Abstract
The clinical use of bioactive material in the field of biomedical tissue engineering has become increasingly of interest in practice. This study investigates how BiodentineTM (BD), a tricalcium silicate cement, in culture media, affects the odonto/osteogenic differentiation potential of in vitro cultured human dental pulp stem cells (hDPSCs). hDPSCs were extracted and characterized for their expression profile by flow cytometry. Then, hDPSCs were cultured in media containing BD for 3 weeks to study the impact of BD on the odonto/osteogenesis pathway, compared to the positive control (osteogenic media) and negative control (cell culture media). Odonto/osteogenic differentiation of hDPSCs treated with BD was assessed by measuring the level of expression of odonto/osteogenic markers by flow cytometry, ELISA and Alizarin red stain. Additionally, the expression profile of the genes involved in the odonto/osteogenesis pathway was investigated, using PCR array. Our results indicate that hDPSCs treatment with BD results in an increased tendency for odonto/osteogenic differentiation. The BD treated group demonstrates a significant increase in the expression of odonto/osteogenic markers, osteocalcin (OCN) (p < 0.005), osteopontin (OPN) (p < 0.0005) and alkaline phosphatase (ALP) (p < 0.0005), and the presentation of calcium deposits by ARS, compared to the negative control by using t-test and ANOVA. Moreover, the BD-treated group is marked by the upregulation of genes related to the odonto/osteogenesis pathway, compared to the control groups, specifically the genes that are involved in the bone morphogenic protein (BMP) (p < 0.05) signaling pathway, the activation of the extracellular matrix-related gene (ECMG) (p < 0.05) and the Ca2+ signaling pathway (p < 0.05), compared to day 1 of treatment by using ANOVA. BD shows a stimulatory effect on the odonto/steogenic capacity of hDPSCs, suggesting BD as a good candidate and a very promising and useful means to be applied in regenerative medicine to regenerate dentine tissue in clinical settings.
1. Introduction
Recently, the clinical application of regenerative medicine in the field of dentistry was verified by using different techniques and different therapeutic procedures in order to restore the main functions of dental tissues [1]. One such technique is using stem cells in therapeutic applications by injecting them solely or in combination with other molecules [2]. Hence, the injected cells are responsible for the regeneration of the damaged or missed dental tissues, thereby restoring their biological function and structure [1]. Dental pulp stem cells (DPSCs) are the first type of dental stem cells that were isolated from the pulp of human third molars [3]. Since then, mesenchymal stem cells (MSCs) of dental origin have been isolated from different dental tissues, such as (SHED) stem cells from human exfoliated deciduous teeth [4], (PDLSCs) periodontal ligament stem cells [5], (SCAP) stem cells of apical papilla tissue of impacted third molars [6], (DFSCs) dental follicle stem cells [7] and stem cells from periapical cysts [8]. All these cells are derived from the same dental origin, but they vary in terms of MSC surface markers, and in their differentiation potential for different cell-lineages [9].
Human-derived dental pulp stem cells (hDPSCs) are considered an attractive and very promising source of stem cells for use in clinical applications for many reasons. First, teeth are easily collected at any dental clinic. Second, no ethical approval is required for teeth collection, because extracted teeth are considered medical waste [4]. Third, hDPSCs are tested and examined in a high manner. Fourth, these cells, just like MSCs, possess immunosuppressive activity [10]. Fifth, no change on their differentiation potential toward different lineages is observed after cryopreservation [9,11,12]. Sixth, they possess a high mineralization potential, which is characterized by the volume of the mineralized matrix produced through their differentiation process, indicating their high potential to regenerate and repair pulp and bone tissues [13]. Therefore, hDPSCs have high potential to differentiate into odonto/osteoblasts, making them a good candidate for future therapeutic applications to regenerate dentin and bone defects and restore damaged tooth structures [14].
The combinatorial strategy of using stem cells together with scaffolds and bioactive factors is a promising tissue-engineering approach that can be applied for the regeneration of dental and bone tissues [15,16,17,18]. The accompanying scaffolds are made of biomaterials, such as biodegradable metals, bioactive ceramics, and biodegradable polymers, serving as 3D structures that promote such cellular processes as proliferation, migration and differentiation [19]. Additionally, those scaffolds can be modified using different treatments to enhance the secretion of biological molecules. For instance, the release of odonto/osteogenic factors as biological molecules results in increasing the activity of both cell types: endogenous and injected cells [19,20,21].
In recent years, BiodentineTM (BD) has gained attention as a novel ‘dentin substitute’. This new biologically active material claims to improve such properties as physical qualities and handling, including its other wide range of applications, such as endodontic repair and pulp capping in restorative dentistry [22,23,24,25]. Different studies have examined the role of BD in enhancing the odontogenic and osteogenic differentiation potential of DPSCs [22,26,27,28]. However, the molecular signaling cascades that are activated through the differentiation process are not very well understood and remain to be elucidated. Since no previous studies have evaluated these networks, our study aims to investigate the odonto/steogenic potential of hDPSCs combined with the bioactive material BiodentineTM by studying the signaling networks at the molecular level: gene and protein levels. This study aids in revealing a variety of molecular mechanisms underlying gene expression regulation for dentin and bone regeneration through different signaling networks, which can either be used in in vitro differentiation studies, or be translated for clinical manifestation.
2. Materials and Methods
2.1. Preparation of Biodentine™
To prepare 2 mg/mL of Biodentine™, a Biodentine™ (Septodont, France) capsule was mingled with its own solution, provided by the manufacturer, as previously described [22,29,30]. Following that, a step of dryness and sterilization of the mix was performed by using an oven at 60 °C for 15 min. Then, a pestle and mortar were used to grind the dried mixture to a powder. To prepare the concentration, 2 mg/mL of the prepared dried powder was added to 50 mL of the cell culture alpha-modification of Eagle’s Medium (α-MEM, Gibco, Waltham, MA, USA), supplemented with 2 mM L-glutamine (Invitrogen, Waltham, MA, USA), 100 mg/mL streptomycin (Invitrogen, Waltham, MA, USA), 100 units’/mL penicillin (Invitrogen, Waltham, MA, USA) and 0.25 mg/mL Amphotericin B (Invitrogen, Waltham, MA, USA). In addition, 5% concentration of human platelet lysate (PL) was used. The medium was vortexed until it was completely suspended. Following that, the medium was filtered twice by using a 70 μM cell strainer (BD Biosciences, NJ, USA). A cell culture medium without Biodentine™ was used as a blank control. Figure 1 provides a summary of the work plan.
