment for motion correction in x-ray micro-tomography. Dev. X-ray Tomogr. X 2016, 9967, 996710. [Google Scholar]
- Zhang, J.; Hu, J.; Jiang, Z.; Zhang, K.; Liu, P.; Wang, C.; Yuan, Q.; Pianetta, P.; Liu, Y. Automatic 3D image registration for nano-resolution chemical mapping using synchrotron spectro-tomography. J. Synchrotron Radiat. 2021, 28, 278–282. [Google Scholar] [CrossRef]
- Guzzi, F.; Kourousias, G.; Gianoncelli, A.; Pascolo, L.; Sorrentino, A.; Billè, F.; Carrato, S. Material Concerning a Publication on an Autograd-Based Method for Ptychography, Implemented within the SciComPty Suite. 2021. Available online: https://doi.org/10.5281/zenodo.5113938 (accessed on 19 July 2021).
- Han, R.; Bao, Z.; Zeng, X.; Niu, T.; Zhang, F.; Xu, M.; Gao, X. A joint method for marker-free alignment of tilt series in electron tomography. Bioinformatics 2019, 35, i249–i259. [Google Scholar] [CrossRef] [Green Version]
- Guzzi, F.; Kourousias, G.; Billè, F.; Pugliese, R.; Gianoncelli, A.; Carrato, S. A parameter refinement method for Ptychography based on Deep Learning concepts. arXiv 2021, arXiv:2105.08058. [Google Scholar]
- Donoho, D.L. The Curses and Blessings of Dimensionality. In Proceedings of the American Math, Society Lecture-Math Challenges of the 21st Century, Los Angeles, CA, USA, 7–12 August 2000; pp. 1–33. [Google Scholar]
- Guizar-Sicairos, M.; Fienup, J.R. Phase retrieval with transverse translation diversity: A nonlinear optimization approach. Opt. Express 2008, 16, 7264. [Google Scholar] [CrossRef]
- Guizar-Sicairos, M.; Thurman, S.T.; Fienup, J.R. Efficient subpixel image registration algorithms. Opt. Lett. 2008, 33, 156–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, J.D.; Houston, M.; Luebke, D.; Green, S.; Stone, J.E.; Phillips, J.C. GPU Computing. Proc. IEEE 2008, 96, 879–899. [Google Scholar] [CrossRef]
- Nickolls, J.; Dally, W.J. The GPU Computing Era. IEEE Micro 2010, 30, 56–69. [Google Scholar] [CrossRef]
- Pratx, G.; Xing, L. GPU computing in medical physics: A review. Med. Phys. 2011, 38, 2685–2697. [Google Scholar] [CrossRef]
- Palenstijn, W.J.; Bédorf, J.; Sijbers, J.; Batenburg, K.J. A distributed ASTRA toolbox. Adv. Struct. Chem. Imaging 2016, 2, 19. [Google Scholar] [CrossRef] [Green Version]
- van Aarle, W.; Palenstijn, W.J.; De Beenhouwer, J.; Altantzis, T.; Bals, S.; Batenburg, K.J.; Sijbers, J. The ASTRA Toolbox: A platform for advanced algorithm development in electron tomography. Ultramicroscopy 2015, 157, 35–47. [Google Scholar] [CrossRef] [Green Version]
- Matenine, D.; Goussard, Y.; Després, P. GPU-accelerated regularized iterative reconstruction for few-view cone beam CT. Med. Phys. 2015, 42, 1505–1517. [Google Scholar] [CrossRef]
- Vogelgesang, M.; Chilingaryan, S.; Rolo, T.d.; Kopmann, A. UFO: A Scalable GPU-based Image Processing Framework for On-line Monitoring. In Proceedings of the 2012 IEEE 14th International Conference on High Performance Computing and Communication 2012 IEEE 9th International Conference on Embedded Software and Systems, Liverpool, UK, 25–27 June 2012; pp. 824–829. [Google Scholar]
- Biguri, A.; Lindroos, R.; Bryll, R.; Towsyfyan, H.; Deyhle, H.; khalil Harrane, I.E.; Boardman, R.; Mavrogordato, M.; Dosanjh, M.; Hancock, S.; et al. Arbitrarily large tomography with iterative algorithms on multiple GPUs using the TIGRE toolbox. J. Parallel Distrib. Comput. 2020, 146, 52–63. [Google Scholar] [CrossRef]
- Palenstijn, W.; Batenburg, K.; Sijbers, J. Performance improvements for iterative electron tomography reconstruction using graphics processing units (GPUs). J. Struct. Biol. 2011, 176, 250–253. [Google Scholar] [CrossRef] [Green Version]
- Pelt, D.M.; Gürsoy, D.; Palenstijn, W.J.; Sijbers, J.; De Carlo, F.; Batenburg, K.J. Integration of TomoPy and the ASTRA toolbox for advanced processing and reconstruction of tomographic synchrotron data. J. Synchrotron Radiat. 2016, 23, 842–849. [Google Scholar] [CrossRef]
- Chghaf, M.; Gac, N. Student Session: Data distribution on a multi-GPU node for TomoBayes CT reconstruction. In Proceedings of the 2020 IEEE 26th International Conference on Embedded and Real-Time Computing Systems and Applications (RTCSA), Gangnueng, Korea, 19–21 August 2020; pp. 1–2. [Google Scholar]
- Palenstijn, W.J.; Bédorf, J.; Batenburg, J. A distributed SIRT implementation for the ASTRA Toolbox. In Proceedings of the 13th International Meeting on Fully Three-Dimensional Image Reconstruction in Radiology and Nuclear Medicine 2015 (Fully3D 1), Newport, RI, USA, 31 May–4 June 2015; pp. 166–169. [Google Scholar]
- Gürsoy, D.; De Carlo, F.; Xiao, X.; Jacobsen, C. Tomopgy GPU Notes. 2021. Available online: https://tomopy.readthedocs.io/en/latest/faq.html#do-tomopy-astra-and-ufo-support-all-gpus (accessed on 9 July 2021).
