Featured Application
The use by medical of a neural network classification system for ECG signals with preprocessing steps to recognize atrial fibrillation.
Abstract
Today, cardiovascular disease is the leading cause of death in developed countries. The most common arrhythmia is atrial fibrillation, which increases the risk of ischemic stroke. An electrocardiogram is one of the best methods for diagnosing cardiac arrhythmias. Often, the signals of the electrocardiogram are distorted by noises of varying nature. In this paper, we propose a neural network classification system for electrocardiogram signals based on the Long Short-Term Memory neural network architecture with a preprocessing stage. Signal preprocessing was carried out using a symlet wavelet filter with further application of the instantaneous frequency and spectral entropy functions. For the experimental part of the article, electrocardiogram signals were selected from the open database PhysioNet Computing in Cardiology Challenge 2017 (CinC Challenge). The simulation was carried out using the MatLab 2020b software package for solving technical calculations. The best simulation result was obtained using a symlet with five coefficients and made it possible to achieve an accuracy of 87.5% in recognizing electrocardiogram signals.
1. Introduction
The number of people who suffer from cardiac diseases is increasing every day. This disease is the leading cause of death in developed countries [1,2,3]. Electrocardiography is a method of recording and studying the electric fields that are generated during the work of the heart. An ECG is the result of electrocardiography [4,5] and is a graphical record of the electrical activity of the heart produced by depolarization and repolarization of the atria and ventricles. The electrocardiogram (ECG) is a non-invasive technique used to detect cardiovascular disease. The ECG is described by the waveforms of the P, QRS, and T waves, which are associated with each heart-rate function. The P wave displays the process of depolarization of the atrial myocardium; the QRS complex displays depolarization of the ventricles; the ST segment, and the T wave displays the processes of repolarization of the ventricular myocardium [6]. Figure 1 shows an example of an electrocardiogram waveform with P, Q, R, S, and T characteristics, as well as standard electrocardiogram intervals are PQ intervals, ST intervals, and QRS complex.
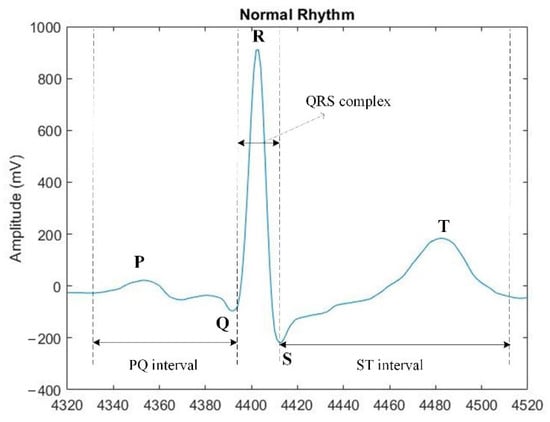
Figure 1.
Example of an electrocardiogram signal.
Atrial fibrillation is a major risk factor for ischemic stroke [7]. The main criteria for the presence of atrial fibrillation are the absence of P waves, the presence of atrial fibrillation waves, different R-R intervals, the heart-rate (HR) is constant or accelerated, and the QRS complex is less than 0.12 s [8]. Since the P wave is not detected during atrial fibrillation, the interval between QRS complexes increases and there is no possibility to calculate the PQ and QT intervals. Calculating HR or QRS time from digital ECG signals is problematic. Therefore, it is necessary to pay attention to the absence of the P-peak and the presence of different intervals between R-R peaks when determining atrial fibrillation.
In some modern electrocardiographs, various signal filters are used, which allow obtainment of a higher quality of the electrocardiogram, while introducing some distortions in the form of the received signal. Low-pass filters in the range of 0.5–1 Hz can reduce the effect of the floating contour while introducing distortions in the shape of the ST segment [4]. A low-frequency anti-tremor filter in the 35 Hz range suppresses artifacts associated with muscle activity. A notch filter in the range of 50–60 Hz neutralizes line pickups [5].
Today, learning algorithms are becoming more accurate, but recognition systems created based on artificial intelligence in medicine, and in particular, in cardiology, are not able to achieve a 100% accurate result [9]. For this reason, it is relevant to search for ways to increase this indicator. One of the possible ways to increase this indicator is the preliminary processing of ECG signals. Finding peaks in signals is an important step in many automatic ECG processing systems. In this work, a neural network classification system for ECG signals is proposed for determining atrial fibrillation with a preprocessing stage. At the stage of preprocessing, signals are selected by the number of heartbeat counts, as well as wavelet analysis to clean up noise and isolate the R-peak and spectral analysis to isolate the P-peak. Automatic detection of atrial fibrillation from ECG signals will allow doctors to determine if a patient needs cardiac care.
