2.1. Dataset
The EEG repository built in [
21] contains 200 records from 200 patients that, given their structure, cannot be processed by machine learning algorithms. Each EEG record was acquired under the electrode positioning system 10–20, considering a sampling rate of 200 samples per second for 21 channels and an approximate duration of 30 min. The 200 EEG records were diagnosed by a pediatric neurologist with 20 years of experience reading this kind of exam.
Besides, each EEG was decomposed channel by channel, and 672 segments diagnosed as abnormal were extracted and described using a set of feature extractors. Each segment had 200 samples. This same process was carried out for a set of 672 segments considered normal. Thus, the dataset was built with 142 features extracted from 1344 EEG segments. Since all the descriptors were applied to all the segments, the dataset did not contain columns with null data.
The descriptors used to extract the features from the EEG signals are described below.
Statistical features allow summarizing the values that describe a segment of EEG signal in a single value. The measures of this type that will be applied in the construction of the dataset are min, max, mean, median, low median, high median, variance, and standard deviation.
Entropy is considered a family of statistical measures that quantify the variant complexity in a system. In this study, we evaluated three different ways of measuring Entropy:
The signal is viewed as a function of time, and energy represents its size. The energy can be measured in different ways, but the area under the curve is the most common measure to describe the size of a signal. It measures the signal strength, and this concept can be applied to any signal or vector.
The fractal dimension corresponds to a noninteger dimension of a geometric object. Based on this principle, fractal dimension analysis is used to analyze biomedical signals. In this approach, the waveform is considered a geometric figure [
22].
This type of analysis provides a quick mechanism to calculate the fractal dimension bypassing the series in a binary sequence. For example, the following describes the equation that calculates the Petrosian fractal dimension [
22]:
This exponent is a measure of the predictability of the signal. It is a scalar between 0 and 1 which measures long-range correlations of a time series [
23].
The zero-crossing rate is a statistical feature that describes the number of times that a signal crosses the horizontal axis.
The Hjort parameters describe statistical properties in the time domain [
12]. Usually, these are used to analyze electroencephalography signals.
Activity, also known as the variance or mean power, measures the squared standard deviation of the amplitude.
Mobility measures the standard deviation of the slope concerning the standard deviation of the amplitude.
This parameter is associated with the wave shape.
The discrete wavelet transform allows the analysis of a signal in a specific segment. The procedure consists of expressing a continuous signal to expand coefficients of the internal product between the particular segment and a mother wavelet function. As a result, the wavelet transform’s discretization changes from a continuous mapping to a finite set of values. This process is done by changing the integral in the definition by an approximation with summations. Hence, the discretization represents the signal in terms of elementary functions accompanied by coefficients.
The mother wavelet functions include a set of scale functions. The parent functions represent the fine details of the signal, while the scale functions calculate an approximation. Thus, considering the above, a function or signal can be described as a summation of wavelet functions and scale functions.
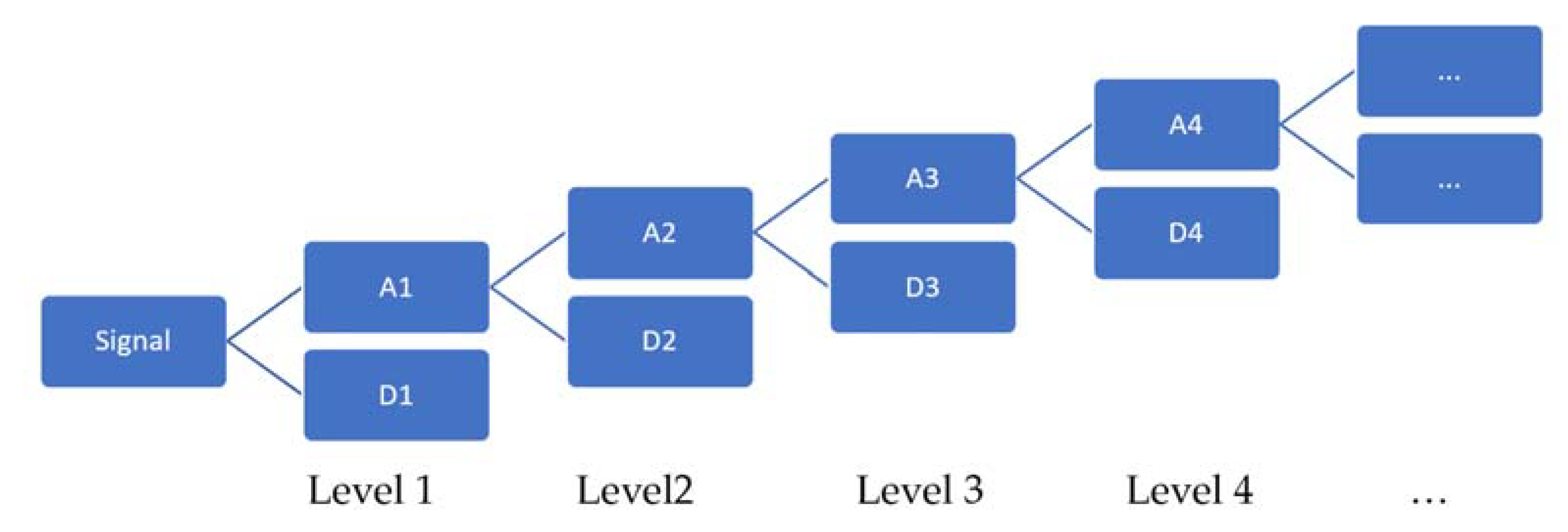
A signal can be decomposed into various levels from the time domain to the frequency domain in wavelet analysis. The decomposition is done from the detail coefficients as well as the approximation coefficients.
