Probiotic Potential of a Novel Vitamin B2-Overproducing Lactobacillus plantarum Strain, HY7715, Isolated from Kimchi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and LAB Strains Isolation
2.2. Riboflavin Overproducing LAB Isolation and Identification
2.3. Quantitative Analysis of Vitamin B2
2.4. Expression Analysis of Riboflavin Biosynthesis Genes
2.5. Sequence Comparison of RFN Element Present Upstream of the Rib Operon
2.6. Stability Study of HY7715 Riboflavin Overproduction Characteristics
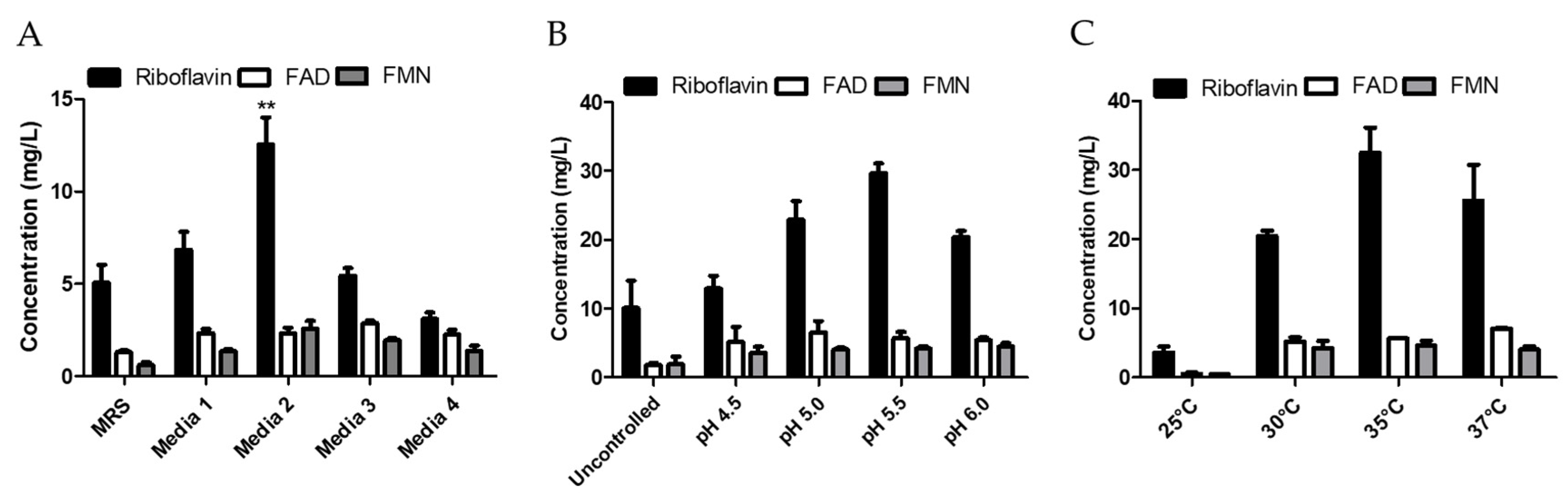
2.7. Optimization of Media Composition for Improving Riboflavin Production
2.8. Optimization of Culture Conditions to Improve Riboflavin Production
2.9. Measurement of Riboflavin Production in Co-Culture Conditions
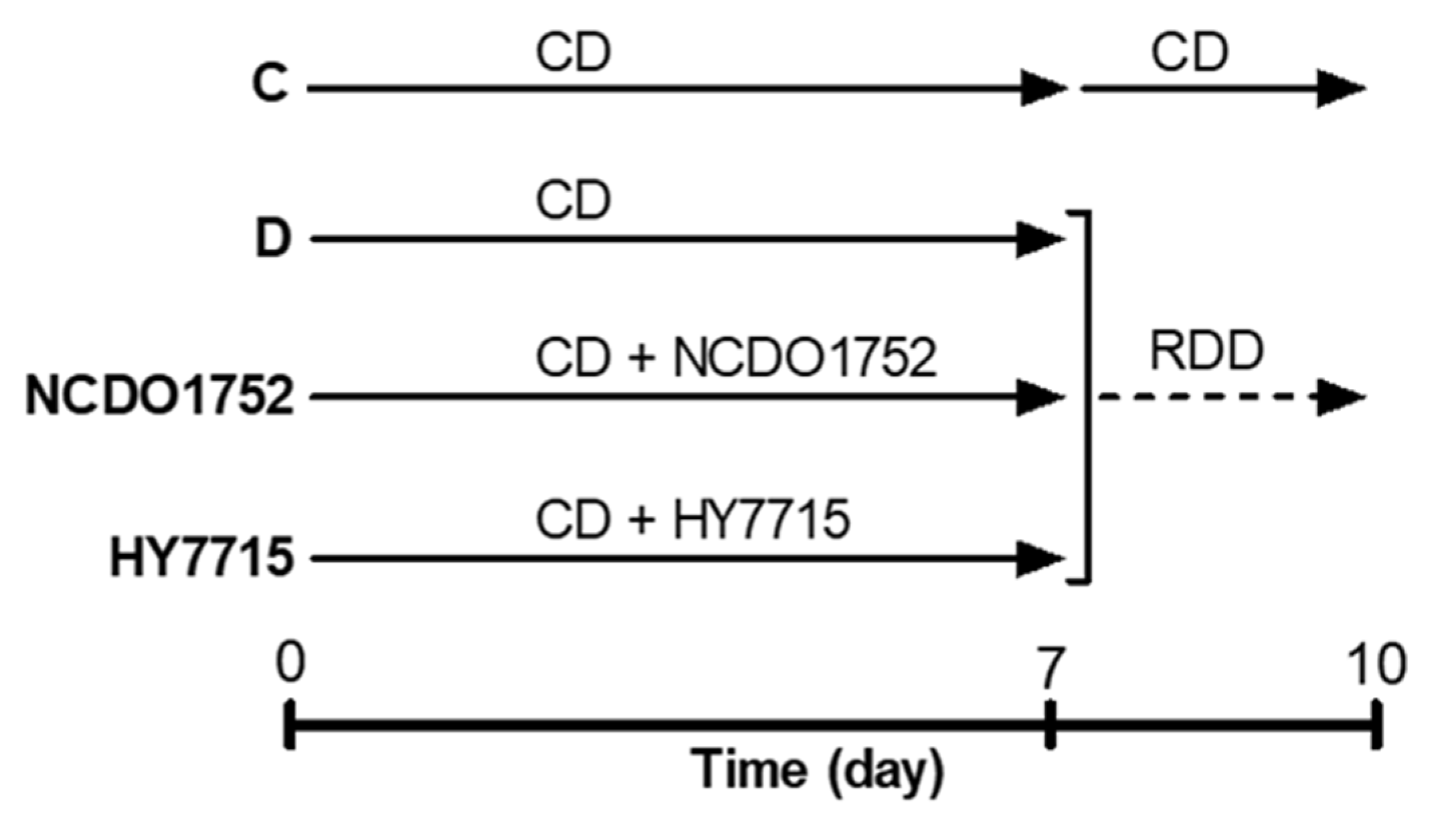
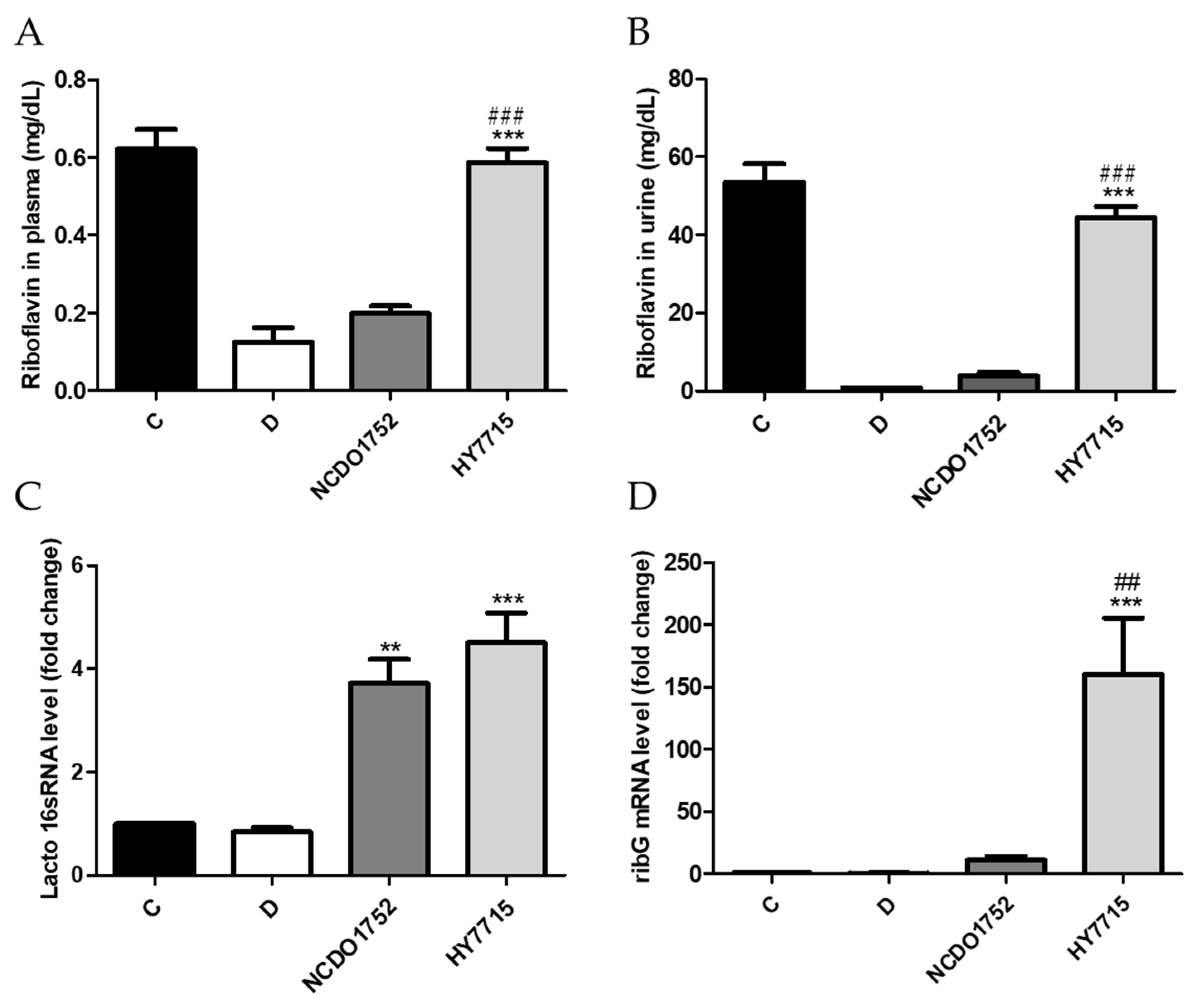
2.10. Prevention of Riboflavin Deficiency In Vivo
2.11. Intestinal Cell Adhesion Assay
2.12. Survivability in the Simulated Gastrointestinal Tract
2.13. Safety Evaluation
2.14. Statistical Analyses
3. Results
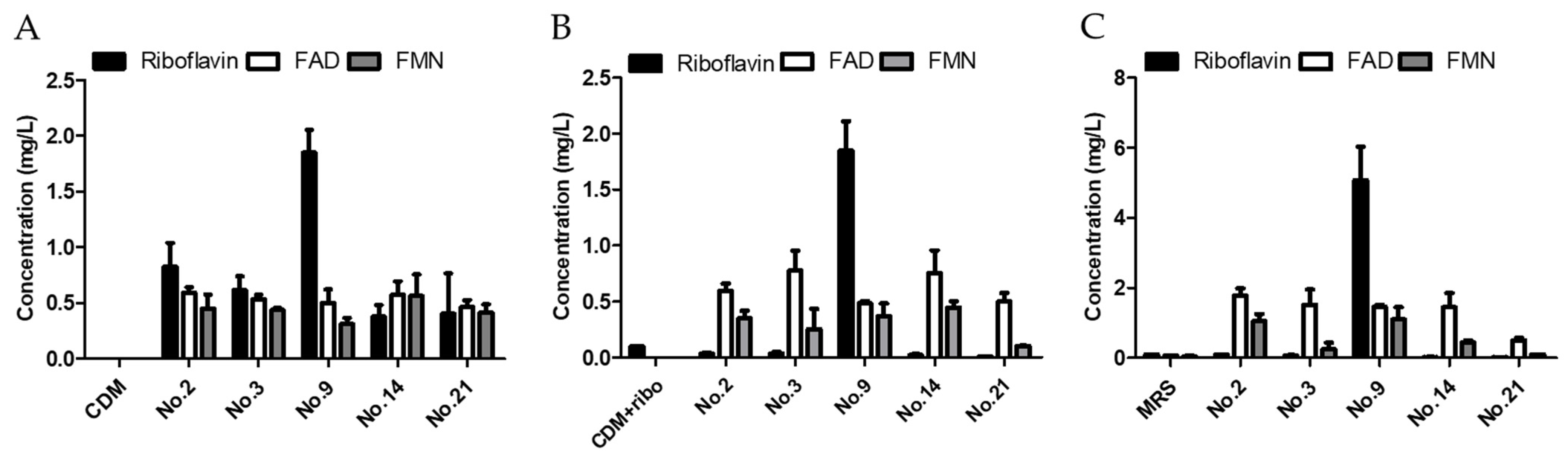
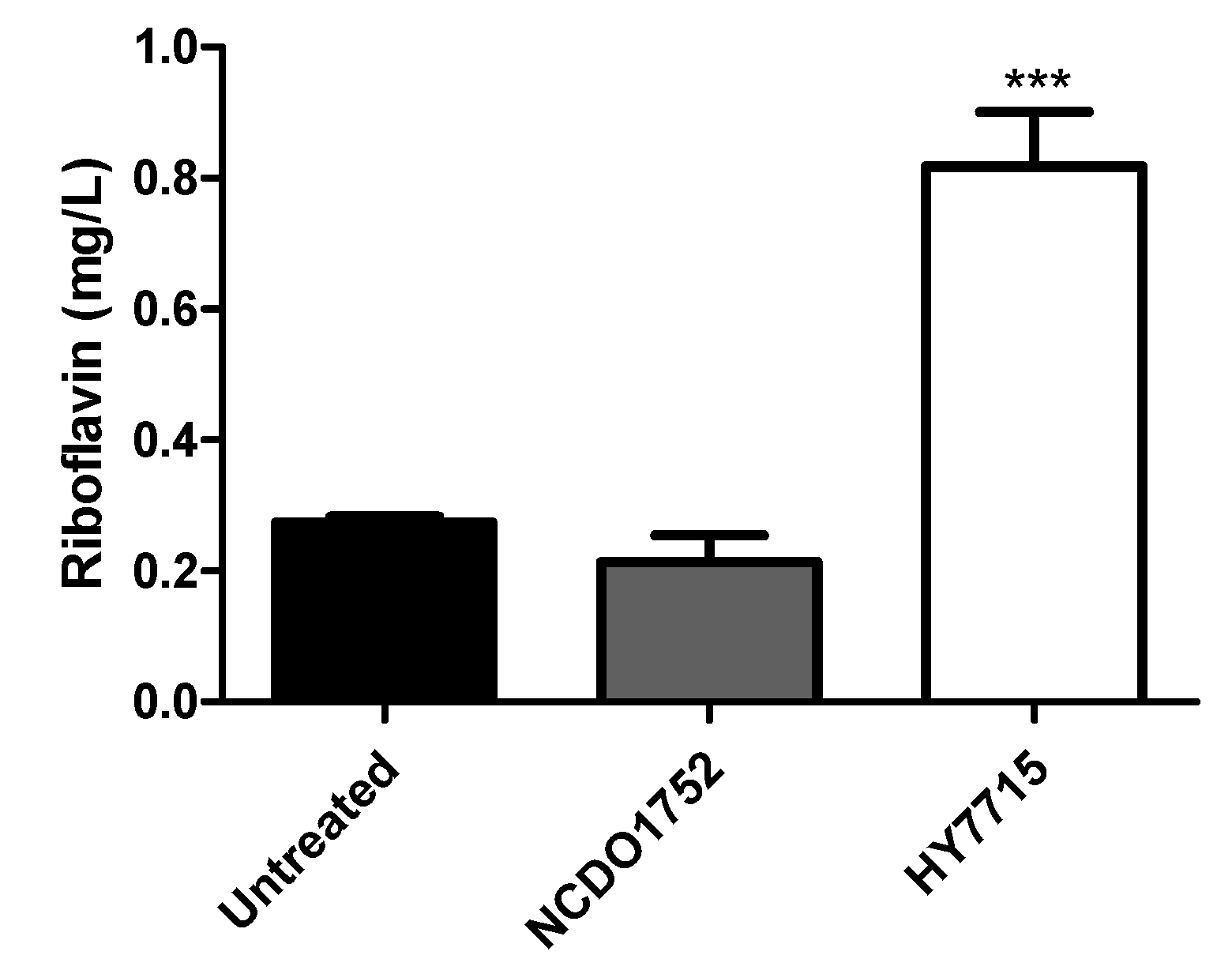
3.1. Isolation of Riboflavin over Production LAB from Kimchi
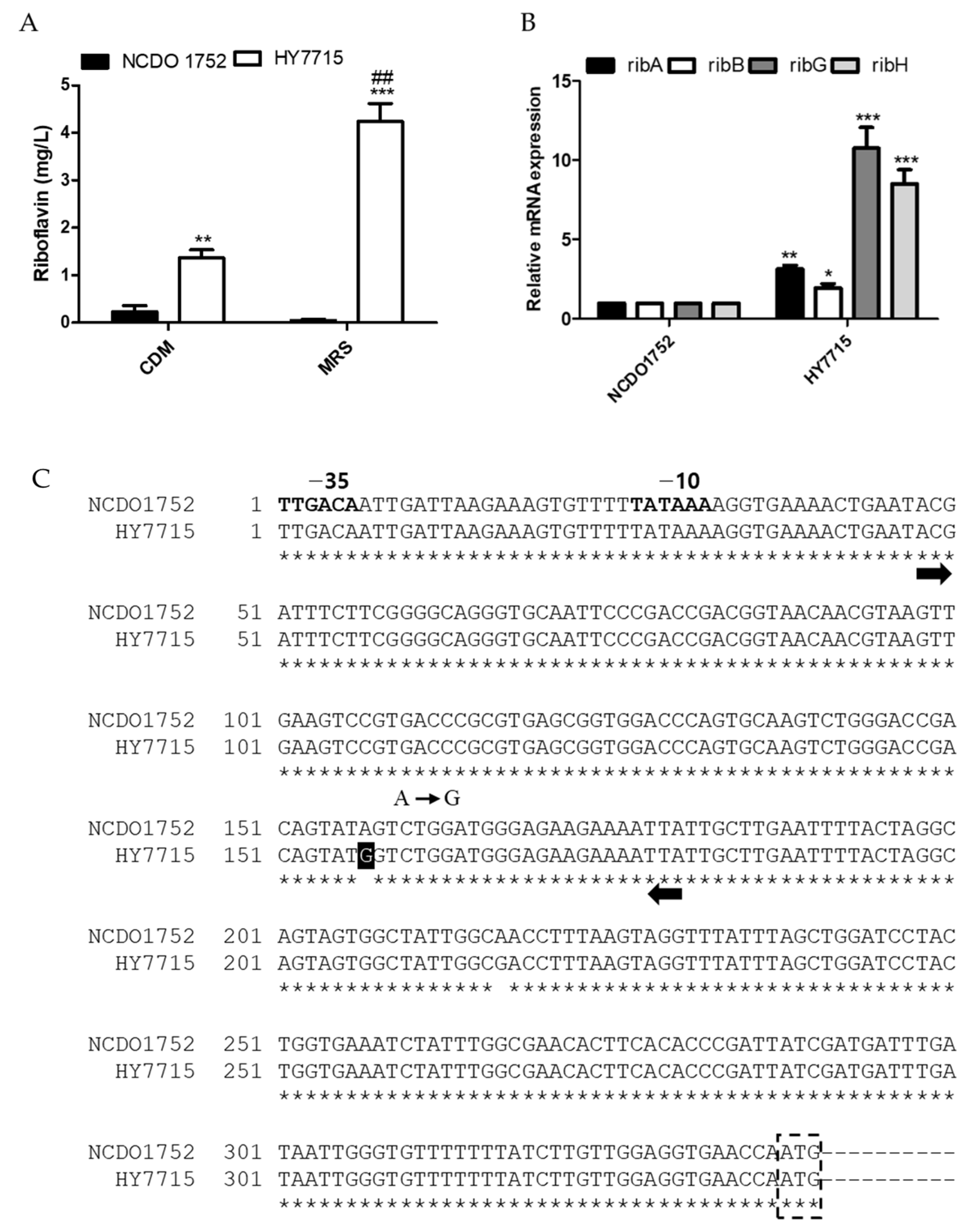
3.2. Analysis of Genetic Factors Leading to Riboflavin Overproduction
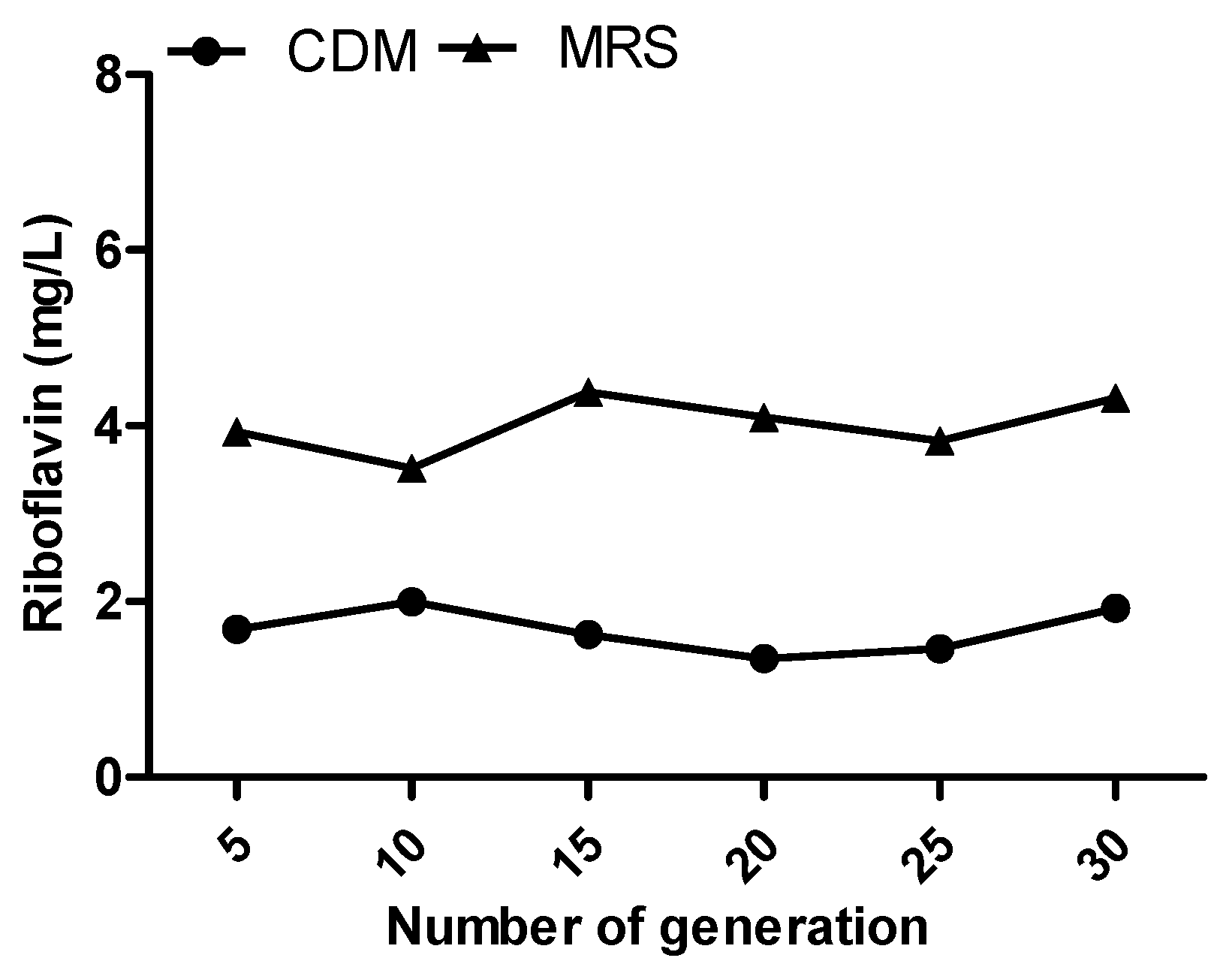
3.3. Stability Study of Riboflavin Overproduction Phenotype in HY7715
3.4. Optimization of Media Composition and Growth Conditions for Riboflavin Production
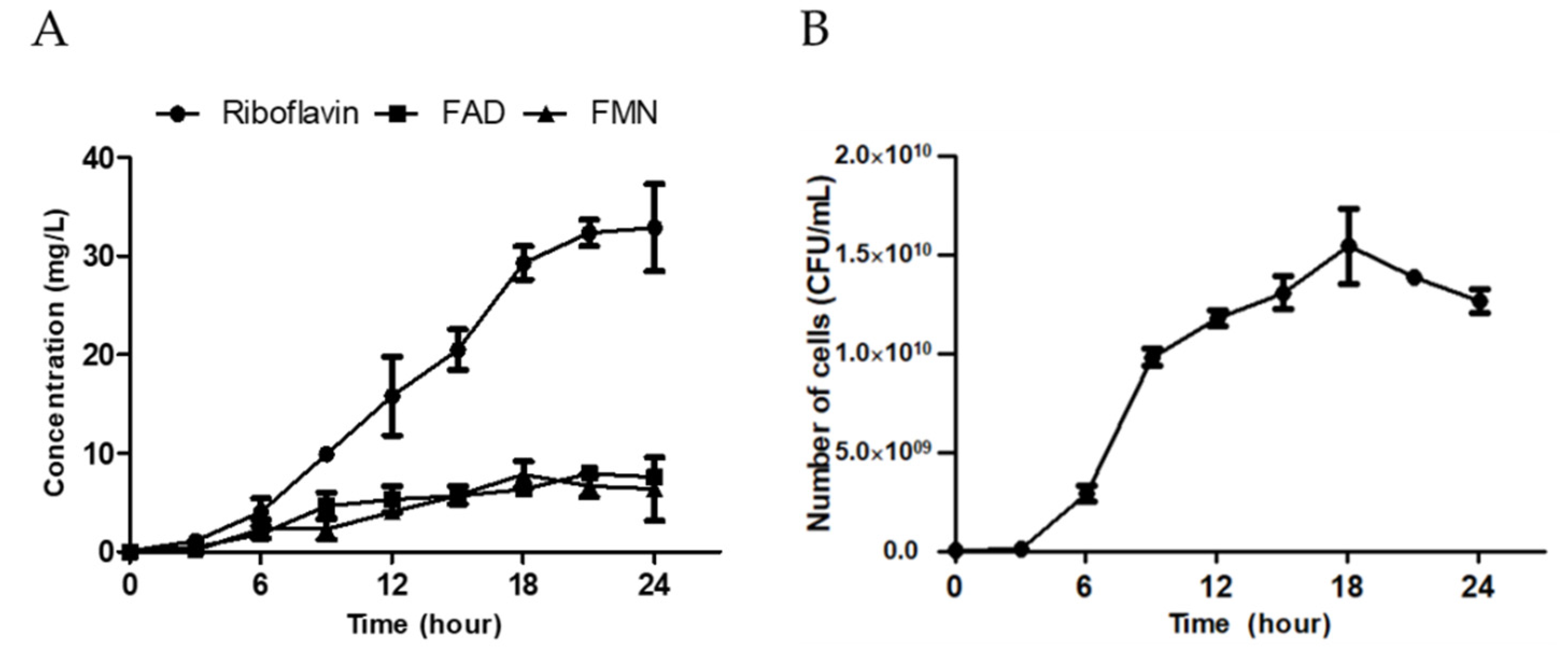
3.5. Mass Cultivation of HY7715
3.6. Riboflavin Production of HY7715 in Co-Culture with Caco-2 Cells
3.7. Confirmation of Riboflavin Production of HY7715 In Vivo Using an Animal Model
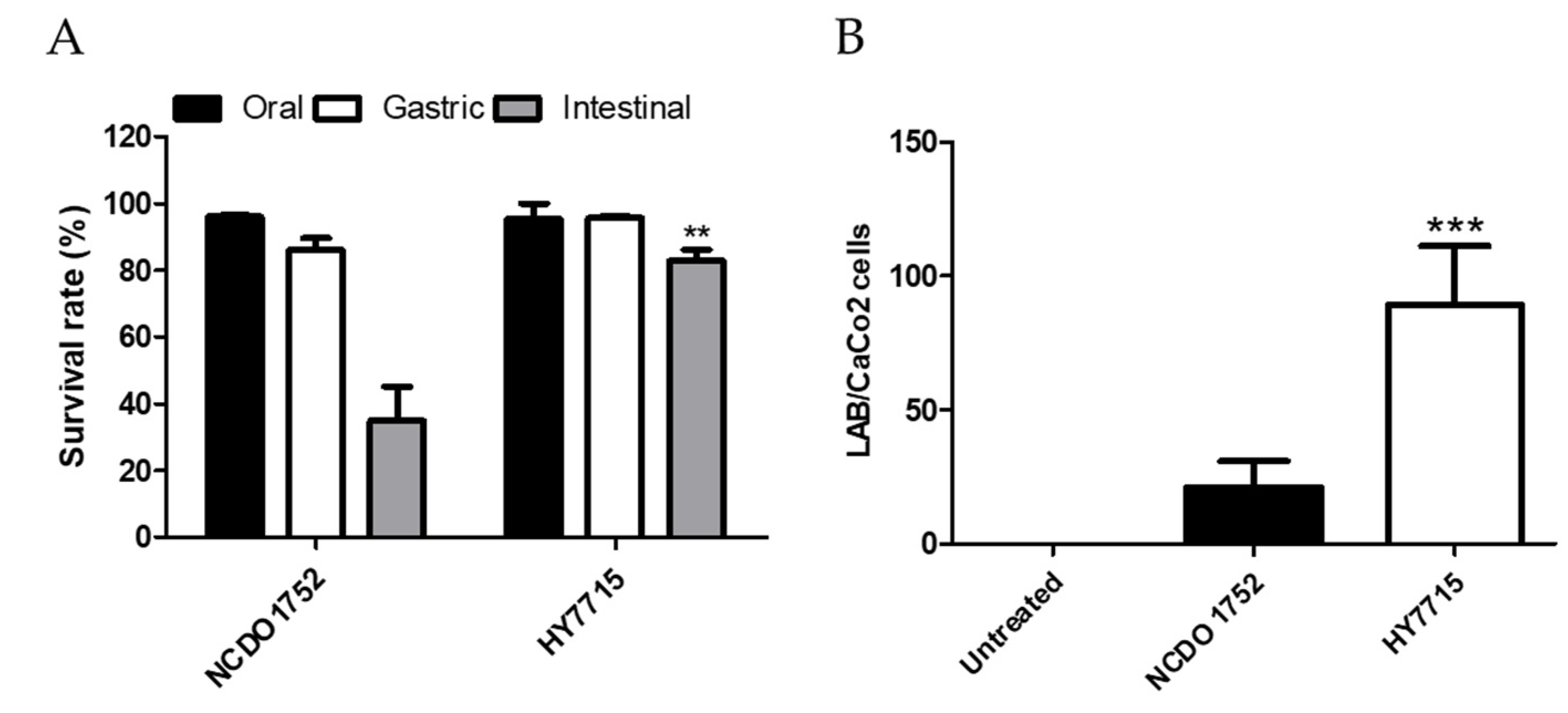
3.8. Probiotic Properties of HY7715
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Udhayabanu, T.; Manole, A.; Rajeshwari, M.; Varalakshmi, P.; Houlden, H.; Ashokkumar, B. Riboflavin responsive mitochondrial dysfunction in neurodegenerative diseases. J. Clin. Med. 2017, 6, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saedisomeolia, A.; Ashoori, M. Riboflavin in human health: A review of current evidences. Adv. Food Nutr. Res. 2018, 83, 57–81. [Google Scholar] [CrossRef]