2. Related Research
Today, medicine is considered one of the promising areas for the introduction of artificial intelligence, because the analysis of medical signals is the most common research method in this area. An example is an algorithm for identifying patients with atrial fibrillation in sinus rhythm based on convolutional neural networks (CNN) in [10]. The work [11] presents a hybrid approach with the use of Empirical Mode Decomposition and CNN to classify ECG signals. In [12], the author has implemented the extraction of ECG signal features based on wavelet transform with further classification using Long Short-Term Memory (LSTM). The work [13] presents a method for detecting atrial fibrillation using LSTM. The method achieved an accuracy of 98.51%. The essence of the approach proposed in the work is the separation of ECG signals with a sliding window and the loading of the obtained blocks into the decision-making system. ECG signals were taken from the MIT-BIH database Atrial Fibrillation Database. Dataset was 10 h long and each signal contained 100 heartbeats (R-R peak). The authors point out the slow learning speed and high requirements for computing resources. For the simulation, the authors used the Quadro M5000 GPU (Nvidia, Santa Clara, CA, USA).
This work is of scientific interest, however, due to the different methodology and resources used, it cannot be compared with the proposed system for neural network determination of atrial fibrillation on ECG signals with wavelet-based preprocessing.
Finding peaks in signals is an important step in many signal processing applications. Automatic peak detection using neural network classification systems is difficult due to the physiological variability of P waves and QRS complexes, as well as the presence of various types of noise, including muscle noise, artifacts due to electrode movement, power-line noise, and baseline deviations.
Software QRS complex recognition is an integral part of modern computerized ECG monitoring systems. An algorithm for their recognition is presented in [14] and is based on optimized filtering and simple threshold setting since optimized filtering is considered a factor in achieving good timing accuracy. The most widely known method for detecting a single R-peak is the Pan-Tomkins method, which uses three types of processing steps: linear digital filtering, nonlinear transformation, and decision rule algorithms [15]. The work [16] presents an algorithm for detecting QRS using multi-stage morphological filtering to suppress impulse noise. In work [17], to determine the QRS complex, it is proposed to create an estimated QRS signal using the parameters extracted from the original ECG signal.
For automatic disease diagnosis systems based on ECG signals, accurate determination of the P wave is critical. In work [18], one of the first methods of processing ECG signals to isolate the P-peak in its flow using wavelet transform and subsequent training of the neural network is described. In [3], a system for determining the P and T waves based on the wavelet transform is presented.
The main methods of processing ECG signals for noise reduction are digital filtering, adaptive filtering, wavelet filtering. The paper [19] describes a method for processing ECG signals using an adaptive wavelet transform based on the Poincare section and the Shannon method. There are also methods for processing signals online in real-time for use in pacemakers. The paper [20] describes a processing method using biorthogonal wavelet transform based on a linear phase structure for noise removal, feature extraction, and compression of the ECG signal. There are also methods for computerized detection of fibrillation. Work [21] describes a method for detecting fibrillation using an eight-layer neural convolutional network, which requires only basic data normalization without preliminary processing and extraction of features from raw ECG samples. Work [7] describes a method for constructing classifiers based on several sets of functions (a set of Andreotti, Zabikhi functions, an aggregated set of functions, and a set of Dutt functions) and using a random forest classification method.
3. Materials and Methods
3.1. Neural Network System for Atrial Fibrillation Recognition by ECG Signal
Any digital signal is distorted by noises of varying nature. Noise on the ECG signals makes it difficult to analyze the data, both for the specialist and for systems based on artificial intelligence. Signal preprocessing allows the ECG to be prepared for further classification. This paper proposes a system for determining atrial fibrillation by ECG signals, which includes four stages presented in Figure 2. The first stage consists of the preprocessing of signals to isolate signals with the same count of heartbeats. At the second stage, noise is removed from the ECG signals using a discrete wavelet transform. At the third stage, the P-peak is isolated using spectral analysis. The fourth step is to classify signals using the LSTM network.
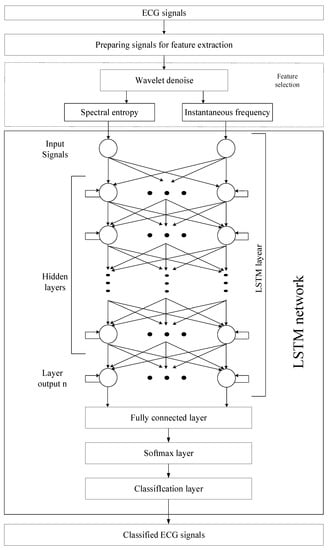
Figure 2.
Neural network classification system with the pre-processing stage for ECG signals.
3.2. Method for Pre-Processing of ECG Signals
As part of the preprocessing of the ECG signal, its length is checked. Each ECG signal has a specific number of heartbeats and several samples. The length of ECG signals is measured in the number of samples that make up the signal. The ECG signal database can contain signals with a different number of samples. For the correct operation of the neural network classification system, the number of samples must be the same for all signals. To select ECG signals with one length, the following steps are necessary. At the first stage, the ECG signals must be divided into groups with the same number of samples. The second step is to select a group consisting of the largest number of signals of the same length. The third step removes signals consisting of fewer and more samples than in the selected group.