Figure 1.
Flow chart of work plan.
2.2. Sample Collection
Human third molar samples were collected from healthy donors (18, 19 and 21 years), according to the Institutional Review Board (IRB) guidelines from the Cell Therapy Center/The University of Jordan (IRB/06/2018) and approved on 13 March 2018. All human participants signed their informed consent before their tooth donation.
2.3. Cell Culture of hDPSCs
hDPSCs were isolated by using the explant method, as described previously [3]. Briefly, the teeth were disinfected with phosphate buffer saline (1×) (PBS, Gibco, Waltham, MA, USA) thrice, followed by incubation for five minutes. The teeth were excavated with hand piece to remove the pulp tissue. The pulp tissue sample was cut into 1–2 mm fragments, and individual pieces were placed in a 6-well culture plate with alpha MEM medium (a-MEM, Gibco, Waltham, MA, USA), supplemented with 2 mM L-glutamine (Invitrogen, Waltham, MA, USA), 100 mg/mL streptomycin (Invitrogen, Waltham, MA, USA), 100 units’/mL penicillin (Invitrogen, Waltham, MA, USA) and 0.25 mg/mL Amphotericin B (Invitrogen, Waltham, MA, USA). For monolayer generation of the primary culture, 5% concentration of human platelet lysate (PL) was used. The cells were incubated at 37 °C in a 5% CO2 incubator, and the culture medium was changed every three days until the cells reached 70–80% confluence. The medium was exchanged every 3 days until the outgrowth of the hDPSCs was observed. The morphological appearance was observed under the inverted microscope (Zeiss, Oberkochen, Germany).
2.4. Characterization of hDPSCs by Flow Cytometry
The derived hDPSCs were isolated and characterized, according to their expression of MSC surface markers. Briefly, hDPSCs at P3 were collected by using 0.25% trypsin EDTA (Gibco, Waltham, MA, USA). Then, the cells were stained with the human mesenchymal stem cell characterization kit (BD stemflow kit, BD Biosciences, NJ, USA). Cells were stained and incubated for 30 min at room temperature with the following fluorescein-labeled antibodies: CD44, PerCP-Cy™5.5, FITC CD90, PE CD105, APC CD73 and PE-negative cocktail (which contains CD34, CD45, CD19, HLA-DR and CD11b), as well as their isotype controls as per the manufacturer’s recommendations. Following that, the cells were centrifuged at 300× g for 5 min, followed by re-suspension with PBS. The expression profile was analyzed by FACS DIVA software version 7, using FACS Canto II (BD, Biosciences, NJ, USA).
2.5. Odonto/Osteogenic Differentiation of hDPSCs with BiodentineTM
To study the impact of BiodentineTM on the odonto/osteogenic signaling pathway, hDPSCs were treated with BiodentineTM for 3 weeks. At first, hDPSCs (P3) were cultured in 6-well plates at a seeding density of 1 × 105 cells/well in alpha MEM supplemented with 5% platelet lysate (PL) until the cells reached 60–70% confluence. After that, the derived cells were treated with either 2 mg/mL BiodentineTM as the test group and osteogenic media (osteogenesis differentiation kit, Gibco, Waltham, MA, USA) as the positive control, whereas un-induced cells cultured in their growth media (alpha MEM +5%PL) were used as a negative control group. Media were exchanged thrice per week.
2.6. PCR Array
In order to evaluate the effect of BiodentineTM on the odonto/osteogenic signaling pathway, PCR array was performed after 3 weeks of treatment.
RNA was extracted from hDPSCs using the TRIzol–hybrid method (Qiagen, Germantown, MD, USA) from all three groups (treated with BiodentineTM, positive and negative). A total of 0.5 μg extracted RNA was converted to cDNA by using the RT2 First Strand Kit (Qiagen, Germantown, MD, USA). Then, cDNA samples were diluted and amplified with the RT2 SYBR® green master mix of (PAHS-058Z, RT2 Profiler™ PCR Array Human osteogenesis pathway, Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions, and loaded onto the 96-well array. The amplification conditions were as follows: 95 °C for 10 min, then 40 cycles of 95 °C for 15 s and 60 °C for one min. Samples were run on the CFX 96 C1000 system (Biorad, Hercules, CA, USA). Data were analyzed automatically, according to the SABiosciences company (Qiagen, Germantown, MD, USA) web portal, www.SABiosciences.com/pcrarraydataanalysis.php, by using the method, according to the following equations:
GOI: Gene of interest
HKG: Housekeeping gene
The expression levels of the genes were normalized to the following housekeeping genes: beta-2-microglobulin (B2M), hypoxanthine phosphoribosyl transferase 1 (HPRT1), and actin beta (ACTB). For data analysis, the differential expression level of the odonto/osteoblastic genes was identified, using Student’s t-test (two-tailed, unpaired). A cut-off point of 2 was used as a threshold to determine the statistical significance of the upregulated or downregulated genes (p < 0.05). Day 1 of each biological replicate was used as a reference sample to compare gene fold regulation for all treatment groups, of the same biological replicate.
2.7. Flow Cytometry for Odonto/Osteogenic Markers
Flow cytometry was performed to detect the expression of the odonto/osteogenic markers, Osteopontin (OPN) and Osteocalcin (OCN).
After 3 weeks of treatment, hDPSCs were collected by using 0.25% Trypsin EDTA (Gibco, USA), followed by an incubation step with cold methanol and kept at −20 °C until use. Then, the cells were centrifuged at 300× g for 5 min, re-suspended in the stain buffer (BD Biosciences, NJ, USA) followed by the addition of antibodies OPN-APC (R&D systems, Minneapolis, MN, USA) and OCN-PE (R&D systems, USA) for 40 min on the shaker. Then, the cells were washed by adding 1 mL of cell wash (BD Biosciences, NJ, USA) and centrifuged at 300× g for 5 min. Finally, the samples were re-suspended in 300 µL PBS and analyzed by FACS Canto II FACS Diva 7 software (BD Biosciences, NJ, USA).
2.8. ELISA for Odonto/Osteogenic Markers
To measure the expression level of the odonto/osteogenic markers at protein level, the following human ELISA kits were used: alkaline phosphatase assay kit (ALP, Abcam, Eugene, OR, USA), and human osteopontin Elisa Kit (OPN, R&D, Minneapolis, MN, USA), according to the manufacturer’s instructions after 3 weeks of treatment.
