Transcription of Listeria monocytogenes Key Virulence Genes on Tomato, Cucumber and Carrot
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain, Inoculum and Sample Preparation
2.2. Sampling and Microbiological Analyses
2.3. In Vitro and In Situ Gene Transcription Assay
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef]
- Vadia, S.; Arnett, E.; Haghighat, A.C.; Wilson-Kubalek, E.M.; Tweten, R.K.; Seveau, S. The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog. 2011, 7, e1002356. [Google Scholar] [CrossRef]
- Alberti-Segui, C.; Goeden, K.R.; Higgins, D.E. Differential function of Listeria monocytogenes listeriolysin O and phospholipases C in vacuolar dissolution following cell-to-cell spread. Cell. Microbiol. 2007, 9, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Rajabian, T.; Gavicherla, B.; Heisig, M.; Mueller-Altrock, S.; Goebel, W.; Gray-Owen, S.D.; Ireton, K. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol. 2009, 11, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Sabet, C.; Lecuit, M.; Cabanes, D.; Cossart, P.; Bierne, H. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 2005, 73, 6912–6922. [Google Scholar] [CrossRef]
- Gaballa, A.; Guariglia-Oropeza, V.; Wiedmann, M.; Boor, K.J. Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol. Mol. Biol. Rev. 2019, 83, e00034-19. [Google Scholar] [CrossRef]
- Henderson, L.O.; Gaballa, A.; Orsi, R.H.; Boor, K.J.; Wiedmann, M.; Guariglia-Oropeza, V. Transcriptional profiling of the L. monocytogenes PrfA regulon identifies six novel putative PrfA-regulated genes. FEMS Microbiol. Lett. 2020, 367, fnaa193. [Google Scholar] [CrossRef] [PubMed]
- Olesen, I.; Thorsen, L.; Jespersen, L. Relative transcription of Listeria monocytogenes virulence genes in liver pates with varying NaCl content. Int. J. Food Microbiol. 2010, 141, S60–S68. [Google Scholar] [CrossRef] [PubMed]
- Bae, D.; Crowley, M.R.; Wang, C. Transcriptome analysis of Listeria monocytogenes grown on a ready-to-eat meat matrix. J. Food Prot. 2011, 74, 1104–1111. [Google Scholar] [CrossRef]
- Rantsiou, K.; Greppi, A.; Garosi, M.; Acquadro, A.; Mataragas, M.; Cocolin, L. Strain dependent expression of stress response and virulence genes of Listeria monocytogenes in meat juices as determined by microarray. Int. J. Food Microbiol. 2012, 152, 116–122. [Google Scholar] [CrossRef]
- Rantsiou, K.; Mataragas, M.; Alessandria, V.; Cocolin, L. Expression of virulence genes of Listeria monocytogenes in food. J. Food Saf. 2012, 32, 161–168. [Google Scholar] [CrossRef]