3.3. Removing Noise from ECG Signals Using a Discrete Wavelet Transform
Any digital signal is distorted by noises of varying nature. To isolate signal features, it is necessary to clean noise from them. An ECG signal that is distorted by noise can be written as:
where is the ECG signal, is the ECG signal without noise distortion, is the noise on the ECG signal.
Wavelet transform is a common way to remove noise from a signal [22]. To clean the noise from the ECG signals, a discrete wavelet transform (DWT) of the symlet family was used, which is a Daubechies wavelet with the least asymmetry and a compact carrier. The detail factor for each case is set empirically.
There are three stages of using wavelet transform to clean the noise from the ECG signal. The first step is to obtain noisy wavelet coefficients using the DWT of a noisy signal. The second stage is the choice of thresholding. The third stage is an inverse wavelet transform to obtain a purified signal [23]. The DWT of the ECG signal is:
where is the number of samples on the ECG signal, is the ECG signal distorted by noise, is a symlet, and variables can take on the values , .
To obtain DWT, a low-pass analysis filter with an impulse response and a high-pass analysis filter with an impulse response are used. As a result of filtering, approximating and detailing coefficients are obtained [24].
With each DWT application, the number of samples on the ECG signal is halved by Table 1 [24,25]. Each subsequent decomposition is carried out in terms of the low-frequency component by Figure 3.

Table 1.
Length of ECG signals in DWT, where is the initial number of samples on the ECG signal.
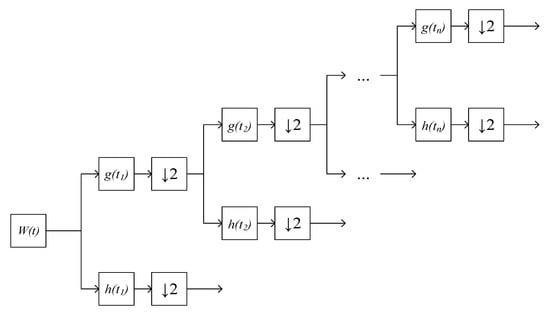
Figure 3.
DWT ECG signal, where is an ECG signal distorted by noise, is a decimation, is a low-pass analysis filter, is a high-pass analysis filter.
The next step in clearing noise from the ECG signal is to select a threshold function and thresholding. The thresholding is a value that determines whether there is noise in the signal. If the value of the wavelet coefficient at a certain moment is greater than the value of the threshold, then it is considered the value of the signal, if less, then the noise [24]. To determine the threshold limit of the ECG signal, the minimax threshold was used [26]:
The wavelet coefficients are transformed by thresholding, using the threshold functions. The functions of hard- and soft-threshold determination are most often used [24]. To determine the noise on the ECG signal, the soft-threshold function was used [26]:
where is the value of the wavelet coefficient.
The reverse DWT for obtaining a cleaned ECG signal is as follows [24]:
where is the ECG signal without noise distortion, and are approximating and detailing coefficients after processing by the threshold function. The reverse DWT of the ECG signal is performed according to Figure 4.
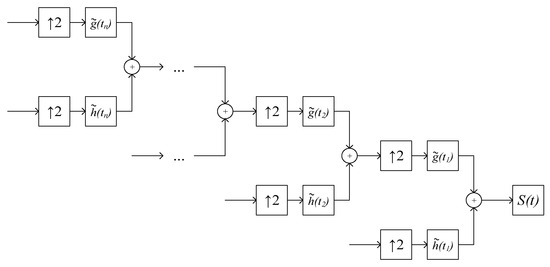
Figure 4.
Reverse DWT ECG signal, where is a noise-free ECG signal, is interpolation, is a low-pass synthesis filter, is a high-pass synthesis filter.
3.4. Isolation of the P-Peak Feature Using Spectral Analysis
The instantaneous frequency and spectral entropy functions were selected to isolate the P-peak. To calculate it, it is necessary to calculate the amplitude spectrum of the process using the Fourier transform, then normalize the amplitude spectrum so that the sum of its readings becomes equal to 1 and calculate the entropy using Shannon’s formula. Changes in spectral entropy over time are associated with changes in the waveform, which allows its use to distinguish features on the ECG signal. Since the ECG signal consists of a finite number of samples, Shannon’s formula for calculating the spectral entropy of the ECG signal is:
where is the amount of information, is the number of possible events, is the value of the i-th samples on the ECG signal. However, it is more correct to use the Fourier transform when working with stationary signals. Therefore, for more accurate identification of the P-peak on the ECG signal, several signal-processing methods are required. With a deviation in the work of the heart, changes in the frequency of the ECG signals occur. To determine such changes, an instantaneous frequency is used, since this method allows the researcher to take into account the nature of the process, which changes over time [27].
As the ECG signal is non-stationary, in order to calculate the instantaneous frequency, the researcher can refer to the works of Carson and Fry [28] and Van de Pol [29]:
where is an ECG signal. Formula (9) describes the rate of change in the phase of the ECG signal, i.e., the instantaneous frequency shows how often the peaks appear and disappear.