- Gregor, J.; Benson, T. Computational Analysis and Improvement of SIRT. IEEE Trans. Med. Imaging 2008, 27, 918–924. [Google Scholar] [CrossRef]
- Luu, M.B.; Van Riessen, G.A.; Abbey, B.; Jones, M.W.; Phillips, N.W.; Elgass, K.; Junker, M.D.; Vine, D.J.; McNulty, I.; Cadenazzi, G.; et al. Fresnel coherent diffractive imaging tomography of whole cells in capillaries. New J. Phys. 2014, 16, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Evangelidis, G.D.; Psarakis, E.Z. Parametric Image Alignment Using Enhanced Correlation Coefficient Maximization. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 30, 1858–1865. [Google Scholar] [CrossRef] [Green Version]
- van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the Advances in Neural Information Processing Systems 32: NeurIPS 2019, Vancouver, BC, Canada, 8–14 December 2019; pp. 8024–8035. [Google Scholar]
- Jaderberg, M.; Simonyan, K.; Zisserman, A.; Kavukcuoglu, K. Spatial Transformer Networks. In Proceedings of the Advances in Neural Information Processing Systems 28: Annual Conference on Neural Information Processing Systems 2015, Montreal, QC, Canada, 7–12 December 2015; pp. 2017–2025. [Google Scholar]





| CPU | Intel(R) Xeon(R) CPU E5-2643 v4 @ 3.40 GHz 24 hyper-threading core, 20 available (virtualisation) |
| GPU | 2× Nvidia Tesla k80, 4 available processors |
| Virtualisation system | proxmox-ve: 6.1-2 (kernel: 5.3.13-1-pve) |
| Virtual machine OS | Ubuntu 18.04 LTS (kernel 5.0.0-29-generic) |
| Python | 3.9.5 Anaconda |
| CUDA | 11.1 |
| PyTorch [55] | 1.9 |
| Scikit-Image [54] | 0.18.1 |
| ASTRA Toolbox [42] | 1.9.9-dev1 |
| TomoPy [7] | 1.10.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzzi, F.; Kourousias, G.; Gianoncelli, A.; Pascolo, L.; Sorrentino, A.; Billè, F.; Carrato, S. Improving a Rapid Alignment Method of Tomography Projections by a Parallel Approach. Appl. Sci. 2021, 11, 7598. https://doi.org/10.3390/app11167598
Guzzi F, Kourousias G, Gianoncelli A, Pascolo L, Sorrentino A, Billè F, Carrato S. Improving a Rapid Alignment Method of Tomography Projections by a Parallel Approach. Applied Sciences. 2021; 11(16):7598. https://doi.org/10.3390/app11167598
Chicago/Turabian StyleGuzzi, Francesco, George Kourousias, Alessandra Gianoncelli, Lorella Pascolo, Andrea Sorrentino, Fulvio Billè, and Sergio Carrato. 2021. "Improving a Rapid Alignment Method of Tomography Projections by a Parallel Approach" Applied Sciences 11, no. 16: 7598. https://doi.org/10.3390/app11167598
APA StyleGuzzi, F., Kourousias, G., Gianoncelli, A., Pascolo, L., Sorrentino, A., Billè, F., & Carrato, S. (2021). Improving a Rapid Alignment Method of Tomography Projections by a Parallel Approach. Applied Sciences, 11(16), 7598. https://doi.org/10.3390/app11167598