- Mataragas, M.; Rovetto, F.; Bellio, A.; Alessandria, V.; Rantsiou, K.; Decastelli, L.; Cocolin, L. Differential gene expression profiling of Listeria monocytogenes in Cacciatore and Felino salami to reveal potential stress resistance biomarkers. Food Microbiol. 2015, 46, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Duodu, S.; Holst-Jensen, A.; Skjerdal, T.; Cappelier, J.M.; Pilet, M.F.; Loncarevic, S. Influence of storage temperature on gene expression and virulence potential of Listeria monocytogenes strains grown in a salmon matrix. Food Microbiol. 2010, 27, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Pilevar, Z.; Hosseini, H.; Abdollahzadeh, E.; Shojaee-Aliabadi, S.; Tajedin, E.; Yousefi, M.; Bahrami, A.; Khosroshahi, N.K. Effect of Zataria multiflora Boiss. essential oil, time, and temperature on the expression of Listeria monocytogenes virulence genes in broth and minced rainbow trout. Food Control 2020, 109, 106863. [Google Scholar] [CrossRef]
- Alessandria, V.; Rantsiou, K.; Dolci, P.; Zeppa, G.; Cocolin, L.A. Comparison of gene expression of Listeria monocytogenes in vitro and in the soft cheese Crescenza. Int. J. Dairy Technol. 2013, 66, 83–89. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Molfeta, C.; Panagiotopoulou, O.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Expression of Listeria monocytogenes key virulence genes during growth in liquid medium, on rocket and melon at 4, 10 and 30 °C. Food Microbiol. 2016, 55, 7–15. [Google Scholar] [CrossRef]
- Kang, J.; Burall, L.; Mammel, M.K.; Datta, A.R. Global transcriptomic response of Listeria monocytogenes during growth on cantaloupe slices. Food Microbiol. 2019, 77, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.P.; Petrone, G.; Di Biase, A.; Ammendolia, M.G.; Superti, F.; Seganti, L. Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophagelike cells. Microb. Pathog. 2000, 29, 137–144. [Google Scholar] [CrossRef]
- Conte, M.P.; Petrone, G.; Di Biase, A.M.; Longhi, C.; Penta, M.; Tinari, A.; Superti, F.; Fabozzi, G.; Visca, P.; Seganti, L. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect. Immun. 2002, 70, 4369–4378. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garner, M.R.; James, K.E.; Callahan, M.C.; Wiedmann, M.; Boor, K.J. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl. Environ. Microbiol. 2006, 72, 5384–5395. [Google Scholar] [CrossRef] [PubMed]
- Zilelidou, E.A.; Milina, V.; Paramithiotis, S.; Zoumpopoulou, G.; Poimenidou, S.V.; Mavrogonatou, E.; Kletsas, D.; Papadimitriou, K.; Tsakalidou, E.; Skandamis, P.N. Differential modulation of Listeria monocytogenes fitness, in vitro virulence and transcription of virulence-associated genes in response to the presence of different microorganisms. Appl. Environ. Microbiol. 2020, 86, e01165-20. [Google Scholar] [CrossRef] [PubMed]