Firstly, the spun media were collected after treating cells with BiodentineTM, osteogenic differentiation media (positive control) and, as a negative control, the cell culture media from un-induced cell cultures. Secondly, the expression level of ALP and OPN was measured by using the following kits: ALP (Abcam, Eugene, OR, USA) and OPN marker (R&D systems, Minneapolis, MN, USA). The absorbance (O.D.) was measured at 405 nm and 450 nm by using the ELISA plate reader Glomax (Promega, Madison, WI, USA) for ALP and OPN, respectively.
2.9. Alizarin Red Stain ARS
To determine the efficiency of odonto/osteogenic differentiation and the mineralization potential of the treated cells after three weeks of treatments, BiodentineTM, positive and negative controls, and calcium deposits were detected by using Alizarin red stain (Sigma, Saint Louis, MO, USA, 4.20 pH), according to the manufacturer’s instructions. The stained cells were observed under the inverted microscope (Zeiss, Oberkochen, Germany).
2.10. Statistical Analysis
GraphPad Prism and Microsoft Windows Excel were used for data analysis. All experiments were run in triplicate in three independent experiments (n = 3). The results are expressed as means± standard deviations. A paired t-test and one-way ANOVA analysis were used to determine the statistical differences among all assays. (Significance assumed for (p < 0.05).
3. Results
3.1. Characterization of hDPSCs
The derived hDPSCs were characterized for their MSCs expression profile. As shown in Figure 2, hDPSCs-derived cells showed a high expression level of MSC surface markers, CD44, CD73, CD105 and CD90, and negative expression for the hematopoietic stem cell markers.

Figure 2.
Flow cytometric analysis of mesenchymal stem cells (MSC) surface markers expression of DPSCs derived from pulp tissue. DPSC histograms show that cells are positive for MSCs surface markers (A) CD90, (B) CD105, (C) CD73, and (D) CD44, and (E) have negative expression of hematopoietic stem cells surface markers (negative cocktail), including CD45, CD34, CD19, CD14 and HLA-DR.
3.2. Odonto/Osteogenic Differentiation of hDPSCs
To determine the impact that BiodentineTM has on the odonto/osteogenic differentiation potential of hDPSCs, cells were characterized after 3 weeks of induction for the expression of genes that are involved in the odonto/osteogenic signaling pathway (by means of the PCR arrays). Additionally, the functionality of the odontoblast and osteoblast cells that were produced by the treatment was determined by measuring the expression levels of odonto/osteogenic markers by flow cytometry, ELISA and ARS.
3.2.1. PCR Arrays for Odonto/Osteogenic Signaling Pathway
At the gene level (Figure 3), it was found that the investigated 84 genes play major roles in the odontogenesis and osteogenesis pathway, and alteration in the expression level of any of these genes apart from the normal expression level would impact the overall outcome of the pathway. While comparing the effect of the BiodentineTM group with the positive control group, we here report similar expression patterns of the odontogenesis- and osteogenesis-related genes in both groups in terms of fold regulation and p values as compared to day 1 of treatment (Figure 3). As shown in the scatter plots of Figure 3B–D, each treatment group was compared to day 1 of treatment, showing the number of genes that were upregulated, downregulated and unchanged genes. The results of the PCR array were divided into three major signaling pathways of odontogenesis differentiation and ossification, extracellular matrix-related gene clusters during odonto/osteogenic differentiation, and Ca2+ signaling.
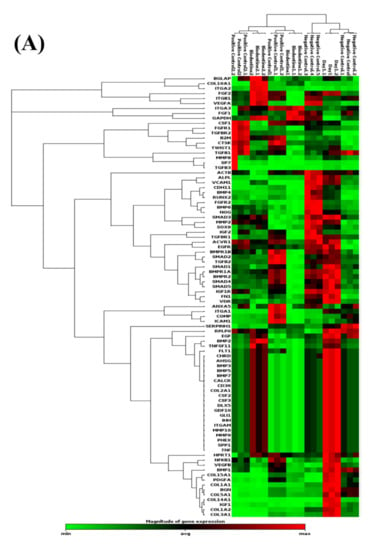
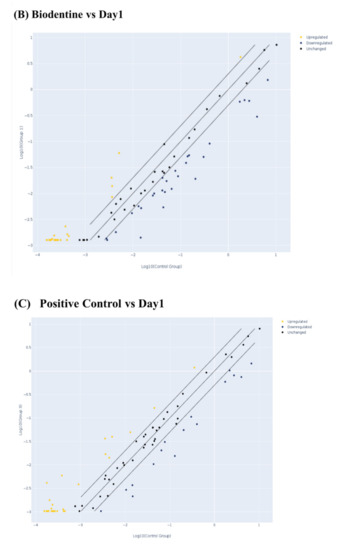
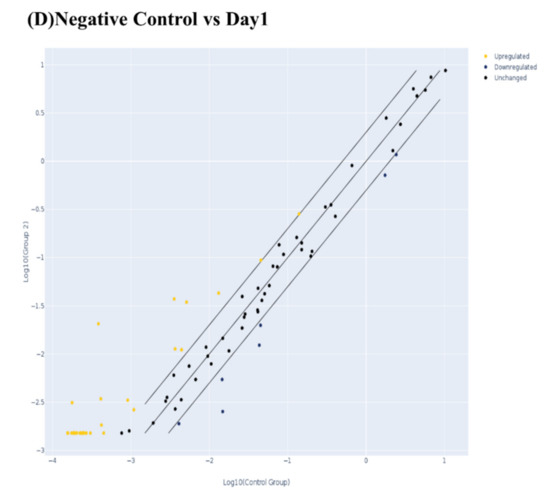
Figure 3.
Evaluation of human odonto/osteogenesis pathway by quantitative PCR array. (A) Heat map of gene expression levels in human odonto/osteogenesis pathway in hDPSCs treated with Biodentine, osteogenic differentiation media (positive control), and uninduced cells, cultured in their own growth media (negative control), compared to day 1 of treatment. Scatter plots of odonto/osteogenesis signaling pathway in hDPSCs treated with (B) Biodentine, (C) osteogenic differentiation media (positive control), and (D) uninduced cells, cultured in their own growth media (negative control), compared to day 1 of treatment. A cut-off point of two-fold change in expression was utilized. Yellow dots indicate the genes that were upregulated; blue dots indicate the genes that were downregulated; and black dots indicate the unchanged genes.