- Pitkin, R.M.; Allen, L.; Bailey, L.B.; Bernfield, M. Dietary reference intakes for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin, and choline. In The National Academies Collection; National Institutes of Health: Washington, DC, USA, 1998. [Google Scholar]
- Mataix, J.; Aranda, P.; Sánchez, C.; Montellano, M.A.; Planells, E.; Llopis, J. Assessment of thiamin (vitamin B1) and riboflavin (vitamin B2) status in an adult mediterranean population. Br. J. Nutr. 2003, 90, 661–666. [Google Scholar] [CrossRef] [Green Version]
- Zempleni, J.; Galloway, J.R.; McCormick, D.B. Pharmacokinetics of orally and intravenously administered riboflavin in healthy humans. Am. J. Clin. Nutr. 1996, 63, 54–66. [Google Scholar] [CrossRef] [Green Version]
- Sherwood, R.A. Methods for Assessment of Vitamin B2; Elsevier Science: Amsterdam, The Netherlands, 2019; pp. 165–172. [Google Scholar] [CrossRef]
- Brody, M.S.; Hille, R. The kinetic behavior of chicken liver sulfite oxidase. Biochemistry 1999, 38, 6668–6677. [Google Scholar] [CrossRef]
- Riboflavin Deficiency in Man. Nutr. Rev. 1950, 8, 133–135. [CrossRef]
- Mazur-Bialy, A.I.; Pochec, E.; Plytycz, B. Immunomodulatory effect of riboflavin deficiency and enrichment—Reversible pathological response versus silencing of inflammatory activation. J. Physiol. Pharmacol. 2015, 66, 793–802. [Google Scholar]
- Mosegaard, S.; Dipace, G.; Bross, P.; Carlsen, J.; Gregersen, N.; Olsen, R.K.J. Riboflavin deficiency-implications for general human health and inborn errors of metabolism. Int. J. Mol. Sci. 2020, 21, 3847. [Google Scholar] [CrossRef] [PubMed]
- Mensink, G.B.; Fletcher, R.; Gurinovic, M.; Huybrechts, I.; Lafay, L.; Serra-Majem, L.; Szponar, L.; Tetens, I.; Verkaik-Kloosterman, J.; Baka, A.; et al. Mapping low intake of micronutrients across Europe. Br. J. Nutr. 2013, 110, 755–773. [Google Scholar] [CrossRef] [Green Version]
- Powers, H.J.; Hill, M.H.; Mushtaq, S.; Dainty, J.R.; Majsak-Newman, G.; Williams, E.A. Correcting a marginal riboflavin deficiency improves hematologic status in young women in the United Kingdom (RIBOFEM). Am. J. Clin. Nutr. 2011, 93, 1274–1284. [Google Scholar] [CrossRef] [Green Version]