- Werbrouck, H.; Vermeulen, A.; Van Coillie, E.; Messens, W.; Herman, L.; Devlieghere, F.; Uyttendaele, M. Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 2009, 134, 140–146. [Google Scholar] [CrossRef]
- Rieu, A.; Guzzo, J.; Piveteau, P. Sensitivity to acetic acid, ability to colonize abiotic surfaces and virulence potential of Listeria monocytogenes EGD-e after incubation on parsley leaves. J. Appl. Microbiol. 2010, 108, 560–570. [Google Scholar] [CrossRef]
- Wałecka, E.; Molenda, J.; Karpíšková, R.; Bania, J. Effect of osmotic stress and culture density on invasiveness of Listeria monocytogenes strains. Int. J. Food Microbiol. 2011, 144, 440–445. [Google Scholar] [CrossRef]
- Pricope-Ciolacu, L.; Nicolau, A.I.; Wagner, M.; Rychli, K. The effect of milk components and storage conditions on the virulence of Listeria monocytogenes as determined by a Caco-2 cell assay. Int. J. Food Microbiol. 2013, 166, 59–64. [Google Scholar] [CrossRef]
- Colas-Meda, P.; Vinas, I.; Oliveira, M.; Anguera, M.; Serrano, J.C.E.; Abadias, M. Exposure to minimally processed pear and melon during shelf life could modify the pathogenic potential of Listeria monocytogenes. Food Microbiol. 2017, 62, 275–281. [Google Scholar] [CrossRef]
- Wałecka-Zacharska, E.; Korkus, J.; Skowron, K.; Wietlicka-Piszcz, M.; Kosek-Paszkowska, K.; Bania, J. Effect of temperatures used in food storage on duration of heat stress induced invasiveness of L. monocytogenes. Microorganisms 2019, 7, 467. [Google Scholar] [CrossRef]
- Alves, A.; Magalhães, R.; Brandão, T.R.S.; Pimentel, L.; Rodríguez-Alcalá, L.M.; Teixeira, P.; Ferreira, V. 2020. Impact of exposure to cold and cold-osmotic stresses on virulence-associated characteristics of Listeria monocytogenes strains. Food Microbiol. 2020, 87, 103351. [Google Scholar] [CrossRef]
- Tan, Q.; Xu, H.; Chen, T.; Li, P.; Aguilar, Z.P.; Xu, D.; Ming, X.; Xu, F.; Wei, H. Differential expression of virulence and stress fitness genes during interaction between Listeria monocytogenes and Bifidobacterium longum. Biosci. Biotechnol. Biochem. 2012, 76, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Dutra, V.; Silva, A.C.; Cabrita, P.; Peres, C.; Malcata, X.; Brito, L. Lactobacillus plantarum LB95 impairs the virulence potential of Gram-positive and Gram-negative food-borne pathogens in HT-29 and Vero cell cultures. J. Med. Microbiol. 