Odontogenic Differentiation and Ossification
The following BMP proteins are genes that are involved in odonto/osteogenic differentiation and ossification, as they are known for their high odonto/osteogenic capacity: 2, 4, 5, 6 and 7. Interestingly, the BiodentineTM-treated group and both control groups showed a significant upregulation in the expression level of BMP2, -3, -5 and -7, whereas only the BD-treated group showed a significant downregulation of BMP4 and -6 (p < 0.05). Meanwhile, BMP3 was upregulated in the BD-treated group in a moderate amount, compared to the other treated groups. As a consequence, our results show that the expression of other odonto/osteogenic markers. SP7, SPP1 and COL2A, were upregulated significantly on all groups, resulting in accelerating the process of matrix mineralization. Additionally, after 21 days of treatment, the BiodentineTM-treated group showed a significant downregulation of ALP and RUNX2 expression levels (p < 0.05) (Table 1).

Table 1.
PCR array results of fold changes and the statistical significance of genes that are involved in odontogenic differentiation and ossification. All groups were compared to day 1 of treatment. Red: upregulated genes, Green: Downregulated genes. The bold numbers represent significant p values.
Overall, none of the three treatment groups showed a significant change in the expression pattern of TGF β genes. Except for TGFβ2 gene, as BD-treated cells showed a remarkable decrease in the expression level of TGFβ2, which stimulates their state in osteogenesis. Apparently, BD stimulates the expression of these genes in a range that enables the treated hDPSCs to differentiate into odontoblasts and osteoblasts.
Extracellular Matrix-Related Gene Clusters during Odonto/Osteogenic Differentiation (ECMG)
To characterize ECMG expression profiles, hDPSCs were compared as three groups; BD-treated, positive control, and negative control for 21 days’ culture time.
Matrix metallopeptidase 2 (MMP2) was significantly downregulated among all treated groups (p < 0.05), whereas the matrix metallopeptidase 9 (MMP9) gene was upregulated among all treated groups; the significant difference was observed in both control groups (p < 0.05), while BD upregulated the expression without any statistical difference.
For collagen synthesis and formation, COL10A was significantly downregulated, except for the BD-treated group, as the downregulation was insignificant (fold change less than two), compared to the other treated groups. COL2A, COL14A1 and COL15A 1 were upregulated in a statistically significant manner in all treatment groups (p < 0.05). Interestingly, COL1A1, COL1A2, COL3A1 and COL5A1 were only downregulated significantly in the BD-treated group, compared to the other groups (p < 0.05).
For other extracellular matrix proteins, the BD-treated group showed significant upregulation of the major ECM proteins, such as MMP9, MMP10, ITGA2, ITGA3, ITGAM, FLT, and PHEX (p < 0.05) (Table 2). These upregulated expression patterns were observed similarly in cells treated with positive osteogenic differentiation media (positive control). Untreated cells (negative control) showed an elevated expression level of the aforementioned ECMG proteins. However, ITGA2 and IRGA3 were highly upregulated in a statistically significant manner compared to the other control groups (p < 0.05), whereas ITGA1 was only downregulated when the cells were treated with BD.

Table 2.
PCR array results of fold changes and the statistical significance of genes which are involved in extracellular matrix-related gene clusters during odonto/osteogenic differentiation. All groups were compared to day 1of treatment. Red: upregulated genes, Green: Downregulated genes. The bold numbers represent significant p values.
Ca2+ Signaling Pathway
The BD-treated group promotes the expression levels of the following growth factors in a significant manner (p < 0.05): IGF1 and 2, VEGF, PDGF, BMP2, EGF, CALCR, COMP, and DLX5 (Table 3). All these growth factors play a major role in the Ca2+ signaling pathways, resulting in an increase of the odonto/osteogenic potential of the derived treated cells.

Table 3.
PCR array results of fold changes and the statistical significance of genes, which are involved in Ca2+ signaling and regulatory genes. All groups were compared to day 1 of treatment. All groups were compared to day 1of treatment. Red: upregulated genes, Green: Downregulated genes. The bold numbers represent significant p values.
3.2.2. Flow Cytometry
The expression of osteopontin (OPN) and osteocalcin (OCN) as major odonto/osteogenic differentiation markers in the odonto/osteogenesis pathway was performed by flow cytometry.
As shown in Figure 4, hDPSCs treated with BD showed a significant increase in the expression level of these two markers, OPN and OCN, compared to the negative control, p < 0.0005 and p < 0.005 respectively. Additionally, the BD-treated cells showed a significant increase in both markers, OPN (p < 0. 05) and OCN (p < 0. 05), compared to the positive control.
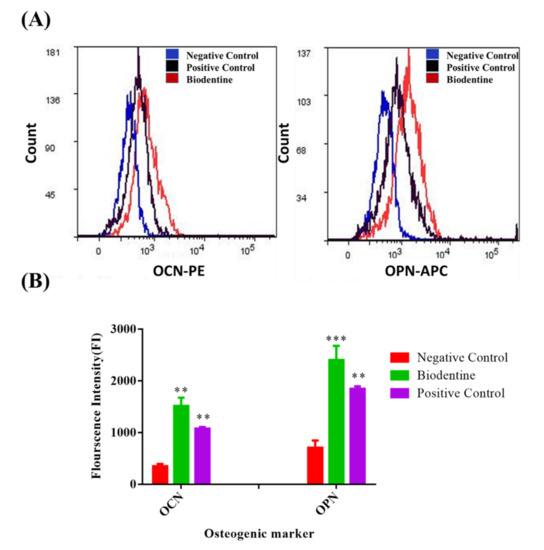
Figure 4.
Flow cytometric results of the odonto/osteogenic differentiation markers. (A) Flow cytometric histograms, and (B) the statistical analysis of the Mean fluorescence Intensity (MFI) of flow cytometric results of the expression level of odonto/osteogenic markers; Osteocalcin (OCN) and Ostepontin (OPN), hDPSCs treated with: Biodentine, osteogenic differentiation media (positive control), compared to the uninduced cells cultured in their own growth media (negative control), after 3 weeks of treatment by flow cytometry. ( *** p < 0.0005, ** p < 0.005).
3.2.3. ELISA
To evaluate the efficiency and maturation stage of odonto/osteoblast formation, ALP and OPN were measured on day 21 of differentiation (Figure 5). The data were consistent with the flow cytometry results; the BD-treated group showed a high secretion level of the measured markers ALP and OPN from day 1 to day 21 of treatment. Both ALP and OPN expression levels were elevated significantly (p < 0.0005 and p < 0.003, respectively) compared to day 1 of treatment.