3.5. LSTM Processing of ECG Data
LSTM networks have been specifically designed to find patterns over time [30]. Since ECG signals are sequences of peaks, the ability to memorize characteristic fragments of time series is critical when using LSTM in this area. Long-term and short-term memory is the main reason that the LSTM network is used as the basic structure of ECG signal recognition systems. In the present study, the deep LSTM network was used to classify ECG signals.
Passing a signal through a standard LSTM network structure involves four stages. The first stage is the passage of the signal through the sigmoidal layer, which is designed to determine the desired information. The second stage consists of passing through the sigmoidal layer, which determines whether this or that information is relevant and transmits a signal to the hyperbolic layer, which defines a new vector of candidates. The third stage is to save a new vector of candidates. The fourth stage consists of passing through the sigmoidal layer, which is designed to determine the required information, and the hyperbolic tangent to display information in the range [−1; 1]. Figure 5 shows a typical LSTM structure.
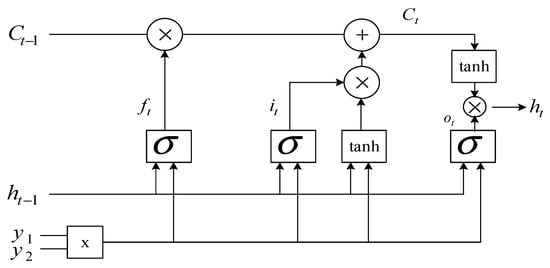
Figure 5.
Standard LSTM structure.
To classify ECG signals, it is proposed to use an LSTM network with two input layers and , which are combined into a two-dimensional vector . All calculations are performed according to the standard LSTM structure, which is shown in Figure 3, where is the input two-dimensional vector, is the output vector from the previous LSTM () block, is the output vector of the LSTM block, is the state vector from the previous LSTM () block, is the state vector of the LSTM block, is the sigmoid activation function, is the hyperbolic tangent activation function, is the multiplication operator, is the addition operator.
The forget gate vector is the result of a computational step through the sigmoidal layer, which is intended to determine the desired information. The resulting vector determines what information needs to be “memorized” and is calculated by the formula:
where is a matrix of parameters, is a vector of parameters. Update gate vector checks the relevance of the information using the sigmoidal activation function:
where is a matrix of parameters, is a vector of parameters.
The next step is to define a new state vector.
where is a matrix of parameters, is a vector of parameters, is the multiplication operator.
The output gate vector that is a candidate for leaving the LSTM network is calculated by the formula:
where is a matrix of parameters, is a vector of parameters. The definition of the output vector is made according to the formula:
The rest of the LSTM network layers are standard and are used in neural networks to classify signals.
4. Results
For modeling, ECG signals were selected from the international open database PhysioNet Computing in Cardiology Challenge 2017 (CinC Challenge) [31]. This database contains over 10,000 ECG records; it is freely available from AliveCor and is a random sample of patient records of no more than one minute in duration. The Physionet Computing in Cardiology Challenge 2017 database consists of 8528 ECG signals for training and 3658 ECG signals for validation. The base consists of four types of single-channel signals: 5152 normal signals (N), 771 signals with cardiac fibrillation (A), 46 noisy signals (~) and 2557 other signals (O). The simulations were performed using two categories of signals, namely signals without heart defects (N) and signals with atrial fibrillation (A). These signal categories were selected to study the signs of atrial fibrillation on ECG signals for more correct LSTM learning. A total of 1000 signals were selected from the database CinC Challenge for the first modeling. For the second experimental simulation, 5925 ECG signals were selected, namely 5152 normal signals (N) and 771 signals with cardiac fibrillation (A). Examples of selected ECG signals from the database CinC Challenge are shown in Figure 6.
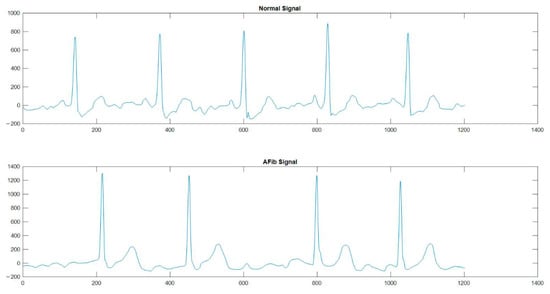
Figure 6.
An example of ECG signals without heart defects (Normal Signal) and signals with atrial fibrillation (AFib signal) from the CinC Challenge database [31].
The simulation was carried out using the MatLab 2020b software package for solving technical calculations. The calculations were performed on a PC with a processor Intel(R) Core (TM) i5-10210U CPU @ 1.60 GHz (8 CPUs), 2.1 GHz.
Database PhysioNet Computing in Cardiology Challenge 2017 (CinC Challenge) consists of ECG signals with different numbers of samples. Figure 7 shows that there are significantly more signals from 9000 samples. For correct training of the neural network, the input ECG signals must contain the same number of samples. Therefore, at the stage of preliminary data processing for the first training, 976 ECG signals with several samples to 9000 were selected. For the second simulation, 5754 ECG signals with 9000 samples were taken at the preprocessing stage.