2016, 65, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Collazo, C.; Abadías, M.; Colás-Medà, P.; Iglesias, M.B.; Granado-Serrano, A.B.; Serrano, J.; Viñas, I. Effect of Pseudomonas graminis strain CPA-7 on the ability of Listeria monocytogenes and Salmonella enterica subsp. enterica to colonize Caco-2 cells after pre-incubation on fresh-cut pear. Int. J. Food Microbiol. 2017, 262, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.B.; Viñas, I.; Colás-Medà, P.; Collazo, C.; Serrano, J.C.; Abadias, M. Adhesion and invasion of Listeria monocytogenes and interaction with Lactobacillus rhamnosus GG after habituation on fresh-cut pear. J. Funct. Foods 2017, 34, 453–460. [Google Scholar] [CrossRef]
- Heisick, J.E.; Wagner, D.E.; Niermand, M.L.; Peeler, J.T. Listeria spp. found on fresh market produce. Appl. Environ. Microbiol. 1989, 55, 1925–1927. [Google Scholar] [CrossRef]
- Vahidy, R. Isolation of Listeria monocytogenes from fresh fruits and vegetables. HortScience 1992, 27, 62. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Mantzourani, K.S.; Katsarou, A.; Cavaiuolo, M.; Ferrante, A.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Estimation of Listeria monocytogenes and Escherichia coli O157:H7 prevalence and levels in naturally contaminated rocket and cucumber samples by deterministic and stochastic approaches. J. Food Prot. 2015, 78, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Arumugaswamy, R.K.; Ali, G.R.; Abd Hamid, S.N. Prevalence of Listeria monocytogenes in foods in Malaysia. Int. J. Food Microbiol. 1994, 23, 117–121. [Google Scholar] [CrossRef]
- Ponniah, J.; Robin, T.; Paie, M.S.; Radu, S.; Ghazali, F.M.; Kqueen, C.Y.; Nishibuchi, M.; Nakaguchi, Y.; Malakar, P.K. Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Control 2010, 21, 774–778. [Google Scholar] [CrossRef]
- Ajayeoba, T.A.; Atanda, O.O.; Obadina, A.O.; Bankole, M.O.; Adelowo, O.O. The incidence and distribution of Listeria monocytogenes in ready-to-eat vegetables in South-Western Nigeria. Food Sci. Nutr. 2016, 4, 59–66. [Google Scholar] [CrossRef]
- Farber, J.M.; Sanders, G.W.; Johnston, M.A. A survey of various foods for the presence of Listeria species. J. Food Prot. 1989, 52, 456–458. [Google Scholar] [CrossRef]
- Park, W.J.; Ryu, H.Y.; Lim, G.Y.; Lee, Y.D.; Park, J.H. Microbial prevalence and quality of organic farm produce from various production sites. Korean J. Food Sci. Technol. 2014, 46, 262–267. [Google Scholar] [CrossRef][Green Version]
- Roth, L.; Simonne, A.; House, L.; Ahn, S. Microbiological analysis of fresh produce sold at Florida farmers’ markets. Food Control 2018, 92, 444–449. [Google Scholar] [CrossRef]