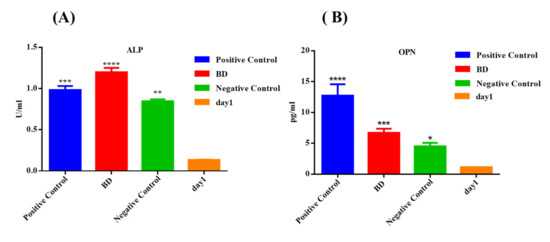
Figure 5.
Measurement of odonto/osteogenic markers concentration (A) ALP and (B) OPN, secreted by DPSCs treated with Biodentine, odonto/osteogenic differentiation media (positive control), and uninduced cells cultured in their own growth media (negative control), after 3 weeks of treatment by ELISA. (**** p < 0.00005, *** p < 0.0005, ** p < 0.005, * p < 0.05).
In comparison to the other control groups, the BD group showed the highest significant elevation in the secretion level of ALP, compared to the positive control (p < 0.0005) and negative control (p < 0.00005). For the OPN marker, the BD-treated group showed a significant increase in the secretion level of OPN, compared to the negative control (p < 0.05), whereas the positive control showed the highest secretion level of the latter marker, compared to the BD-treated group (p < 0.005) (Figure 5).
3.2.4. Alizarin Red Stain (Calcium Deposition Staining)
Calcium deposition is a marker used to detect the maturation stage and mineralization potential of differentiated MSCs into odontogenesis and osteogenesis, thus extended accumulation and the increase of calcium deposits is an indicator for the commitment of cells toward producing mature odontoblast and osteoblasts [7,8]. Calcium deposits were detected by using Alizarin red stain after 3 weeks of treatments (Figure 6). Remarkably, the BD-treated group showed a high detectable level of calcium deposits, similar to the positive control treated group, while the un-induced cells did not show any calcium deposits after staining.
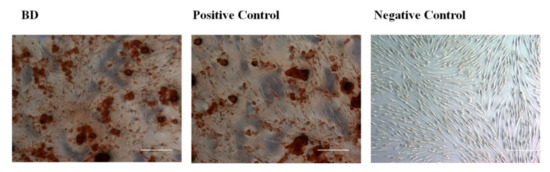
Figure 6.
Calcium deposits staining with Alizarin red stain for DPSCs treated with Biodentine, osteogenic differentiation media (positive control), and uninduced cells cultured in their own growth media (negative control) after 3 weeks of treatment.
4. Discussion
BiodentineTM is a capping material that is designed to stimulate the odontogenesis and dentinogenesis processes in vital pulp therapy. Consequently, such material is required to be biocompatible, with high physiochemical standards, and most importantly, to have the ability to stimulate the formation of dentin and odontoblastic differentiation of dental pulp cells [31]. Hence, the biocompatibility of BD with cellular processes, such as proliferation and differentiation, is mandatory. Many previous studies have examined the biocompatibility of BD with different types of dental stem cells, such as DPSCs [22], SHED [30], SCAP [32] and PDLSCs [29], in addition to evaluating its role in the differentiation processes, such as dentinogenesis, osteogenesis and odontogenesis, for different types of stem cells [26,27,28,33]. However, no previous study has evaluated the signaling pathways that are activated through the dentinogenic differentiation process. Therefore, this study was performed to evaluate the impact of BD on the molecular pathways and signaling networks that are activated through odonto/osteogenic differentiation of hDPSCs at both gene and protein levels.
At gene level, our data show that BD has stimulated the expression level of BMP genes, which are highly involved in teeth development, growth and formation [27,28,29]. The BMP proteins (2, 4, 5, 6 and 7) are products of genes that are involved in odonto/osteogenic differentiation and ossification, as they are known for their high odonto/osteogenic capacity. These genes play essential roles in odontogenic and osteogenic differentiation signaling pathways by increasing the expression of odonto/osteoblastic differentiation markers, such as osterix (SP7), osteopontin (SPP1), ALPL and osteocalcin (BGLAP), RUNX2, the calcium mineralization potential, and collagen forming genes, such as CoL2A [34,35,36]. The BD-treated group has shown a downregulation of the expression levels of BMP4 and 6. This downregulation could be explained by the fact that both BMP4 and 6 are expressed at early stages of the odontogenesis and osteogenesis pathways [37]. In addition, the odonto/osteogenic potency of each BMP might depend on the stage of differentiation of the treated cells. Thus, BD-treated cells might be at different stages of differentiation [38]. For the BMP3 gene, the BD-treated group showed a moderate, insignificant increase in its expression level, compared to other groups, which is reflected in their potency to differentiate into odonto/osteoblasts, as overexpression of BMP3 opposes odontoblast and osteoblast differentiation by activating activin receptor (AcvrIIB)- Smad2/3 signaling [39].
Moreover, our results are consistent with the previously published data regarding the enhancement of expression levels of SP7, SPP1 and COL2A, in addition to the downregulation that was observed in the expression levels of RUNX2 and ALP [32,35]. Both RUNX2 and ALP markers play essential roles in the differentiation pathway, as RUNX2 promotes and regulates odontoblast and osteoblast formation, and ALP is responsible for their development [36,40,41,42]. Additionally, the expression level of ALP is used to monitor the maturation stage of MSCs toward osteogenesis and tooth mineralization; therefore, it is considered an indicator for the commitment of cells to continue odonto/osteo-lineage differentiation [40]. Thus, secreted ALP is accumulated in the media, and this accumulation is a sign of extended mineralization [43,44]. Our results are also in agreement with the previously published data on RUNX2 and ALPL being expressed in early days of differentiation, then declining rapidly in expression with time in a remarkable manner [32,40,41,42]. The downregulation in the expression level of these two genes is consistent with what other groups have reported [37,40], except for the results from Luo et al., where DPSCs were cultured in media containing BD in addition to other mineralizing factors [26].