- Larsson, C.L.; Johansson, G.K. Dietary intake and nutritional status of young vegans and omnivores in Sweden. Am. J. Clin. Nutr. 2002, 76, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Waldmann, A.; Koschizke, J.W.; Leitzmann, C.; Hahn, A. dietary intakes and lifestyle factors of a vegan population in Germany: Results from the German vegan study. Eur. J. Clin. Nutr. 2003, 57, 947–955. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, J.G.; Laiño, J.E.; del Valle, M.J.; Vannini, V.; van Sinderen, D.; Taranto, M.P.; de Valdez, G.F.; de Giori, G.S.; Sesma, F. B-group vitamin production by lactic acid bacteria--Current knowledge and potential applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Juarez Del Valle, M.; Laiño, J.E.; de Moreno de LeBlanc, A.; Savoy de Giori, G.; LeBlanc, J.G. Soyamilk fermented with riboflavin-producing lactobacillus plantarum CRL 2130 reverts and prevents ariboflavinosis in murine models. Br. J. Nutr. 2016, 116, 1229–1235. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Capozzi, V.; Menga, V.; Digesu, A.M.; De Vita, P.; van Sinderen, D.; Cattivelli, L.; Fares, C.; Spano, G. Biotechnological production of vitamin B2-enriched bread and pasta. J. Agric. Food Chem. 2011, 59, 8013–8020. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Capozzi, V.; Arena, M.P.; Spadaccino, G.; Dueñas, M.T.; López, P.; Fiocco, D.; Spano, G. Riboflavin-overproducing strains of lactobacillus fermentum for riboflavin-enriched bread. Appl. Microbiol. Biotechnol. 2014, 98, 3691–3700. [Google Scholar] [CrossRef]
- Ge, Y.Y.; Zhang, J.R.; Corke, H.; Gan, R.Y. Screening and spontaneous mutation of pickle-derived lactobacillus plantarum with overproduction of riboflavin, related mechanism, and food application. Foods 2020, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Burgess, C.M.; Smid, E.J.; Rutten, G.; van Sinderen, D. A general method for selection of riboflavin-overproducing food grade micro-organisms. Microb. Cell Fact. 2006, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Burgess, C.M.; Smid, E.J.; van Sinderen, D. Bacterial vitamin B2, B11 and B12 overproduction: An overview. Int. J. Food Microbiol. 