- Odumeru, J.A.; Mitchell, S.J.; Alves, D.M.; Lynch, J.A.; Yee, A.J.; Wang, S.L.; Styliadis, S.; Farber, J.M. Assessment of the microbiological quality of ready-to-use vegetables for health-care food services. J. Food Prot. 1997, 60, 954–960. [Google Scholar] [CrossRef]
- Ho, J.L.; Shands, K.N.; Friedland, G.; Eckind, P.; Fraser, D.W. An outbreak of type 4b Listeria monocytogenes infection involving patients from eight Boston hospitals. Arch. Intern. Med. 1986, 146, 520–524. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Andritsos, N.D.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Listeria monocytogenes serotype prevalence and biodiversity in diverse food products. J. Food Prot. 2014, 77, 2115–2120. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Gkolfakis, P.; Patlaka, A.; Grounta, A.; Vourli, G.; Paramithiotis, S.; Touloumi, G.; Triantafyllou, K.; Drosinos, E.H. In vitro transcription of Listeria monocytogenes after exposure to human gastric and duodenal aspirates. J. Food Prot. 2020, 83, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Hadjilouka, A.; Mavrogiannis, G.; Mallouchos, A.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Effect of lemongrass essential oil on Listeria monocytogenes gene expression. LWT-Food Sci. Technol. 2017, 77, 510–516. [Google Scholar] [CrossRef]
- Lappa, I.; Dionysopoulou, A.M.; Paramithiotis, S.; Georgiadou, M.; Drosinos, E.H. Dual transcriptional profile of Aspergillus flavus during co-culture with Listeria monocytogenes and AFB1 production: A pathogen: Pathogen interaction. Pathogens 2019, 8, 198. [Google Scholar] [CrossRef]
- Leimeister-Waechter, M.; Domann, E.; Chakraborty, T. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 1992, 174, 947–952. [Google Scholar] [CrossRef]
- Dramsi, S.; Biswas, I.; Maguin, E.; Braun, L.; Mastroeni, P.; Cossart, P. Entry of Listeria monocytogenes into hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol. Microbiol. 1995, 16, 251–261. [Google Scholar] [CrossRef]
- Engelbrecht, F.; Chun, S.K.; Ochs, C.; Hess, J.; Lottspeich, F.; Goebel, W.; Sokolovic, Z. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 1996, 21, 823–837. [Google Scholar] [CrossRef]
- Phelps, C.C.; Vadia, S.; Arnett, E.; Tan, Y.; Zhang, X.; Pathak-Sharma, S.; Gavrilin, M.A.; Seveau, S. Relative roles of listeriolysin O, InlA and InlB in Listeria monocytogenes uptake by host cells. Infect. Immun. 2018, 86, e00555-18. [Google Scholar] [CrossRef] [PubMed]
- De las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vázquez-Boland, J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef]