For collagen synthesis and formation, COL10A was significantly downregulated, except in the BD-treated group, as the downregulation was insignificant (fold change less than two), compared to the other treated groups. The type X collagen gene (COL10A1) influences deposition of other matrix molecules to this region, thereby providing a proper environment for hematopoiesis, mineralization and modeling, which are essential for endochondral ossification [45]. Col10A (Col X) plays a critical role through this process in addition to its structural support of the matrix, as 45% of the total collagen produced represents Col 10A. Moreover, Col10A initiates the biomineralization through the increase in the Ca2+ influx into the matrix [35]. Hence, it is very important during tissue engineering, as it is essential for both MSC-mediated cartilage and bone formation [46]. Additionally, the BD-treated group has shown an increase in the expression levels of Col14 and Col1. These two collagen types have key roles with respect to the architecture of the extracellular matrix, and bone quality. Moreover, these genes have a direct relationship with other molecules, such as IFG molecules, which are important to start the induction of new bone formation [47,48].
On the other hand, the BD-treated group has shown downregulation of the expression of COL1A1, COL1A2, COL3A1 and COL5A1. This could be explained by the fact that Collagen 1 and 3 are considered early markers for odontogenic and osteogenic differentiation in which these genes are important for the phenotypical appearance of odontoblasts and osteoblasts, and play key roles as regulatory molecules for the differentiation and proliferation of human osteoblastic cells [49], as the organic phase of dental tissues is composed of about 30% collagen, types I, III, and V [50]. Thus, their upregulated expression levels can be detected at early stages of odontogenesis: days 4 to 7 [49]. Regarding the expression of collagen genes, our results could be explained by previously published studies, where it was shown that calcium silicate biomaterials have a negative effect on the expression of collagen type I, as these biomaterials can downregulate its expression and stimulate the degradation of collagen fibers in the matrix [51,52].
Furthermore, BD has downregulated the expression of MMP2 and stimulated the expression of MMP9, which have key roles in different cellular mechanisms, such as proliferation, migration, differentiation and the healing process of pulp tissue [23]. These results are similar to the previously published data [40,53].
For other extracellular matrix proteins, BD-treated cells showed upregulation of the major ECM proteins, such as MMP9, MMP10, ITGA2, ITGA3, ITGAM, FLT, and PHEX. These upregulated expression patterns were observed to be similar to other groups: positive and negative control groups. However, the untreated group showed an elevated expression level of the ECMG proteins. This could be explained as previously described [45], that this upregulation could be considered an artifact that appears to be greater or similar to treated groups: either BD or the positive control group [45]. Additionally, based on previously published results, the undifferentiated MSCs highly express these ECM genes; upon the osteogenic differentiation, a downregulation in the expression of the derived genes was detected. This could be a result of either the fact that the viability of cells was decreased at late phases of osteogenic induction, or that the long-term culture of un-induced cells may provoke cells to start their own differentiation. [45,46,47].
For genes that are involved in the Ca2+ signaling pathway, the BD-treated group has a stimulatory effect on the expression levels of these growth factors: IGF1 and 2, VEGF, TGFβ, BMP2, and BMP4. Since this pathway has a very important role in the odontoblast and osteoblast differentiation processes, the high level of cytoplasmic Ca2+ promotes the upregulation of the previously mentioned growth factors. These growth factors are responsible for the communication between cells. The Ca2+ signaling pathway stimulates the expression of several mesenchymal transcription factors that are important for the continuation of tooth development [54]. In addition to that, it will help to improve the understanding of the biological process of odontogenesis and bone regeneration [55].
At the protein level, our results were in agreement with the previously published data that showed the great impact of BD on the secretion level of odonto/osteogenic markers, OCN, OPN, ALP and calcium deposits, which were measured using different assays: flow cytometry, ELISA and ARS staining [26,32,33].
5. Conclusions
Our results conclude that BiodentineTM mediates the odonto/osteogenic differentiation of hDPSCs in vitro by using different signaling pathways as the following: the BMP osteogenic signaling pathway, the activation of the extracellular matrix-related gene (ECMG) and the Ca2+ signaling pathway, in addition to enhancing the secretion of odonto/osteogenic markers, OPN, OCN and ALP. Therefore, BiodentineTM can provide a very promising and useful means for applying regenerative medicine to regenerate dentine tissue in clinical settings.
However the effect of BD on the odonto/osteogenic differentiation process and its signaling networks is still contradictory. Therefore, more studies are required in order to find out the mechanism of action of BD by which it can affect the differentiation potential of these cells. Moreover, more in vitro studies on different cell types must be carried out to approve the role of BD on the odonto/osteogenesis pathways in addition to other cytotoxicity and biocompatibility studies since the main use of BD is based on the modulation of inflammation and enhancement of the regeneration of pulp tissue. Finally, in vivo studies are important to make sure that this binomial product (DPSCs-BiodentineTM) can display the same results when used in clinical applications.
Author Contributions