Figure 1 describes the different encoding paths for n levels of decomposition. The upper level of the tree represents the temporal representation. As the decomposition levels increase, an increase in the compensation in the time–frequency resolution is obtained. Finally, the last level of the tree describes the representation of the signal frequency.
The fast Fourier transform computes a short version of the discrete Fourier transform of a signal by decomposing the original signal into different frequencies (smaller transforms). The decomposed signals are used to calculate the resulting transform signal. FFT is used to convert a signal from the time domain to a representation in the frequency domain or vice versa.
Features extracted from the fast Fourier transform calculation are as follows:
The spectral centroid is a statistical measure used to describe the spectrum’s shape in digital signal processing. This centroid defines the spectrum as a probability distribution and represents where the center of mass of the spectrum is located.
The Spectral Flatness defines the ratio of the geometric mean to the arithmetic mean of a power spectrum.
The crest factor defines how extreme the peaks are in a signal.
Matched filters are basic signal analysis tools used to extract known waveforms from a signal that has been contaminated with noise. For example, in the context of the detection of epileptic spikes, given a signal that describes the brain activity (EEG), the matched filter seeks a well-known pattern of epilepsy s(t); then, if the signal contains an epileptiform pattern, the signal is described by the brain activity with the abnormality generating Otherwise, the signal only contains the normal brain activity
Considering the above, 21 descriptors were applied on the normal and abnormal EEG segments, and their wavelet coefficients (5) were generated from the original segments generating 126 features. The 21 descriptors are min, max, mean, median, high median, low median, variance, standard deviation, Shannon entropy, approximate entropy, Renyi entropy, kurtosis, skewness, energy, Higuchi fractal dimension, Petrosian fractal dimension, Hurst exponent, zero-crossing rate, Hjort activity, Hjort mobility, and Hjort complexity. Besides, the fast Fourier transform (FFT) was calculated, and 15 descriptors were applied to the result of FFT: min, max, mean, median, high median, low median, variance, standard deviation, Shannon entropy, kurtosis, skewness, energy, spectral centroid, spectral flatness, and crest factor. The matched filter was also applied to the original segments, and a Boolean feature was generated with the results. Then, we obtained 142 features: 126 features calculated from the original segment and 5 wavelet coefficients (21 × 6), 15 features extracted from the FFT calculation, and the matched filter.
Considering the number of segments that could be analyzed in a single EEG record (1 EEG with 21 channels and 30 min of duration could generate more than 37,800 segments of 200 samples), it is necessary to reduce the number of features not only to reduce the complexity of describing the segments but also to avoid the introduction of noise and redundant information into the classification process and increase the stability of the classifiers.
