Lignocellulolytic Potential of the Recently Described Species Aspergillus olivimuriae on Different Solid Wastes
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism and Materials
2.2. Annotation of Protein-Coding Genes for Holocellulose Degrading Enzymes
2.3. Inoculum Preparation
2.4. Solid-State Cultures
2.5. Analytical Methods
2.6. Recovery of the Enzymatic Extract from Solid-State Cultures
2.7. Enzymatic Assays
2.8. Statistical Analysis
3. Results and Discussion
3.1. Chemical Characterization of the Residues
3.2. Fungal Colonization of Solid Substrates and Impact on Their Chemical Composition
3.3. Gene Prediction of Holocellulose Degrading Enzymes in A. olivimuriae
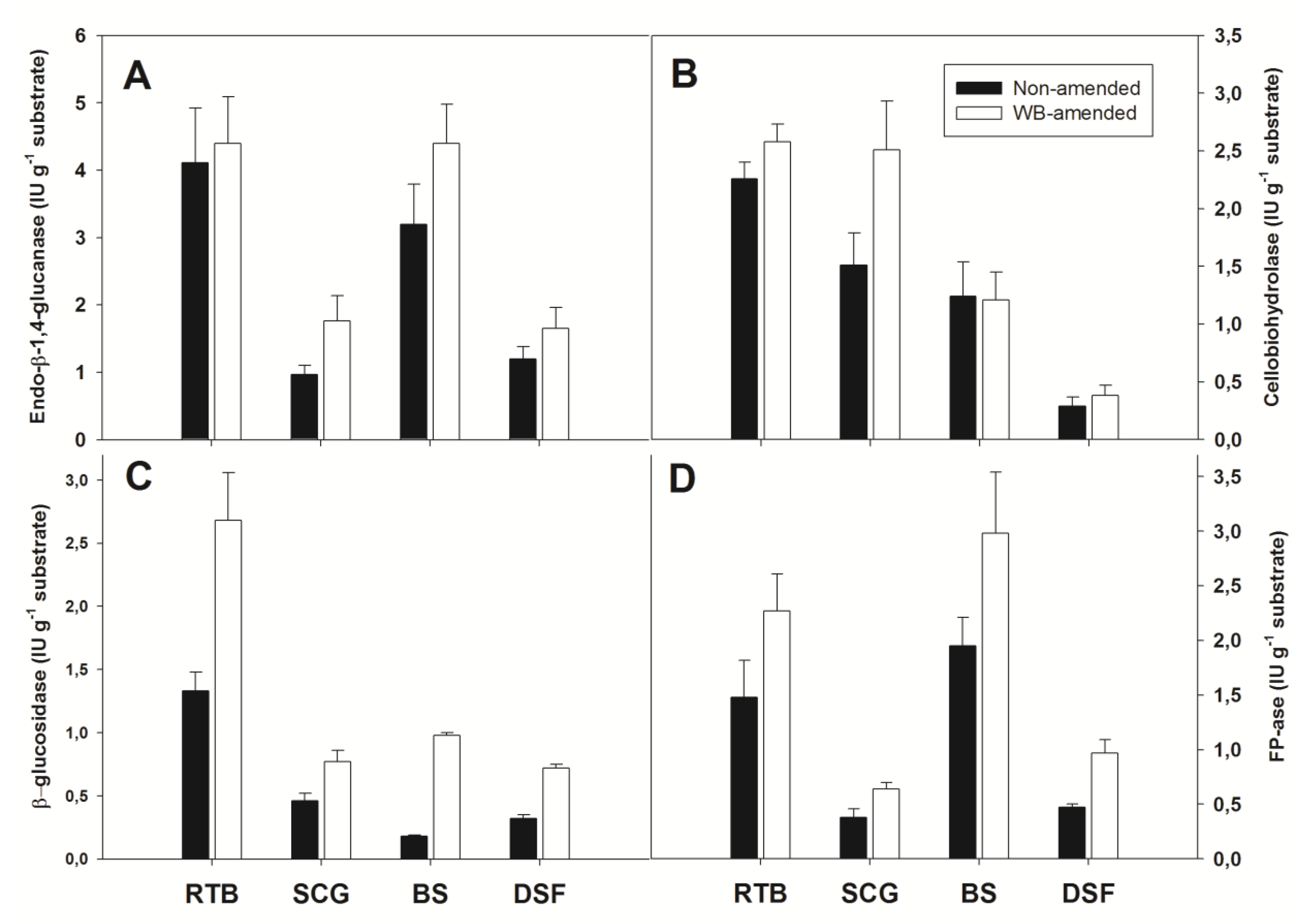
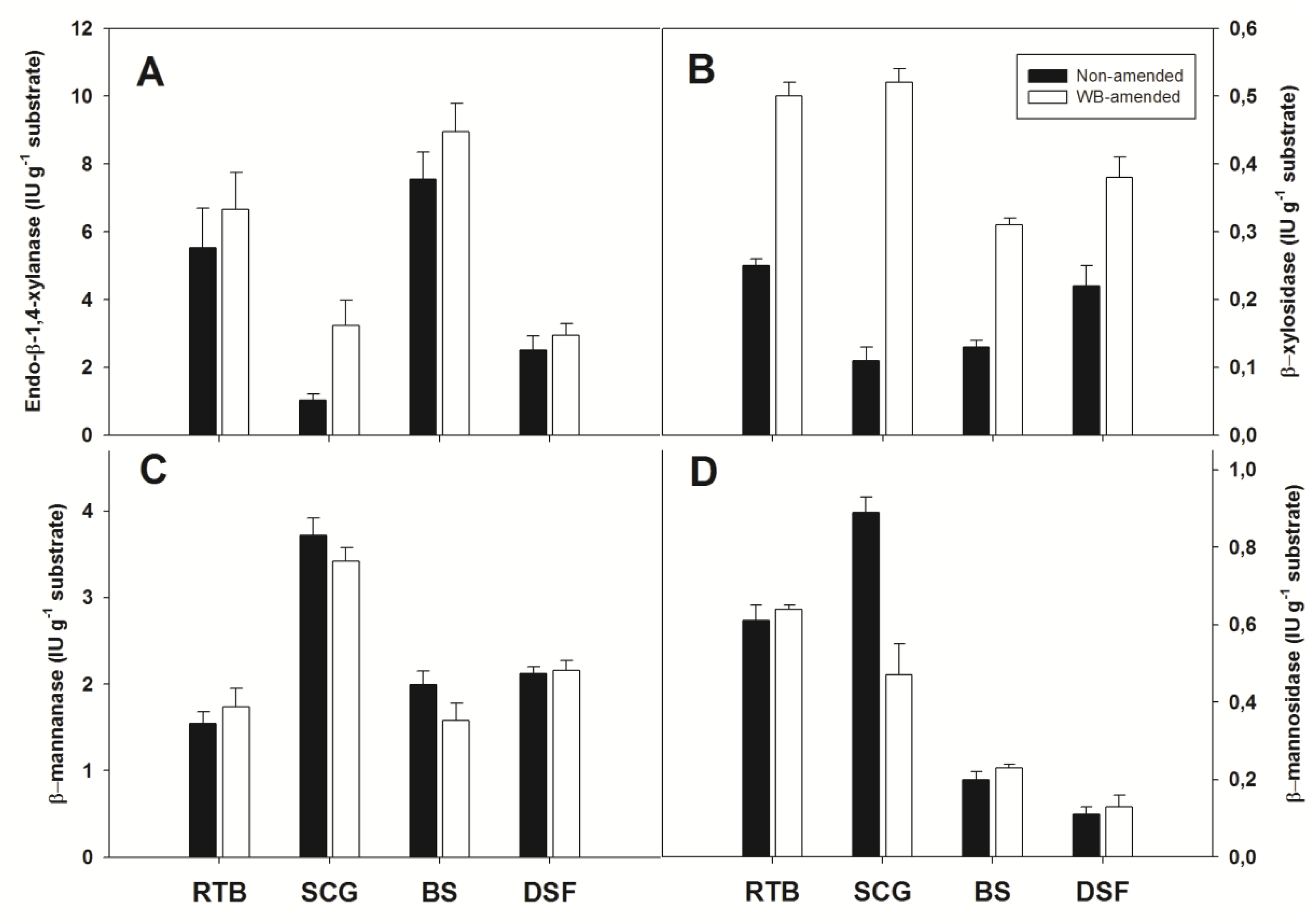
3.4. Solid-State Production of A. olivimuriae Extracellular Glycosyl Hydrolases and Lyases
3.5. Solid-State Production of Lignin-Modifying Enzymes by A. olivimuriae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Symbols and Abbreviations
References
- Troiano, D.; Orsat, V.; Dumont, M.J. Status of filamentous fungi in integrated biorefineries. Renew. Sustain. Energy Rev. 2020, 117, 109472. [Google Scholar] [CrossRef]
- Culleton, H.; McKie, V.; de Vries, R.P. Physiological and molecular aspects of degradation of plant polysaccharides by fungi: What have we learned from Aspergillus? Biotechnol. J. 2013, 8, 884–894. [Google Scholar] [CrossRef]
- Ma, Y.; Ling, T.J.; Su, X.Q.; Jiang, B.; Nian, B.; Chen, L.J.; Liu, L.M.; Zhang, Z.Y.; Wang, D.P.; Mu, Y.Y.; et al. Integrated proteomics and metabolomics analysis of tea leaves fermented by Aspergillus niger, Aspergillus tamarii and Aspergillus fumigatus. Food Chem. 2021, 334, 127560. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, 5. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, P.M.; Andersen, M.R.; Kolenova, K.; Benoit, I.; Gruben, B.S.; Trejo-Aguilar, B.; Visser, H.; van Solingen, P.; Pakula, T.; Seiboth, B.; et al. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fung. Genet. Biol. 2009, 46, S161–S169. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 1–14. [Google Scholar] [CrossRef]