Conceptualization, D.A. and A.A.; formal analysis, D.A. and H.J.; methodology, D.A., R.Z., S.Z. and R.A.; project administration, H.J.; supervision, A.A.; writing—original draft, R.Z. and N.A.; writing—review and editing, D.A. and R.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board from the Cell Therapy Center/The University of Jordan (IRB/06/2018) and approved on 13 March 2018.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mason, C.; Dunnill, P. A brief definition of regenerative medicine. Regen. Med. 2008, 3, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Abou Neel, E.A.; Chrzanowski, W.; Salih, V.M.; Kim, H.-W.; Knowles, J.C. Tissue engineering in dentistry. J. Dent. 2014, 42, 915–928. [Google Scholar] [CrossRef] [Green Version]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. SHED: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, B.-M.; Miura, M.; Gronthos, S.; Bartold, P.M.; Batouli, S.; Brahim, J.; Young, M.; Robey, P.G.; Wang, C.Y.; Shi, S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004, 364, 149–155. [Google Scholar] [CrossRef]
- Sonoyama, W.; Liu, Y.; Yamaza, T.; Tuan, R.S.; Wang, S.; Shi, S.; Huang, G.T.-J. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: A pilot study. J. Endod. 2008, 34, 166–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.-J.; Gronthos, S.; Shi, S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: Their biology and role in regenerative medicine. J. Dent. Res. 2009, 88, 792–806. [Google Scholar] [CrossRef]
- Tatullo, M.; Marrelli, M.; Paduano, F. The regenerative medicine in oral and maxillofacial surgery: The most important innovations in the clinical application of mesenchymal stem cells. Int. J. Med Sci. 2015, 12, 72. [Google Scholar] [CrossRef] [Green Version]
- Mitsiadis, T.; Orsini, G.; Jimenez-Rojo, L. Stem cell-based approaches in dentistry. Eur. Cells Mater. 2015, 30, 248–257. [Google Scholar] [CrossRef]
- Pierdomenico, L.; Bonsi, L.; Calvitti, M.; Rondelli, D.; Arpinati, M.; Chirumbolo, G.; Becchetti, E.; Marchionni, C.; Alviano, F.; Fossati, V. Multipotent mesenchymal stem cells with immunosuppressive activity can be easily isolated from dental pulp. Transplantation 2005, 80, 836–842. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Walboomers, X.F.; Shi, S.; Fan, M.; Jansen, J.A. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Eng. 2006, 12, 2813–2823. [Google Scholar] [CrossRef]
- Huang, G.T.-J.; Yamaza, T.; Shea, L.D.; Djouad, F.; Kuhn, N.Z.; Tuan, R.S.; Shi, S. Stem/progenitor cell–mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng. Part A 2010, 16, 605–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iohara, K.; Murakami, M.; Takeuchi, N.; Osako, Y.; Ito, M.; Ishizaka, R.; Utunomiya, S.; Nakamura, H.; Matsushita, K.; Nakashima, M. A novel combinatorial therapy with pulp stem cells and granulocyte colony—Stimulating factor for total pulp regeneration. Stem Cells Transl. Med. 2013, 2, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Davies, O.; Cooper, P.; Shelton, R.; Smith, A.; Scheven, B. A comparison of the in vitro mineralisation and dentinogenic potential of mesenchymal stem cells derived from adipose tissue, bone marrow and dental pulp. J. Bone Miner. Metab. 2015, 33, 371–382. [Google Scholar] [CrossRef]
- Henkel, J.; Woodruff, M.A.; Epari, D.R.; Steck, R.; Glatt, V.; Dickinson, I.C.; Choong, P.F.; Schuetz, M.A.; Hutmacher, D.W. Bone regeneration based on tissue engineering conceptions—A 21st century perspective. Bone Res. 2013, 1, 216–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bose, S.; Roy, M.; Bandyopadhyay, A. Recent advances in bone tissue engineering scaffolds. Trends Biotechnol. 2012, 30, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Kinane, D.; Stathopoulou, P.; Papapanou, P. Periodontal Diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef]
- Grayson, W.L.; Bunnell, B.A.; Martin, E.; Frazier, T.; Hung, B.P.; Gimble, J.M. Stromal cells and stem cells in clinical bone regeneration. Nat. Rev. Endocrinol. 2015, 11, 140. [Google Scholar] [CrossRef]
- Loeffler, J.; Duda, G.N.; Sass, F.A.; Dienelt, A. The metabolic microenvironment steers bone tissue regeneration. Trends Endocrinol. Metab. 2018, 29, 99–110. [Google Scholar] [CrossRef]
- Gao, C.; Peng, S.; Feng, P.; Shuai, C. Bone biomaterials and interactions with stem cells. Bone Res. 2017, 5, 17059. [Google Scholar] [CrossRef]
- Moioli, E.K.; Clark, P.A.; Xin, X.; Lal, S.; Mao, J.J. Matrices and scaffolds for drug delivery in dental, oral and craniofacial tissue engineering. Adv. Drug Deliv. Rev. 2007, 59, 308–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Z.; Li, D.; Kohli, M.R.; Yu, Q.; Kim, S.; He, W.-x. Effect of Biodentine™ on the proliferation, migration and adhesion of human dental pulp stem cells. J. Dent. 2014, 42, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Amano, K.; Iohara, K.; Ito, M.; Imabayashi, K.; Into, T.; Matsushita, K.; Nakamura, H.; Nakashima, M. Matrix metalloproteinase-3 accelerates wound healing following dental pulp injury. Am. J. Pathol. 2009, 175, 1905–1914. [Google Scholar] [CrossRef] [Green Version]
- Palma, P.J.; Marques, J.A.; Santos, J.; Falacho, R.I.; Sequeira, D.; Diogo, P.; Caramelo, F.; Ramos, J.C.; Santos, J.M. Tooth discoloration after regenerative endodontic procedures with calcium silicate-based cements—An ex vivo study. Appl. Sci. 2020, 10, 5793. [Google Scholar] [CrossRef]
- Sequeira, D.B.; Oliveira, A.R.; Seabra, C.M.; Palma, P.J.; Ramos, C.; Figueiredo, M.H.; Santos, A.C.; Cardoso, A.L.; Peça, J.; Santos, J.M. Regeneration of pulp-dentin complex using human stem cells of the apical papilla: In vivo interaction with two bioactive materials. Clin. Oral Investig. 2021, 1–13. [Google Scholar] [CrossRef]
- Luo, Z.; Kohli, M.R.; Yu, Q.; Kim, S.; Qu, T.; He, W.-X. Biodentine induces human dental pulp stem cell differentiation through mitogen-activated protein kinase and calcium-/calmodulin-dependent protein kinase II pathways. J. Endod. 2014, 40, 937–942. [Google Scholar] [CrossRef]
- Laurent, P.; Camps, J.; About, I. BiodentineTM induces TGF-β1 release from human pulp cells and early dental pulp mineralization. Int. Endod. J. 2012, 45, 439–448. [Google Scholar] [CrossRef]
- Loison-Robert, L.S.; Tassin, M.; Bonte, E.; Berbar, T.; Isaac, J.; Berdal, A.; Simon, S.; Fournier, B.P. In vitro effects of two silicate-based materials, Biodentine and BioRoot RCS, on dental pulp stem cells in models of reactionary and reparative dentinogenesis. PLoS ONE 2018, 13, e0190014. [Google Scholar] [CrossRef] [Green Version]