2009, 133, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Russo, P.; Dueñas, M.T.; López, P.; Spano, G. Lactic acid bacteria producing B-group vitamins: A great potential for functional cereals products. Appl. Microbiol. Biotechnol. 2012, 96, 1383–1394. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Russo, P.; Capozzi, V.; López, P.; Fiocco, D.; Spano, G. Probiotic abilities of riboflavin-overproducing lactobacillus strains: A novel promising application of probiotics. Appl. Microbiol. Biotechnol. 2014, 98, 7569–7581. [Google Scholar] [CrossRef]
- Daliri, E.B.; Ofosu, F.K.; Chelliah, R.; Lee, B.H.; Oh, D.H. Challenges and perspective in integrated multi-omics in gut microbiota studies. Biomolecules 2021, 11, 300. [Google Scholar] [CrossRef]
- Levit, R.; de Giori, G.S.; de Moreno de LeBlanc, A.; LeBlanc, J.G. Evaluation of the effect of soymilk fermented by a riboflavin-producing lactobacillus plantarum strain in a murine model of colitis. Benef. Microbes 2017, 8, 65–72. [Google Scholar] [CrossRef]
- Levit, R.; Savoy de Giori, G.; de Moreno de LeBlanc, A.; LeBlanc, J.G. Effect of riboflavin-producing bacteria against chemically induced colitis in mice. J. Appl. Microbiol. 2018, 124, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.Y.; Thakur, K.; Feng, J.Y.; Cai, J.S.; Zhang, J.G.; Hu, F.; Russo, P.; Spano, G.; Wei, Z.J. Riboflavin-overproducing lactobacilli for the enrichment of fermented soymilk: Insights into improved nutritional and functional attributes. Appl. Microbiol. Biotechnol. 2020, 104, 5759–5772. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, M.G.; Howland, K.; Martin, C.; Bonner, A.B. A novel HPLC method for the concurrent analysis and quantitation of seven water-soluble vitamins in biological fluids (plasma and urine): A validation study and application. Sci. World J. 2012, 2012, 359721. [Google Scholar] [CrossRef] [Green Version]
- Yoshimatsu, H.; Yonezawa, A.; Yamanishi, K.; Yao, Y.; Sugano, K.; Nakagawa, S.; Imai, S.; Omura, T.; Nakagawa, T.; Yano, I.; et al. Disruption of Slc52a3 gene causes neonatal lethality with riboflavin deficiency in mice. Sci. Rep. 2016, 6, 27557. [Google Scholar] [CrossRef]