- Kanayama, N.; Tohru, S.; Keiichi, K. Purification and characterization of an alkaline manganese peroxidase from Aspergillus terreus LD-1. J. Biosci. Bioeng. 2002, 93, 405–410. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, Z.; Wang, F.; Li, J.; Hu, K.; Du, Z. Lignin degradation in corn stover catalyzed by lignin peroxidase from Aspergillus oryzae broth: Effects of conditions on the kinetics. Renew. Energy 2019, 130, 32–40. [Google Scholar] [CrossRef]
- Jurjevic, Z.; Peterson, S.W.; Horn, B.W. Aspergillus section Versicolores: Nine new species and multilocus DNA sequence-based phylogeny. IMA Fung. 2012, 3, 59–79. [Google Scholar] [CrossRef]
- Hubka, V.; Novàkovà, A.; Kolàrík, M.; Jurjevic, Ž.; Peterson, S.W. Revision of Aspergillus section Flavipedes: Seven new species and proposal of section Jani sect. nov. Mycologia 2015, 107, 169–208. [Google Scholar] [CrossRef]
- Arzanlou, M.; Samadi, R.; Frisvad, J.C.; Houbraken, J.; Ghosta, Y. Two novel Aspergillus species from hypersaline soils of the national park of lake Urmia, Iran. Mycol. Prog. 2016, 15, 1081–1092. [Google Scholar] [CrossRef][Green Version]
- Martinelli, L.; Zalar, P.; Gunde-Cimerman, N.; Azua-Bustos, A.; Sterflinger, K.; Pinar, G. Aspergillus atacamensis and A. salisburgensis: Two new halophilic species from hypersaline/arid habitats with a phialosimplex-like morphology. Extremophiles 2017, 21, 755–773. [Google Scholar] [CrossRef]
- Crognale, S.; Pesciaroli, L.; Felli, M.; Petruccioli, M.; D’Annibale, A.; Bresciani, A.; Peterson, S.W. Aspergillus olivimuriae sp. nov., a halotolerant species isolated from olive brine. Int. J. Syst. Evolut. Microbiol. 2019, 69, 2899–2906. [Google Scholar] [CrossRef]