- Abuarqoub, D.; Aslam, N.; Jafar, H.; Abu Harfil, Z.; Awidi, A. Biocompatibility of biodentine™® with periodontal ligament stem cells: In vitro study. Dent. J. 2020, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Hasweh, N.; Awidi, A.; Rajab, L.; Hiyasat, A.; Jafar, H.; Islam, N.; Hasan, M.; Abuarqoub, D. Characterization of the biological effect of BiodentineTM on primary dental pulp stem cells. Indian J. Dent. Res. 2018, 29, 787. [Google Scholar] [PubMed]
- Chiang, Y.-C.; Chang, H.-H.; Wong, C.-C.; Wang, Y.-P.; Wang, Y.-L.; Huang, W.-H.; Lin, C.-P. Nanocrystalline calcium sulfate/hydroxyapatite biphasic compound as a TGF-β1/VEGF reservoir for vital pulp therapy. Dent. Mater. 2016, 32, 1197–1208. [Google Scholar] [CrossRef]
- Saberi, E.; Farhad-Mollashahi, N.; Aval, F.S.; Saberi, M. Proliferation, odontogenic/osteogenic differentiation, and cytokine production by human stem cells of the apical papilla induced by biomaterials: A comparative study. Clin. Cosmet. Investig. Dent. 2019, 11, 181. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Xu, C.; Du, R.; Wen, Y.; Chang, J.; Huan, Z.; Zhu, Y. Effects of silicate-based composite material on the proliferation and mineralization behaviors of human dental pulp cells: An in vitro assessment. Dent. Mater. J. 2018, 37, 889–896. [Google Scholar] [CrossRef] [Green Version]
- Luu, H.H.; Song, W.X.; Luo, X.; Manning, D.; Luo, J.; Deng, Z.L.; Sharff, K.A.; Montag, A.G.; Haydon, R.C.; He, T.C. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J. Orthop. Res. 2007, 25, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Wei, A.; Whittaker, S.; Williams, L.A.; Tao, H.; Ma, D.D.; Diwan, A.D. The role of BMP-7 in chondrogenic and osteogenic differentiation of human bone marrow multipotent mesenchymal stromal cells in vitro. J. Cell. Biochem. 2010, 109, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Chen, G.; Li, Y.-P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef] [PubMed]
- Bigdeli, N.; de Peppo, G.M.; Lennerås, M.; Sjövall, P.; Lindahl, A.; Hyllner, J.; Karlsson, C. Superior osteogenic capacity of human embryonic stem cells adapted to matrix-free growth compared to human mesenchymal stem cells. Tissue Eng. Part A 2010, 16, 3427–3440. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Ishizuya, T.; Kintou, N.; Wada, Y.; Katagiri, T.; Wozney, J.M.; Rosen, V.; Yoshiki, S. Effects of BMP-2, BMP-4, and BMP-6 on osteoblastic differentiation of bone marrow-derived stromal cell lines, ST2 and MC3T3-G2/PA6. Biochem. Biophys. Res. Commun. 1996, 220, 366–371. [Google Scholar] [CrossRef]
- Kokabu, S.; Gamer, L.; Cox, K.; Lowery, J.; Tsuji, K.; Raz, R.; Economides, A.; Katagiri, T.; Rosen, V. BMP3 suppresses osteoblast differentiation of bone marrow stromal cells via interaction with Acvr2b. Mol. Endocrinol. 2012, 26, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Rathinam, E.; Govindarajan, S.; Rajasekharan, S.; Declercq, H.; Elewaut, D.; De Coster, P.; Martens, L. Transcriptomic profiling of human dental pulp cells treated with tricalcium silicate–based cements by RNA sequencing. Clin. Oral Investig. 2020, 25, 3181–3195. [Google Scholar] [CrossRef] [PubMed]
- Doshi, R.; Kulkarni, U.; Shinde, S.; Sabane, A.; Patil, A. Role of genes in odontogenesis. J. Adv. Med. Med. Res. 2016, 14, 1–9. [Google Scholar] [CrossRef]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef] [Green Version]
- Lian, J.B.; Stein, G.S. Concepts of osteoblast growth and differentiation: Basis for modulation of bone cell development and tissue formation. Crit. Rev. Oral Biol. Med. 1992, 3, 269–305. [Google Scholar] [CrossRef] [Green Version]
- Castano-Izquierdo, H.; Álvarez-Barreto, J.; Dolder, J.V.D.; Jansen, J.A.; Mikos, A.G.; Sikavitsas, V.I. Preculture period of mesenchymal stem cells in osteogenic media influences their in vivo bone forming potential. J. Biomed. Mater. Res. Part A 2007, 82, 129–138. [Google Scholar] [CrossRef]
- Colnot, C.I.; Helms, J.A. A molecular analysis of matrix remodeling and angiogenesis during long bone development. Mech. Dev. 2001, 100, 245–250. [Google Scholar] [CrossRef]
- Knuth, C.; Andres Sastre, E.; Fahy, N.; Witte-Bouma, J.; Ridwan, Y.; Strabbing, E.; Koudstaal, M.; van de Peppel, J.; Wolvius, E.; Narcisi, R. Collagen type X is essential for successful mesenchymal stem cell-mediated cartilage formation and subsequent endochondral ossification. Eur. Cells Mater. 2019, 38, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Bautista, C.M.; Mohan, S.; Baylink, D.J. Insulin-like growth factors I and II are present in the skeletal tissues of ten vertebrates. Metabolism 1990, 39, 96–100. [Google Scholar] [CrossRef]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [Green Version]
- Kirkham, G.; Cartmell, S. Genes and proteins involved in the regulation of osteogenesis. Top. Tissue Eng. 2007, 3, 1–22. [Google Scholar]
- Sandhu, S.V.; Gupta, S.; Bansal, H.; Singla, K. Collagen in health and disease. J. Orofac. Res. 2012, 2, 153–159. [Google Scholar] [CrossRef]
- Leiendecker, A.P.; Qi, Y.-P.; Sawyer, A.N.; Niu, L.-N.; Agee, K.A.; Loushine, R.J.; Weller, R.N.; Pashley, D.H.; Tay, F.R. Effects of calcium silicate–based materials on collagen matrix integrity of mineralized dentin. J. Endod. 2012, 38, 829–833. [Google Scholar] [CrossRef]
- Widbiller, M.; Lindner, S.; Buchalla, W.; Eidt, A.; Hiller, K.-A.; Schmalz, G.; Galler, K. Three-dimensional culture of dental pulp stem cells in direct contact to tricalcium silicate cements. Clin. Oral Investig. 2016, 20, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Egusa, H.; Iida, K.; Kobayashi, M.; Lin, T.Y.; Zhu, M.; Zuk, P.A.; Wang, C.J.; Thakor, D.K.; Hedrick, M.H.; Nishimura, I. Downregulation of extracellular matrix-related gene clusters during osteogenic differentiation of human bone marrow-and adipose tissue-derived stromal cells. Tissue Eng. 2007, 13, 2589–2600. [Google Scholar] [CrossRef] [PubMed]
- Thesleff, I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J. Cell Sci. 2003, 116, 1647–1648. [Google Scholar] [CrossRef] [Green Version]
- Hayrapetyan, A.; Jansen, J.A.; van den Beucken, J.J. Signaling pathways involved in osteogenesis and their application for bone regenerative medicine. Tissue Eng. Part B Rev. 2015, 21, 75–87. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).