2.2. The Ensemble Feature Selection Approach
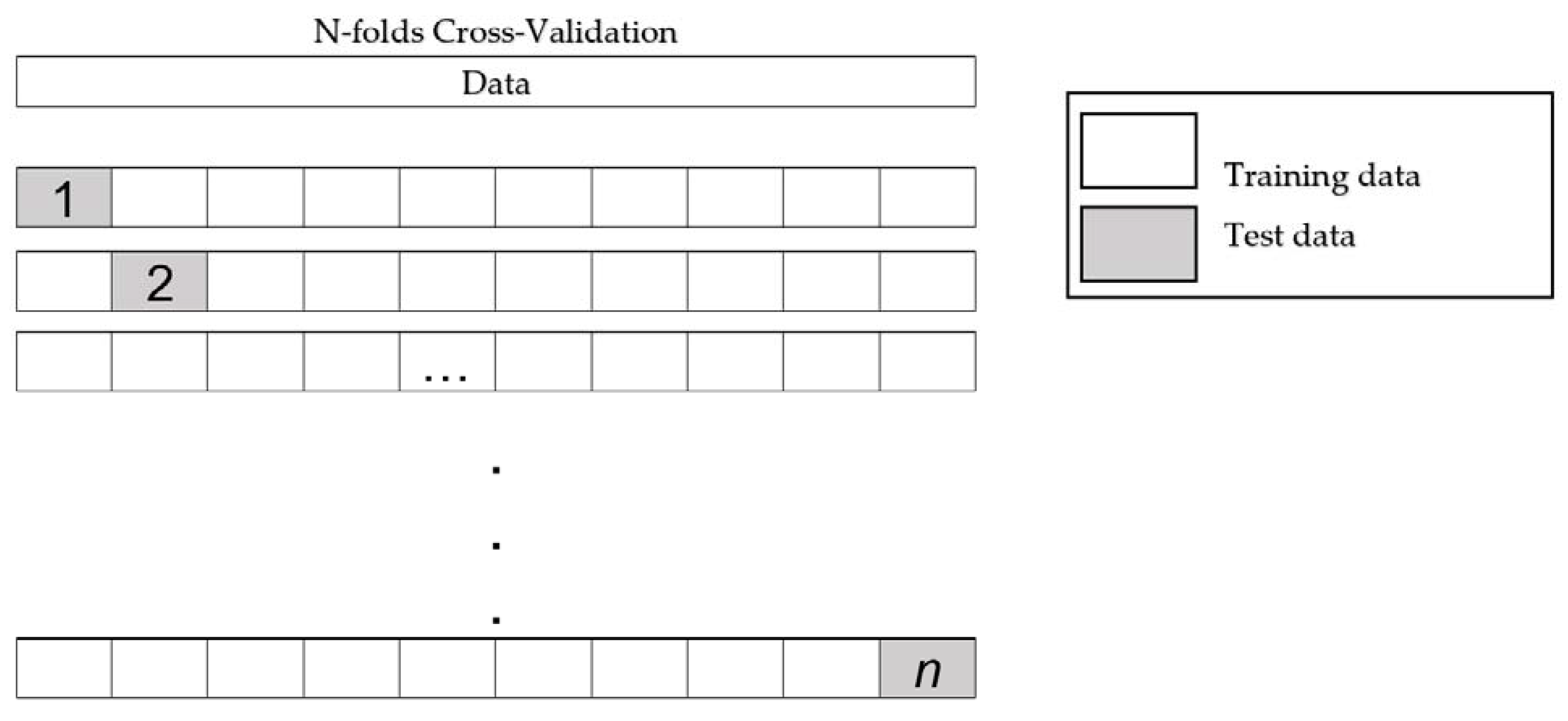
A dataset could contain three types of features: relevant, redundant, and noise. The category of the feature selection (FS) method: filter, wrapper, or embedded, is defined by the mechanism that evaluates the relevance of the features: statistical tests or cross-validation. The analysis performed by FS methods defines a ranking of feature relevance in the filter-based techniques, a subset of relevant features in wrapper methods, or a subset of features with a learning model in the embedded methods. The rankings of features generated by filter methods are used to select the k highest-ranked features.
Considering ensemble learning, the consensus of several experts improves the creation of a decision in a context [
24]. Thus, we decided to use the results of our previous research, where we built a framework of ensemble feature selection [
25]. This considers the pooling of
n FS algorithms by aggregating their results in a unique subset of relevant features. This scheme is described in
Figure 2 and defined in [
26] as a heterogeneous centralized ensemble, where single methods represent each FS method used to select a subset of relevant features, outcomes of single methods are the subset generated, pooling is the process to aggregate all subsets of relevant features, and relevant features are the result of the pooling process.
The EFS method described in [
25] uses an importance index (II) to aggregate the subsets generated by the
n FS algorithms. First, the subsets of features generated by each FS method build a set SUM with all selected features. Then, for each feature in the subset SUM, the importance index is computed according to Equation (15). Thus, the number of times that feature
i is presented in the subset SUM (
) is divided by
n to calculate its importance index. Finally, the EFS selects the features with an importance index greater than a threshold defined by the user.
The main objective of the ensemble feature selection approach is to reach a consensus among several FS methods to generate a subset of relevant features capable of representing the advantages of all used methods and face the biases of the single methods by compensating their disadvantages with the benefits of the others. Thereby, the result of EFS is a subset of relevant features that could improve the performance of subsequent analyses, such as classification processes.
Although the EFS implemented in the framework could be configured with different FS algorithms, in this study, we used five FS algorithms, three based on rankings of features (ANOVA, chi-squared, and mutual information), one wrapper (importance of features calculated by decision trees), and one embedded (recursive feature elimination—RFE). Each single FS algorithm generated a subset of relevant features, which the EFS aggregates.