- Singhania, R.R.; Patel, A.K.; Soccol, C.R.; Pandey, A. Recent advances in solid-state fermentation. Biochem. Eng. J. 2009, 44, 13–18. [Google Scholar] [CrossRef]
- El-Bakry, M.; Abraham, J.; Cerda, A.; Barrena, R.; Ponsá, S.; Gea, T.; Sánchez, A. From wastes to high value added products: Novel aspects of SSF in the production of enzymes. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1999–2042. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Segato, F.; Damásio, A.R.; de Lucas, R.C.; Squina, F.M.; Prade, R.A. Genomics review of holocellulose deconstruction by aspergilli. Microbiol. Mol. Biol. Rev. 2014, 78, 588–613. [Google Scholar] [CrossRef]
- Cruz, R.; Cardoso, M.M.; Fernandes, L.; Oliveira, M.; Mendes, E.; Baptista, P.; Morais, S.; Casal, S. Espresso coffee residues: A valuable source of unextracted compounds. J. Agric. Food Chem. 2012, 60, 7777–7784. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Brozzoli, V.; Bartocci, S.; Terramoccia, S.; Contò, G.; Federici, F.; D’Annibale, A.; Petruccioli, M. Stoned olive pomace fermentation with Pleurotus species and its evaluation as a possible animal feed. Enzyme Microb. Technol. 2010, 46, 223–228. [Google Scholar] [CrossRef]
- Ainsworth, E.; Gillespie, K. Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin-Ciocalteu reagent. Nat. Protoc. 2007, 2, 875–877. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Paredes, C.; Roig, A.; Bernal, M.P.; Sanchez-Monedero, M.A.; Cegarra, J. Evolution of organic matter and nitrogen during co-composting of olive mill wastewater with solid organic wastes. Biol. Fertil. Soils 2000, 32, 222–227. [Google Scholar] [CrossRef]
- Scotti, C.T.; Vergoignan, C.; Feron, G.; Durand, A. Glucosamine measurement as indirect method for biomass estimation of Cunninghamella elegans grown in solid state cultivation conditions. Biochem. Eng. J. 2001, 7, 1–5. [Google Scholar] [CrossRef]