- Kathariou, S.; Metz, P.; Hof, H.; Goebel, W. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J. Bacteriol. 1987, 169, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P.; Vicente, M.F.; Mengaud, J.; Baquero, F.; Perez-Diaz, J.C.; Berche, P. Listeriolysin O is essential for virulence of Listeria monocytogenes: Direct evidence obtained by gene complementation. Infect. Immun. 1989, 57, 3629–3636. [Google Scholar] [CrossRef] [PubMed]
- Domann, E.; Wehland, J.; Niebuhr, K.; Haffner, C.; Leimeister-Wachter, M.; Chakraborty, T. Detection of PrfA-independent promoter responsible for listeriolysin gene expression in mutant Listeria monocytogenes strains lacking the PrfA regulator. Infect. Immun. 1993, 61, 3073–3075. [Google Scholar] [CrossRef]
- Grundling, A.; Gonzalez, M.D.; Higgins, D.E. Requirement of the Listeria monocytogenes broad-range phospholipase PC-PLC during infection of human epithelial cells. J. Bacteriol. 2003, 185, 6295–6307. [Google Scholar] [CrossRef] [PubMed]
- Dussurget, O. New insights into determinants of Listeria monocytogenes virulence. Int. Rev. Cell Mol. Biol. 2008, 270, 1–38. [Google Scholar]
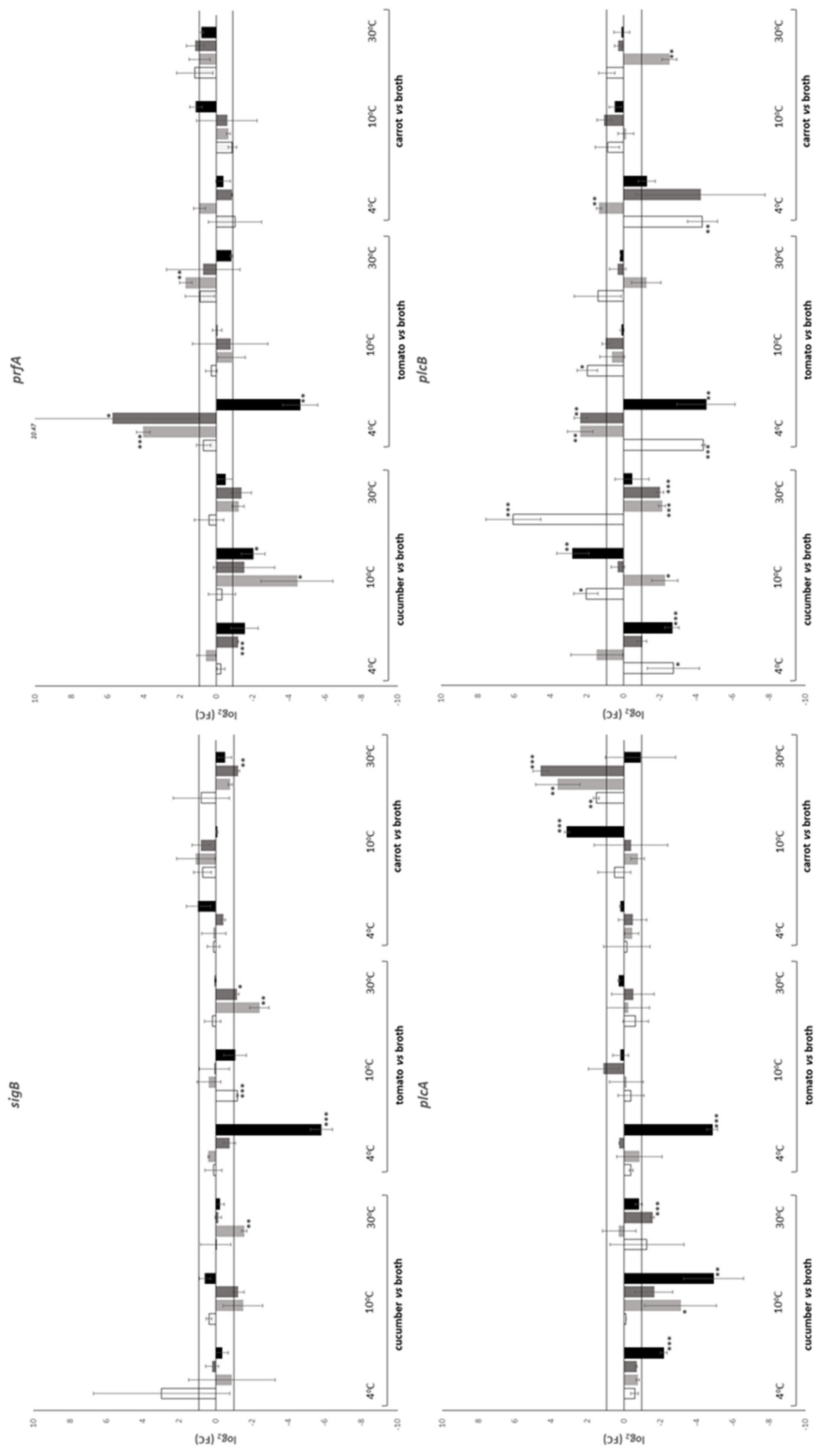
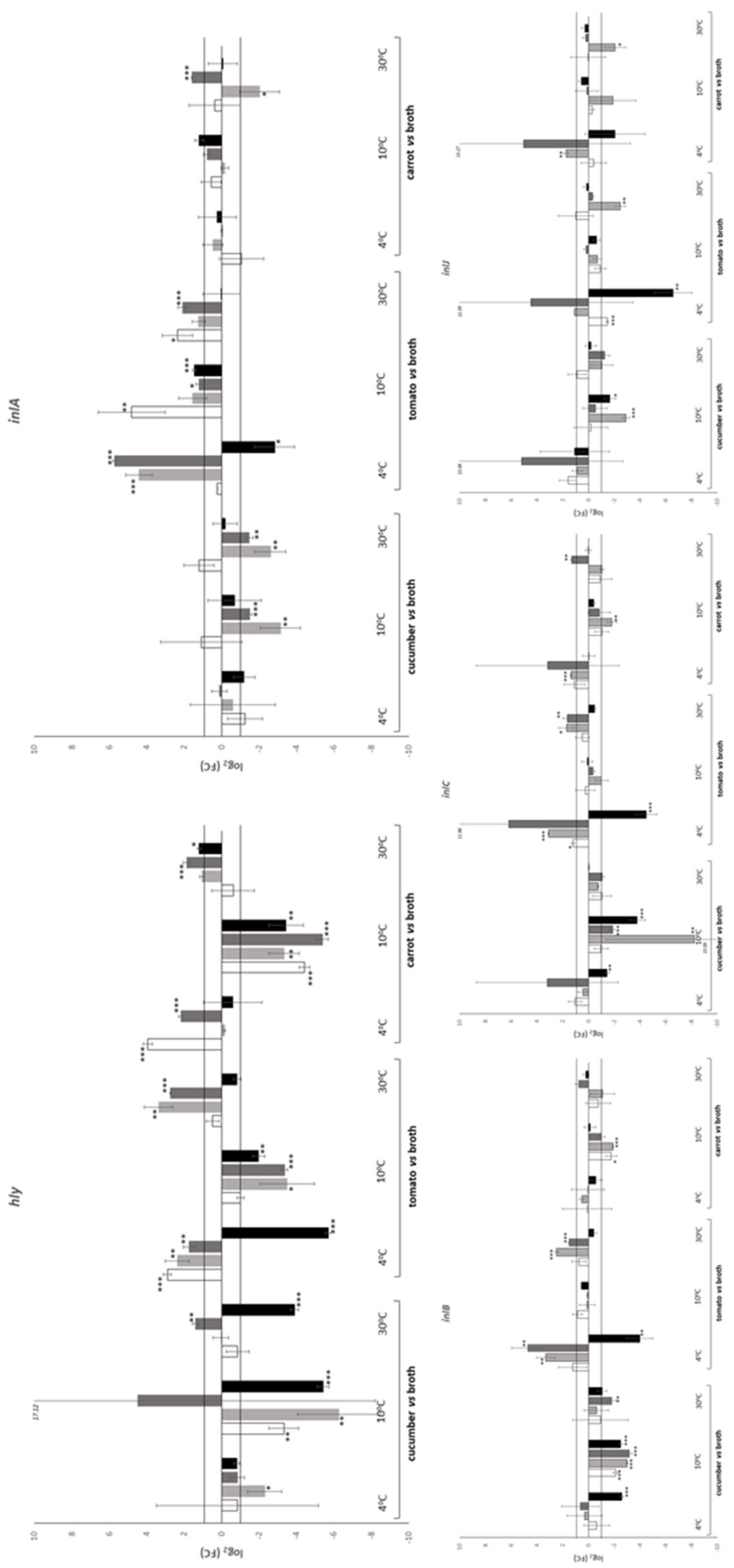
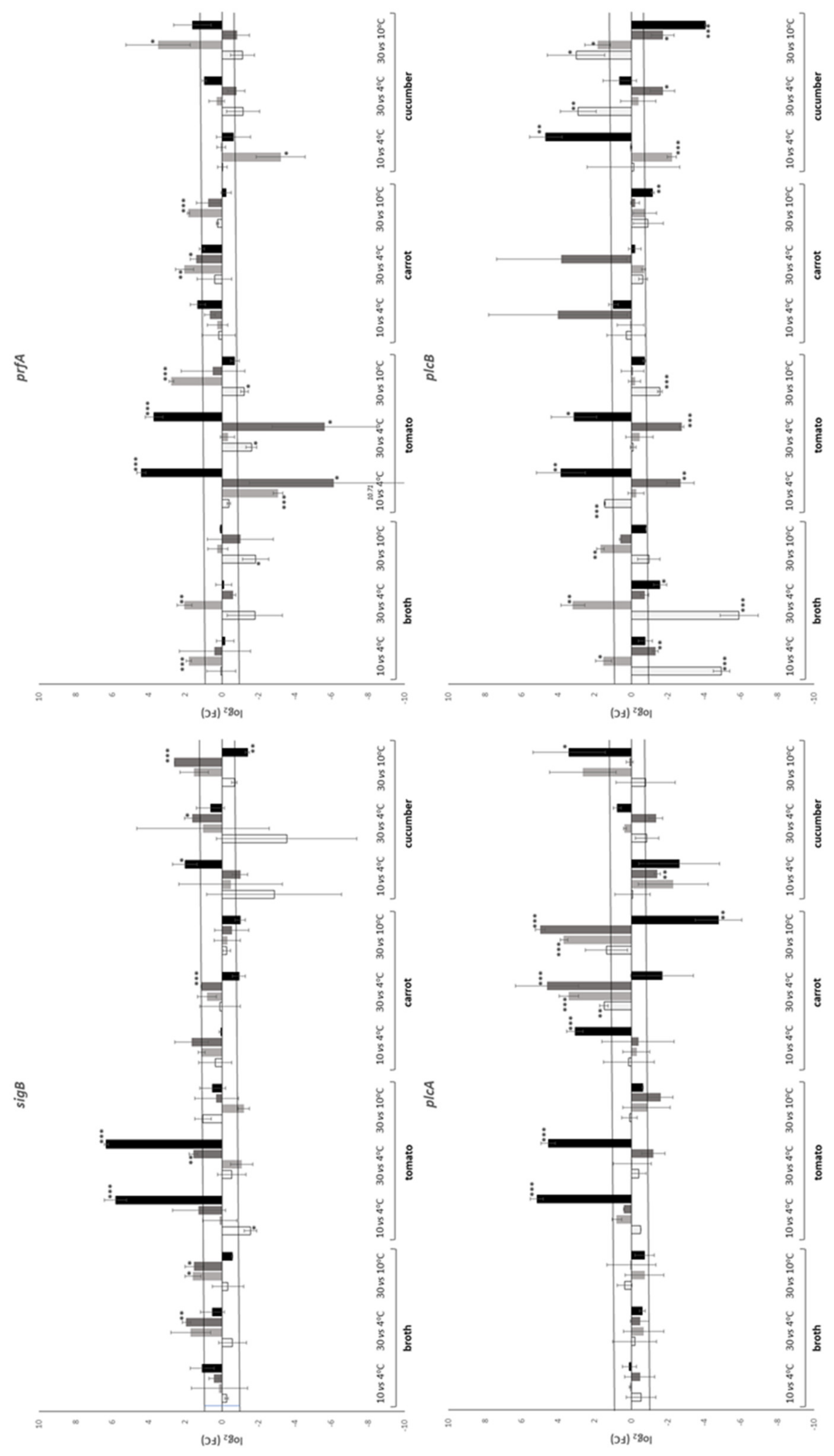
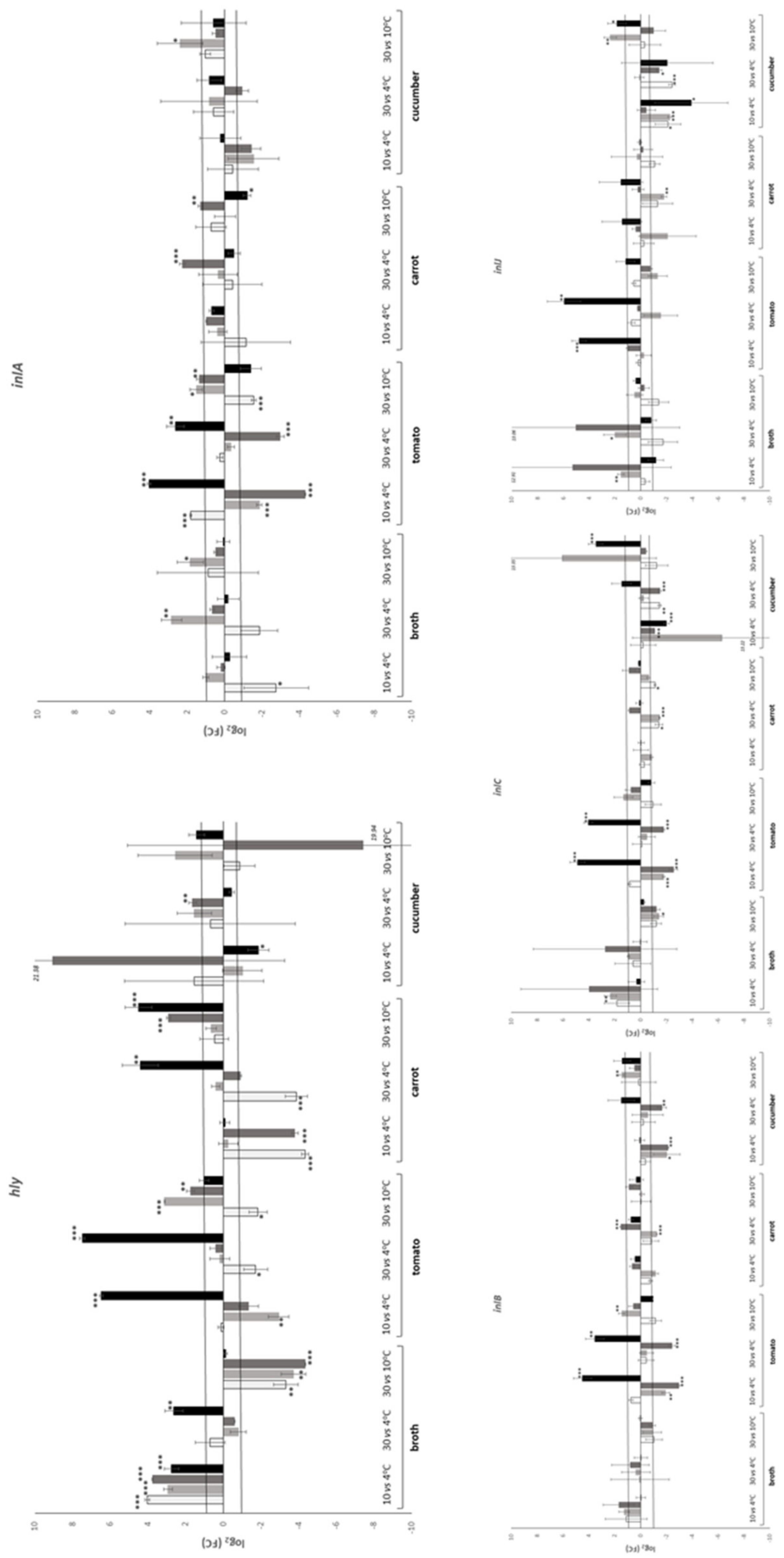
| Temperature (°C) | Time (h) | Substrate | |||
|---|---|---|---|---|---|
| BHI Broth (log CFU mL−1) | Cucumber (log CFU g−1) | Carrot (log CFU g−1) | Tomato (log CFU g−1) | ||
| 4 | 0 | 7.23 (0.20) a | 7.15 (0.22) a | 7.23 (0.25) bc | 7.11 (0.24) a |
| 0.5 | 7.35 (0.37) a | 7.31 (0.31) a | 7.33 (0.37)c | 7.17 (0.47) a | |
| 6.0 | 7.34 (0.40) a | 7.26 (0.33) a | 6.56 (0.42) a | 7.23 (0.52) a | |
| 24.0 | 7.42 (0.33) a | 6.79 (0.20) a | 6.60 (0.24) a b | 6.94 (0.59) a | |
| 10 | 0 | 7.20 (0.10) a | 7.22 (0.31) a | 7.34 (0.21) a | 7.13 (0.19) a |
| 0.5 | 7.36 (0.11) a | 7.09 (0.38) a | 7.19 (0.37) a | 7.36 (0.14) a | |
| 6.0 | 7.69 (0.13) b | 6.92 (0.14) a | 7.10 (0.41) a | 7.36 (0.51) a | |
| 24.0 | 8.12 (0.22) c | 6.99 (0.15) a | 6.89 (0.51) a | 7.21 (0.38) a | |
| 30 | 0 | 7.20 (0.13) a | 7.52 (0.35) a | 7.38 (0.27) a b | 7.34 (0.21) a |
| 0.5 | 7.65 (0.10) b | 7.77 (0.12) ab | 7.51 (0.33) b | 7.44 (0.27) a | |
| 6.0 | 8.19 (0.27) c | 8.28 (0.30) b | 6.73 (0.39) a | 7.20 (0.37) a | |
| 24.0 | 9.00 (0.12) d | 8.25 (0.32) b | 6.86 (0.40) ab | 7.20 (0.40) a | |
| Effect of Substrate | Effect of Temperature | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| log2(FC) | total | cucumber | tomato | carrot | total | BHI broth | cucumber | tomato | carrot |
| sigB | |||||||||
| <−1 | 6 (16.7) | 1 (8.3) | 4 (33.3) | 1 (8.3) | 2 (4.3) | 0 (0.0) | 1 (8.3) | 1 (8.3) | 0 (0.0) |
| −1 to 1 | 30 (83.3) | 11 (91.7) | 8 (66.7) | 11 (91.7) | 36 (74.9) | 9 (75.0) | 8 (66.7) | 8 (66.7) | 11 (91.7) |