- Yang, Y.W.; Chen, M.K.; Yang, B.Y.; Huang, X.J.; Zhang, X.R.; He, L.Q.; Zhang, J.; Hua, Z.C. Use of 16S RRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl. Environ. Microbiol. 2015, 81, 6749–6756. [Google Scholar] [CrossRef] [Green Version]
- Byun, R.; Nadkarni, M.A.; Chhour, K.L.; Martin, F.E.; Jacques, N.A.; Hunter, N. Quantitative analysis of diverse lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 2004, 42, 3128–3136. [Google Scholar] [CrossRef] [Green Version]
- Oleksy-Sobczak, M.; Klewicka, E. Optimization of media composition to maximize the yield of exopolysaccharides production by lactobacillus rhamnosus strains. Probiotics Antimicrob. Proteins 2020, 12, 774–783. [Google Scholar] [CrossRef] [Green Version]
- Hwang, C.; Chang, J.; Houng, J.; Tsai, C.; Lin, C.; Tsen, H. Optimization of medium composition for improving biomass production of lactobacillus plantarum Pi06 using the taguchi array design and the box-behnken method. Biotechnol. Bioproc. Eng. 2012, 17, 827–834. [Google Scholar] [CrossRef]
- Aristimuño Ficoseco, C.; Mansilla, F.I.; Maldonado, N.C.; Miranda, H.; Fátima Nader-Macias, M.E.; Vignolo, G.M. Safety and growth optimization of lactic acid bacteria isolated from feedlot cattle for probiotic formula design. Front. Microbiol. 2018, 9, 2220. [Google Scholar] [CrossRef]
- Hernández, A.; Larsson, C.U.; Sawicki, R.; van Niel, E.W.J.; Roos, S.; Håkansson, S. Impact of the fermentation parameters pH and temperature on stress resilience of lactobacillus reuteri DSM 17938. AMB Express 2019, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Fernández de Palencia, P.; Romano, A.; Fernández, M.; Lucas, P.; Spano, G.; López, P. Biogenic amine production by the wine lactobacillus brevis IOEB 9809 in systems that partially mimic the gastrointestinal tract stress. BMC Microbiol. 2012, 12, 247. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, C.N.; Rosenfeldt Nielsen, V.; Hayford, A.E.; Møller, P.L.; Michaelsen, K.F.; Paerregaard, A.; Sandström, B.; Tvede, M.; Jakobsen, M. Screening of probiotic activities of forty-seven strains of lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 1999, 65, 4949–4956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, S.H.; Hong, D.K.; Bang, S.-J.; Heo, K.; Sim, J.-J.; Lee, J.-L. The functional properties of lactobacillus casei HY2782 are affected by the fermentation time. Appl. Sci. 2021, 11, 2481. [Google Scholar] [CrossRef]