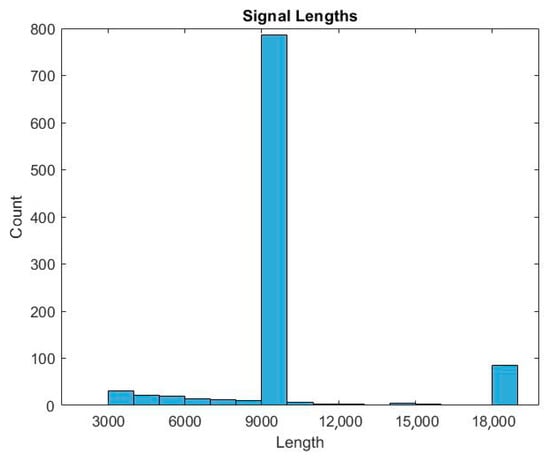
Figure 7.
Histogram of the ratio of the number of samples to the number of signals.
DWT allows splitting the signal into high and low frequencies. Analysis of high frequencies of the ECG signal can determine the presence of peaks. Analysis of low frequencies of the ECG signal allows determining the presence of noise of varying nature [22]. The symlet is an orthogonal wavelet and can be used to reconstruct the signal [23] or to find R-peaks on ECG signals [24,32]. To remove noise from ECG signals and isolate R-peaks on them, the symlet wavelet filter was chosen. The symlet is similar in shape to the QRS complex on the ECG signal. This means that the decomposition coefficients of the ECG signal using the symlet-based DWT have a high correlation with the location of the P-peaks. Their number can be determined based on the analysis of the coefficients of the wavelet decomposition of the ECG signal. Figure 8 shows a graphical display of the symlet and QRS complex. Figure 9 shows examples of the five-level decomposition of an ECG signal based on a symlet. The spikes in coefficients D 2, D 3 and D 4 correspond to heartbeats, from which it is possible to determine the location of the R-peaks of the cardiogram.
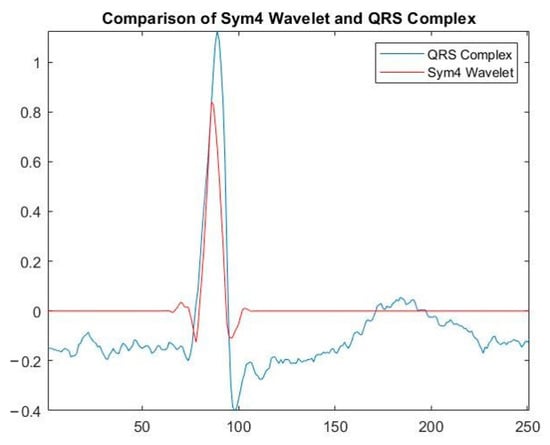
Figure 8.
Symlet repeating the shape of the R-peak on the ECG signal.
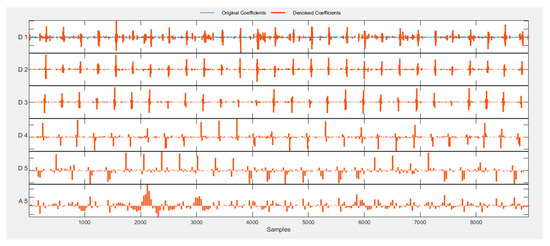
Figure 9.
An example of a five-level decomposition of an ECG signal based on a symlet.
The results from Table 2 were obtained by simulating the proposed system using various symlet. Each value of the table is the result of training the proposed system for neural network determination of atrial fibrillation based on LSTM with signal preprocessing using different coefficients of symlet. The best learning result was obtained using a five-coefficient symlet.

Table 2.
Simulation of the proposed method using a wavelet symlet with different coefficients.
For the correct selection of various signs on the ECG signals, one-dimensional functions must be used. The functions of instantaneous frequency and spectral entropy were selected to isolate the P-peak. Fourier series and integral Fourier transform are the basis of harmonic signal analysis. However, when analyzing ECG signals, these transformations do not provide the possibility of analyzing peaks, understanding the local properties of the signal and its frequency characteristics. Therefore, for these purposes, we used the characteristics of the Fourier spectrum. Instantaneous frequency calculates a spectrogram using short-term Fourier transforms versus window time. Spectral entropy estimates entropy based on a power spectrogram. The time output of the function corresponds to the center of the time windows.
The selected ECG signals from the CinC Challenge database were divided into signals for training and signals for testing in a percentage ratio of 90:10. For training, the architecture of the LSTM neural network was assembled. The network consisted of two input layers, to which preprocessed signals were applied, and one hundred hidden recurrent layers. Preprocessing of signals using a symlet and subsequent application of spectral analysis functions made it possible to reduce the length of ECG signals to 255 HC. Table 3 presents the results of modeling various methods for detecting atrial fibrillation. The matrix of inaccuracies as a result of training the proposed system of neural network determination of atrial fibrillation from ECG signals is presented in Figure 10 and Figure 11.