| >1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 10 (20.8) | 3 (25.0) | 3 (25.0) | 3 (25.0) | 1 (8.3) |
| prfA | |||||||||
| <−1 | 4 (11.1) | 3 (25.0) | 1 (8.3) | 0 (0.0) | 7 (14.6) | 1 (8.3) | 1 (8.3) | 5 (41.7) | 0 (0.0) |
| −1 to 1 | 29 (80.6) | 9 (75.0) | 8 (66.7) | 12 (100) | 32 (66.5) | 9 (75.0) | 10 (83.3) | 4 (33.3) | 9 (75.0) |
| >1 | 3 (8.3) | 0 (0.0) | 3 (25.0) | 0 (0.0) | 9 (18.9) | 2 (16.7) | 1 (8.3) | 3 (25.0) | 3 (25.0) |
| plcA | |||||||||
| <−1 | 5 (13.9) | 4 (33.3) | 1 (8.3) | 0 (0.0) | 2 (4.3) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| −1 to 1 | 27 (75.0) | 8 (66.7) | 11 (91.7) | 8 (66.7) | 37 (76.8) | 12 (100) | 10 (83.3) | 10 (83.3) | 5 (41.7) |
| >1 | 4 (11.1) | 0 (0.0) | 0 (0.0) | 4 (33.3) | 9 (18.9) | 0 (0.0) | 1 (8.3) | 2 (16.7) | 6 (50.0) |
| plcB | |||||||||
| <−1 | 9 (25.0) | 5 (41.7) | 2 (16.7) | 2 (16.7) | 12 (25.1) | 4 (33.3) | 4 (33.3) | 3 (25.0) | 1 (8.3) |
| −1 to 1 | 20 (55.6) | 4 (33.3) | 7 (58.3) | 9 (75.0) | 26 (54.1) | 5 (41.7) | 4 (33.3) | 6 (50.0) | 11 (91.7) |
| >1 | 7 (19.4) | 3 (25.0) | 3 (25.0) | 1 (8.3) | 10 (20.8) | 3 (25.0) | 4 (33.3) | 3 (25.0) | 0 (0.0) |
| hly | |||||||||
| <−1 | 13 (36.1) | 5 (41.7) | 4 (33.3) | 4 (33.3) | 10 (20.8) | 3 (25.0) | 1 (8.3) | 3 (25.0) | 3 (25.0) |
| −1 to 1 | 13 (36.1) | 6 (50.0) | 3 (25.0) | 4 (33.3) | 24 (50.0) | 4 (33.3) | 9 (75.0) | 5 (41.7) | 6 (50.0) |
| >1 | 10 (27.8) | 1 (8.3) | 5 (41.7) | 4 (33.3) | 14 (29.2) | 5 (41.7) | 2 (16.7) | 4 (33.3) | 3 (25.0) |
| inlA | |||||||||
| <−1 | 6 (16.7) | 4 (33.3) | 1 (8.3) | 1 (8.3) | 6 (12.5) | 1 (8.3) | 0 (0.0) | 4 (33.3) | 1 (8.3) |
| −1 to 1 | 22 (61.2) | 8 (66.7) | 4 (33.3) | 10 (83.3) | 32 (66.7) | 9 (75.0) | 11 (91.7) | 3 (25.0) | 9 (75.0) |
| >1 | 8 (22.1) | 0 (0.0) | 7 (58.3) | 1 (8.3) | 10 (20.8) | 2 (16.7) | 1 (8.3) | 5 (41.7) | 2 (16.7) |
| inlB | |||||||||
| <−1 | 9 (25.0) | 6 (50.0) | 1 (8.3) | 2 (16.7) | 7 (14.6) | 0 (0.0) | 3 (25.0) | 3 (25.0) | 1 (8.3) |
| −1 to 1 | 23 (63.9) | 6 (50.0) | 7 (58.3) | 10 (83.3) | 36 (75.0) | 12 (100) | 8 (66.7) | 6 (50.0) | 10 (83.3) |
| >1 | 4 (11.1) | 0 (0.0) | 4 (33.3) | 0 (0.0) | 5 (10.4) | 0 (0.0) | 1 (8.3) | 3 (25.0) | 1 (8.3) |
| inlC | |||||||||
| <−1 | 6 (16.7) | 4 (33.3) | 1 (8.3) | 1 (8.3) | 10 (20.8) | 1 (8.3) | 3 (25.0) | 3 (25.0) | 3 (25.0) |
| −1 to 1 | 23 (63.9) | 8 (66.7) | 6 (50.0) | 9 (75.0) | 33 (68.6) | 10 (83.3) | 7 (58.3) | 7 (58.3) | 9 (75.0) |
| >1 | 7 (19.4) | 0 (0.0) | 5 (41.7) | 2 (16.7) | 5 (10.6) | 1 (8.3) | 2 (16.7) | 2 (16.7) | 0 (0.0) |
| inlJ | |||||||||
| <−1 | 5 (13.9) | 2 (16.7) | 2 (16.7) | 1 (8.3) | 6 (12.5) | 0 (0.0) | 5 (41.7) | 0 (0.0) | 1 (8.3) |