- EFSA. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Arellano, K.; Vazquez, J.; Park, H.; Lim, J.; Ji, Y.; Kang, H.J.; Cho, D.; Jeong, H.W.; Holzapfel, W.H. Safety evaluation and whole-genome annotation of lactobacillus plantarum strains from different sources with special focus on isolates from green tea. Probiotics Antimicrob. Proteins 2020, 12, 1057–1070. [Google Scholar] [CrossRef]
- Cho, H.S. Food and nationalism: Kimchi and Korean national identity. Korean J. Int. Realt. 2006, 4, 207–229. [Google Scholar] [CrossRef]
- Lee, K.W.; Shim, J.M.; Kim, D.W.; Yao, Z.; Kim, J.A.; Kim, H.J.; Kim, J.H. Effects of different types of salts on the growth of lactic acid bacteria and yeasts during kimchi fermentation. Food Sci. Biotechnol. 2018, 27, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Park, K.Y.; Jeong, J.K.; Lee, Y.E.; Daily, J.W., 3rd. Health benefits of kimchi (Korean fermented vegetables) as a probiotic food. J. Med. Food 2014, 17, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Coda, R.; De Angelis, M.; Gobbetti, M. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 2013, 33, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Heo, G.Y.; Lee, J.W.; Oh, Y.J.; Park, J.A.; Park, Y.H.; Pyun, Y.R.; Ahn, J.S. Analysis of kimchi microflora using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 2005, 102, 143–150. [Google Scholar] [CrossRef]
- Cho, J.; Lee, D.; Yang, C.; Jeon, J.; Kim, J.; Han, H. Microbial population dynamics of kimchi, a fermented cabbage product. FEMS Microbiol. Lett. 2006, 257, 262–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, K.; Tomar, S.K.; Brahma, B.; De, S. Screening of riboflavin-producing lactobacilli by a polymerase-chain-reaction-based approach and microbiological assay. J. Agric. Food Chem. 2016, 64, 1950–1956. [Google Scholar] [CrossRef]
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002, 30, 3141–3151. [Google Scholar] [CrossRef] [Green Version]