Table 3.
Simulation results of various methods for detecting atrial fibrillation.
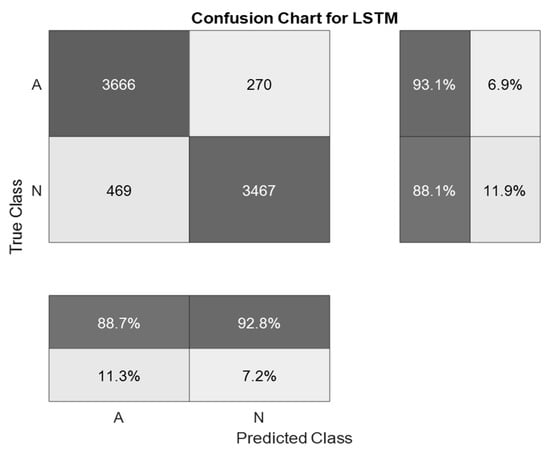
Figure 10.
A confusion matrix is a result of training a system for neural network determination of atrial fibrillation on ECG signals with wavelet-based preprocessing.
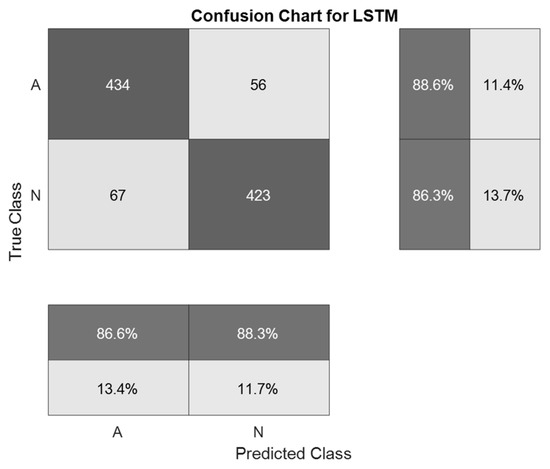
Figure 11.
A confusion matrix is a result of the validation of a system for neural network determination of atrial fibrillation on ECG signals with wavelet-based preprocessing.
The best indicator of the accuracy of ECG signal recognition was obtained using the method proposed in the work and amounted to 87.5%. This accuracy was obtained in the first simulation using 976 ECG signals. The indicator of the accuracy of ECG signal recognition during the second simulation was 87.4%. This result is identical to that obtained in the first simulation. The smallest indicator of recognition accuracy was obtained using a one-dimensional LSTM without preliminary signal processing. The results obtained indicate that the use of pre-processing of ECG signals can significantly increase the recognition accuracy of neural network classification systems.
5. Discussion
The article presents a system for neural network determination of atrial fibrillation on ECG signals with wavelet-based preprocessing. Simulation of the proposed system made it possible to achieve a recognition accuracy of 87.5%, which is significantly higher than the 79.0–82.0% level that can be achieved using known systems [33,34,35].
Work [33] is devoted to the development of a smartphone application for the detection of atrial fibrillation. The method proposed in [33] uses preliminary preprocessing of the ECG signal, which includes the analysis of the R-R and P-peak intervals. Despite the identity of the methods used, the authors of the work indicate the accuracy of the system at 79.0%, which is significantly lower than the result obtained by modeling the system proposed in this work.
The authors of [34] used the training of a recurrent neural network without preliminary signal processing and achieved an accuracy of 82.0%. The recurrent network is the basis of the LSTM network used in our method, which makes it possible to make comparisons.
In [35], the authors used a part of the MIT-BH 2017 database consisting of normal ECG signals, signals with fibrillation, and “others.” A group of noisy signals was not used. For the experiment, the authors used signals with a length of 4 heart counts (4 R-R intervals). The method proposed in [35] did not give a positive result for signals of the “other” category. The results shown for determining the presence of fibrillation from groups with normal signals and signals with fibrillations were 82.0% accuracy, which is also lower than the result of the accuracy of the proposed system for neural network determination of atrial fibrillation on ECG signals.
There are other ways to measure atrial fibrillation. In [36], a more complex methodology is used that requires more computing power. Using multiple convolutional neural networks and LSTM networks is a resource-intensive method. At the same time, the use of pre-processing of the signal before training the neural network can reduce resource costs.
The proposed stages of preliminary processing of ECG signals made it possible to prepare data for further analysis to conduct an automated determination of atrial fibrillation. The average accuracy of a cardiologist’s diagnosis by visual analysis of ECG signals is 65.0–70.0% [37]. The use of the proposed neural network system for determining atrial fibrillation from ECG signals by specialists will make it possible to increase the efficiency of diagnostics in comparison with methods of visual diagnosis. The proposed system for neural network determination of atrial fibrillation on ECG signals can only be used as an additional diagnostic tool by specialists. This system is not a medical device and cannot independently diagnose patients.