| −1 to 1 | 30 (83.4) | 10 (83.3) | 10 (83.3) | 10 (83.3) | 37 (77.0) | 10 (83.3) | 5 (41.7) | 10 (83.3) | 11 (91.7) |
| >1 | 1 (2.7) | 0 (0.0) | 0 (0.0) | 1 (8.3) | 6 (12.5) | 2 (16.7) | 2 (16.7) | 2 (16.7) | 0 (0.0) |
| Total | |||||||||
| <−1 | 63 (19.5) | 34 (31.5) | 17 (15.8) | 12 (11.1) | 62 (14.3) | 10 (9.3) | 19 (17.6) | 22 (20.4) | 11 (10.2) |
| −1 to 1 | 217 (66.9) | 70 (64.8) | 64 (59.2) | 83 (76.8) | 293 (67.7) | 80 (74.0) | 72 (66.7) | 59 (54.6) | 81 (75.0) |
| >1 | 44 (13.6) | 4 (3.7) | 27 (25.0) | 13 (12.1) | 78 (18.0) | 18 (16.7) | 17 (15.7) | 27 (25.0) | 16 (14.8) |
| prfA | sigB | plcA | plcB | hly | inlA | inlB | inlC | inlJ | ||
|---|---|---|---|---|---|---|---|---|---|---|
| prfA | r | |||||||||
| p | ||||||||||
| sigB | r | 0.2548 | ||||||||
| p | 0.0193 | |||||||||
| plcA | r | 0.4970 | 0.3361 | |||||||
| p | 0.0000 | 0.0018 | ||||||||
| plcB | r | 0.4347 | 0.2836 | 0.1373 | ||||||
| p | 0.0000 | 0.0089 | 0.2128 | |||||||
| hly | r | 0.4430 | 0.0654 | 0.2050 | −0.0811 | |||||
| p | 0.0000 | 0.5545 | 0.0614 | 0.4631 | ||||||
| inlA | r | 0.7528 | 0.2293 | 0.3497 | 0.6363 | 0.1316 | ||||
| p | 0.0000 | 0.0359 | 0.0011 | 0.0000 | 0.2327 | |||||
| inlB | r | 0.7959 | 0.2581 | 0.4265 | 0.3001 | 0.4618 | 0.7264 | |||
| p | 0.0000 | 0.0178 | 0.0001 | 0.0056 | 0.0000 | 0.0000 | ||||
| inlC | r | 0.7218 | 0.2803 | 0.4250 | 0.1368 | 0.5274 | 0.5850 | 0.8322 | ||
| p | 0.0000 | 0.0098 | 0.0001 | 0.2147 | 0.0000 | 0.0000 | 0.0000 | |||
| inlJ | r | 0.4045 | 0.4817 | 0.3182 | 0.1934 | 0.3311 | 0.3646 | 0.5134 | 0.6898 | |
| p | 0.0001 | 0.0000 | 0.0032 | 0.0780 | 0.0021 | 0.0006 | 0.0000 | 0.0000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paramithiotis, S.; Kotsakou, C.; Drosinos, E.H. Transcription of Listeria monocytogenes Key Virulence Genes on Tomato, Cucumber and Carrot. Appl. Sci. 2021, 11, 5983. https://doi.org/10.3390/app11135983
Paramithiotis S, Kotsakou C, Drosinos EH. Transcription of Listeria monocytogenes Key Virulence Genes on Tomato, Cucumber and Carrot. Applied Sciences. 2021; 11(13):5983. https://doi.org/10.3390/app11135983
Chicago/Turabian StyleParamithiotis, Spiros, Christina Kotsakou, and Eleftherios H. Drosinos. 2021. "Transcription of Listeria monocytogenes Key Virulence Genes on Tomato, Cucumber and Carrot" Applied Sciences 11, no. 13: 5983. https://doi.org/10.3390/app11135983
APA StyleParamithiotis, S., Kotsakou, C., & Drosinos, E. H. (2021). Transcription of Listeria monocytogenes Key Virulence Genes on Tomato, Cucumber and Carrot. Applied Sciences, 11(13), 5983. https://doi.org/10.3390/app11135983