- Thakur, K.; Tomar, S.K.; De, S. Lactic acid bacteria as a cell factory for riboflavin production. Microb. Biotechnol. 2016, 9, 441–451. [Google Scholar] [CrossRef]
- Noman, A.E.; Al-Barha, N.S.; Sharaf, A.M.; Al-Maqtari, Q.A.; Mohedein, A.; Mohammed, H.H.H.; Chen, F. A Novel strain of acetic acid bacteria gluconobacter oxydans FBFS97 involved in riboflavin production. Sci. Rep. 2020, 10, 13527. [Google Scholar] [CrossRef]
- Brinques, G.B.; do Carmo Peralba, M.; Ayub, M.A. Optimization of probiotic and lactic acid production by lactobacillus plantarum in submerged bioreactor systems. J. Ind. Microbiol. Biotechnol. 2010, 37, 205–212. [Google Scholar] [CrossRef]
- Panda, S.H.; Goli, J.K.; Das, S.; Mohanty, N. Production, optimization and probiotic characterization of potential lactic acid bacteria producing siderophores. AIMS Microbiol. 2017, 3, 88–107. [Google Scholar] [CrossRef]
- Gu, Q.; Li, P. Biosynthesis of vitamins by probiotic bacteria. In Probiotics and Prebiotics in Human Nutrition and Health; Rao, V., Rao, L.G., Eds.; IntechOpen: London, UK, 2016; pp. 135–148. [Google Scholar] [CrossRef] [Green Version]
- Mohedano, M.L.; Hernández-Recio, S.; Yépez, A.; Requena, T.; Martínez-Cuesta, M.C.; Peláez, C.; Russo, P.; LeBlanc, J.G.; Spano, G.; Aznar, R.; et al. Real-time detection of riboflavin production by lactobacillus plantarum strains and tracking of their gastrointestinal survival and functionality in vitro and in vivo using MCherry labeling. Front. Microbiol. 2019, 10, 1748. [Google Scholar] [CrossRef] [Green Version]
- George Kerry, R.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [Green Version]
| Compound | Concentration (g/L) | Compound | Concentration (g/L) |
|---|---|---|---|
| Glucose | 20 | l-glutamine | 0.6 |
| KH2PO4 | 3.1 | l-leucine | 0.4 |
| K2HPO4 | 1.5 | l-alanine | 0.2 |
| MgSO4 | 0.5 | l-lysine | 0.2 |
| MnSO4 | 0.02 | l-phenylalanine | 0.2 |
| FeSO4 | 0.02 | l-proline | 0.2 |
| Tween 80 | 1 | l-serine | 0.2 |
| Inosine | 0.1 | l-cysteine | 0.2 |
| Xanthine | 0.02 | l-arginine | 0.2 |
| Biotin | 0.0005 | l-asparagine | 0.2 |
| Cyanocobalamin | 0.0001 | l-tryptophan | 0.2 |
| Folic acid | 0.0001 | l-valine | 0.2 |
| p-Aminobenzoic acid | 0.0005 | l-histidine | 0.2 |