A promising direction for further research is the construction of more complex systems for the neural network classification of ECG signals, using, together with the analysis of signals, various metadata about patients, such as age, gender, race, genetic predisposition, and other descriptors. One of the next steps for further research is the creation of a mobile application for real-time fibrillation detection. Additionally, further research plans include the complication of the proposed system by using convolutional neural networks to improve the accuracy of determining atrial fibrillation.
Author Contributions
Conceptualization, P.L.; methodology, P.L.; software, M.K.; validation, M.K.; formal analysis, P.L.; investigation, M.K.; resources, U.L.; data curation, P.L.; writing—original draft preparation, M.K.; writing—review and editing, U.L.; visualization, M.K.; supervision, U.L.; project administration, P.L.; funding acquisition, P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Foundation for Basic Research (project no. 19-07-00130 A) and by the Presidential Council for grants (project no. MK-3918.2021.1.6).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors are grateful to the North Caucasus Federal University for supporting the competition of scientific groups and individual scientists of the North Caucasus Federal University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, L.H.; Chen, T.Y.; Lee, S.Y.; Yang, T.H.; Huang, S.Y.; Wu, J.H.; Fang, Q. A wireless ECG acquisition SoC for body sen-sor network. In Proceedings of the 2012 IEEE Biomedical Circuits and Systems Conference (BioCAS), Taiwan, China, 28–30 November 2012; pp. 156–159. [Google Scholar]
- Buxi, D.; Berset, T.; Hijdra, M.; Tutelaers, M.; Geng, D.; Hulzink, J.; Van Noorloos, M.; Romero, I.; Torfs, T.; Van Helleputte, N. Wireless 3-lead ECG system with on-board digital signal processing for ambulatory monitoring. In Proceedings of the 2012 IEEE Biomedical Circuits and Systems Conference (BioCAS), Taiwan, China, 28–30 November 2012; pp. 308–311. [Google Scholar] [CrossRef]
- Martinez, J.P.; Almeida, R.; Olmos, S.; Rocha, A.P.; Laguna, P. A wavelet-based ECG delineator: Evaluation on standard da-tabases. IEEE Trans. Biomed. Eng. 2004, 51, 570–581. [Google Scholar] [CrossRef]
- Halhuber, M.J.; Günther, R.; Ciresa, M. Technique of ECG Recording. In ECG-An Introductory Course A Practical Introduction to Clinical Electrocardiography; Springer: Berlin/Heidelberg, Germany, 1979; pp. 141–145. [Google Scholar]
- Berbari, E.J.; Lander, P. The methods of recording and analysis of the signal averaged ECG. In Signal Averaged Electrocardiography; Springer: Dordrecht, The Netherland, 1993; pp. 49–68. [Google Scholar]
- Thirrunavukkarasu, R.R.; Meeradevi, T.; Ravi, A.; Ganesan, D.; Vadivel, G.P. Detection R Peak in Electrocardiogram Signal Using Daubechies Wavelet Transform and Shannon’s Energy Envelope. In Proceedings of the 2019 5th International Conference on Advanced Computing & Communication Systems (ICACCS), Coimbatore, India, 15–16 March 2019; pp. 1044–1048. [Google Scholar] [CrossRef]
- Lippi, G.; Sanchis-Gomar, F.; Cervellin, G. Global epidemiology of atrial fibrillation: An increasing epidemic and public health challenge. Int. J. Stroke 2020, 16, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Goldberger, Z.D.; Shivikin, A. Clinical Electrocardiography: A Simplified Approach E-Book; Elsevier Health Sciences: London, UK, 2017. [Google Scholar]
- Ahuja, A.S. The impact of artificial intelligence in medicine on the future role of the physician. PeerJ 2019, 7, e7702. [Google Scholar] [CrossRef]
- Huang, M.L.; Wu, Y.S. Classification of atrial fibrillation and normal sinus rhythm based on convolutional neural network. Biomed. Eng. Lett. 2020, 10, 183–193. [Google Scholar] [CrossRef]
- Hasan, N.I.; Bhattacharjee, A. Deep Learning Approach to Cardiovascular Disease Classification Employing Modified ECG Signal from Empirical Mode Decomposition. Biomed. Signal Process. Control. 2019, 52, 128–140. [Google Scholar] [CrossRef]
- Yildirim, Ö. A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput. Biol. Med. 2018, 96, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Shenfield, A.; Kareem, M.; San, T.R.; Fujita, H.; Acharya, U.R. Automated detection of atrial fibrillation using long short-term memory network with RR interval signals. Comput. Biol. Med. 2018, 102, 327–335. [Google Scholar] [CrossRef] [Green Version]
- Mundhe, P.; Pathrikar, A.K. An overview of implementation of efficient QRS Complex detector with FPGA. Int. J. Adv. Res. Comput. Commun. Eng. 2013, 2, 4041–4043. [Google Scholar]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 3, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lian, Y. QRS Detection Based on Multiscale Mathematical Morphology for Wearable ECG Devices in Body Area Networks. IEEE Trans. Biomed. Circuits Syst. 2009, 3, 220–228. [Google Scholar] [CrossRef]
- Abo-Zahhad, M.; Ahmed, S.M.; Zakaria, A. An Efficient Technique for Compressing ECG Signals Using QRS Detection, Estimation, and 2D DWT Coefficients Thresholding. Model. Simul. Eng. 2012, 2012, 1–10. [Google Scholar] [CrossRef]
- Anant, K.S.; Dowla, F.U.; Rodrigue, G.H. Detection of the electrocardiogram P-wave using wavelet analysis. Int. Soc. Opt. Photonics 1994, 2242, 744–750. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Bukkapatnam, S.; Komanduri, R. Nonlinear adaptive wavelet analysis of electrocardiogram signals. Phys. Rev. E 2007, 76, 026214. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Komaragiri, R.; Kumar, M. Design of wavelet transform based electrocardiogram monitoring system. ISA Trans. 2018, 80, 381–398. [Google Scholar] [CrossRef]
- Fujita, H.; Cimr, D. Computer Aided detection for fibrillations and flutters using deep convolutional neural network. Inf. Sci. 2019, 486, 231–239. [Google Scholar] [CrossRef]
- Bnou, K.; Raghay, S.; Hakim, A. A wavelet denoising approach based on unsupervised learning model. EURASIP J. Adv. Signal. Process. 2020, 2020, 1–26. [Google Scholar] [CrossRef]
- Chavan, M.S.; Mastorakis, N.; Chavan, M.N.; Gaikwad, M.S. Implementation of SYMLET wavelets to removal of Gaussian additive noise from speech signal. In Proceedings of the Recent Researches in Communications, Automation, Signal Processing, Nano-technology, Astronomy and Nuclear Physics: 10th WSEAS International Conference on Electronics, Hardware, Wireless and Optical Communications (EHAC’11), Cambridge, UK, 20–22 February 2011; p. 37. [Google Scholar]
- Luo, G.; Zhang, D. Wavelet Denoising. Adv. Wavelet Theory Appl. Eng. Phys. Technol. 2012, 634. [Google Scholar] [CrossRef] [Green Version]
- Zarei, A.; Asl, B.M. Automatic Detection of Obstructive Sleep Apnea Using Wavelet Transform and Entropy-Based Features from Single-Lead ECG Signal. IEEE J. Biomed. Health Informatics 2018, 23, 1011–1021. [Google Scholar] [CrossRef]
- Donoho, D.L.; Johnstone, J.M. Ideal adaption by wavelet shrinkage. Biometrika 1994, 81, 425–455. [Google Scholar] [CrossRef]
- Boashash, B. Estimating and interpreting the instantaneous frequency of a signal. I. Fundamentals. Proc. IEEE 1992, 80, 520–538. [Google Scholar] [CrossRef]
- Carson, J.R.; Fry, T.C. Variable Frequency Electric Circuit Theory with Application to the Theory of Frequency-Modulation. Bell Syst. Tech. J. 1937, 16, 513–540. [Google Scholar] [CrossRef]
- Van der Pol, B. The fundamental principles of frequency modulation. J. Inst. Electr. Eng. Part III Radio Commun. Eng. 1946, 93, 153–158. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- PhysioNet Computing in Cardiology Challenge 2017 (CinC Challenge). Available online: http://physionet.org/challenge/2017/ (accessed on 13 March 2021).
- Mahmoodabadi, S.Z.; Ahmadian, A.; Abolhasani, M.D.; Eslami, M.; Bidgoli, J.H. ECG Feature Extraction Based on Multi-resolution Wavelet Transform. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 1–4 September 2005; pp. 3902–3905. [Google Scholar]
- Andreotti, F.; Carr, O.; Pimentel, M.A.F.; Mahdi, A.; De Vos, M. Comparing Feature Based Classifiers and Convolutional Neural Networks to Detect Arrhythmia from Short Segments of ECG. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017. [Google Scholar] [CrossRef]
- Billeci, L.; Costi, M.; Lombardi, D.; Chiarugi, F.; Varanini, M. Automatic Detection of Atrial Fibrillation and Other Ar-rhythmias in ECG Recordings Acquired by a Smartphone Device. Electronics 2018, 7, 199. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Li, W. Atrial Fibrillation Detection and ECG Classification based on CNN-BiLSTM. arXiv 2020, arXiv:2011.06187. [Google Scholar]
- Jin, Y.; Qin, C.; Huang, Y.; Zhao, W.; Liu, C. Multi-domain modeling of atrial fibrillation detection with twin attentional convolutional long short-term memory neural networks. Knowl. Based Syst. 2020, 193, 105460. [Google Scholar] [CrossRef]
- Zheng, Z.; Chen, Z.; Hu, F.; Zhu, J.; Tang, Q.; Liang, Y. An automatic diagnosis of arrhythmias using a combination of CNN and LSTM technology. Electronics 2020, 9, 121. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).