| Nicotinic acid | 0.0005 | l-glycine | 0.2 |
| Calcium pentothenate | 0.0005 | l-threonine | 0.2 |
| Pyridoxine | 0.0005 | l-isoleucine | 0.2 |
| Myo-inositol | 0.0005 | l-methionine | 0.1 |
| l-ascorbate | 0.0001 | l-tyrosine | 0.1 |
| Sequence | Tm (°C) | Reference | |
|---|---|---|---|
| ribA | F: CGATGACTAGTGAACACGAT R: CCCGGGATGATAGAAATCAG | 55.0 55.1 | NC_012984.1 |
| ribB | F: TTAGATGGGCACATTGTTCA R: TAAACTGATCAACAGACGCA | 55.0 54.9 | |
| ribG | F: ATCCAATCGTGGGTGGTAAT R: TATCGCCGTTTTTAGGGTGA | 53.4 53.8 | |
| ribH | F: TAAGATTGGGATTGTCGTGG R: TAGCGCTTGGTTGTAATCAT | 55.1 55.1 |
| Ingredients | Concentration (g/L) | |||
|---|---|---|---|---|
| Media 1 | Media 2 | Media 3 | Media 4 | |
| Glucose | 20 | 20 | 20 | 20 |
| Yeast extract | 40 | 30 | 10 | 0 |
| Soy peptone | 0 | 10 | 30 | 40 |
| Sodium citrate | 5 | 5 | 5 | 5 |
| K2HPO4 | 2 | 2 | 2 | 2 |
| l-ascorbate | 0.05 | 0.05 | 0.05 | 0.05 |
| Tween 80 | 1 | 1 | 1 | 1 |
| MgSO4 | 0.05 | 0.05 | 0.05 | 0.05 |
| MnSO4 | 0.1 | 0.1 | 0.1 | 0.1 |
| l-cysteine | 0.01 | 0.01 | 0.01 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-Y.; Choi, E.-J.; Lee, J.-H.; Yoo, M.-S.; Heo, K.; Shim, J.-J.; Lee, J.-L. Probiotic Potential of a Novel Vitamin B2-Overproducing Lactobacillus plantarum Strain, HY7715, Isolated from Kimchi. Appl. Sci. 2021, 11, 5765. https://doi.org/10.3390/app11135765
Kim J-Y, Choi E-J, Lee J-H, Yoo M-S, Heo K, Shim J-J, Lee J-L. Probiotic Potential of a Novel Vitamin B2-Overproducing Lactobacillus plantarum Strain, HY7715, Isolated from Kimchi. Applied Sciences. 2021; 11(13):5765. https://doi.org/10.3390/app11135765
Chicago/Turabian StyleKim, Joo-Yun, Eun-Jung Choi, Jae-Ho Lee, Myeong-Seok Yoo, Keon Heo, Jae-Jung Shim, and Jung-Lyoul Lee. 2021. "Probiotic Potential of a Novel Vitamin B2-Overproducing Lactobacillus plantarum Strain, HY7715, Isolated from Kimchi" Applied Sciences 11, no. 13: 5765. https://doi.org/10.3390/app11135765
APA StyleKim, J.-Y., Choi, E.-J., Lee, J.-H., Yoo, M.-S., Heo, K., Shim, J.-J., & Lee, J.-L. (2021). Probiotic Potential of a Novel Vitamin B2-Overproducing Lactobacillus plantarum Strain, HY7715, Isolated from Kimchi. Applied Sciences, 11(13), 5765. https://doi.org/10.3390/app11135765