- Sakurai, Y.; Lee, T.H.; Shiota, H. On the convenient method for glucosamine estimation in koji. Agric. Biol. Chem. 1977, 41, 619–624. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Giovannozzi-Sermanni, G.; D’Annibale, A.; Perani, C.; Porri, A.; Pastina, F.; Minelli, V.; Vitale, N.; Gelsomino, A. Characteristics of paper handsheets after combined biological pretreatments and conventional pulping of wheat straw. Tappi J. 1994, 77, 151–158. [Google Scholar]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Wood, T.M.; Bhat, K.M. Methods for measuring cellulase activities. Methods Enzymol. 1988, 160, 87–112. [Google Scholar]
- Meijer, M.; Houbraken, J.A.M.P.; Dalhuijsen, S.; Samson, R.A.; De Vries, R.P. Growth and hydrolase profiles can be used as characteristics to distinguish Aspergillus niger and other black aspergilli. Stud. Mycol. 2011, 69, 19–30. [Google Scholar] [CrossRef]
- Heerd, D.; Yegin, S.; Tari, C.L.; Fernandez-Lahore, M. Pectinase enzyme-complex production by Aspergillus spp. in solid-state fermentation: A comparative study. Food Bioprod. Process. 2012, 90, 102–110. [Google Scholar] [CrossRef]
- Vilariño, C.; Del Giorgio, J.F.; Hours, R.A.; Cascone, O. Spectrophotometric method for fungal pectinesterase activity determination. LWT-Food Sci. Technol. 1993, 26, 107–110. [Google Scholar] [CrossRef]
- del Pilar Castillo, M.; Ander, P.; Stenstrom, J. Lignin and manganese peroxidase activity in extracts from straw solid substrate fermentations. Biotechnol Techn. 1997, 11, 701–706. [Google Scholar] [CrossRef]
- Balandrán-Quintana, R.R.; Mercado-Ruiz, J.N.; Mendoza-Wilson, A.M. Wheat bran proteins: A review of their uses and potential. Food Rev. Int. 2015, 31, 279–293. [Google Scholar] [CrossRef]
- Bartłomiej, S.; Justyna, R.K.; Ewa, N. Bioactive compounds in cereal grains—occurrence, structure, technological significance and nutritional benefits—A review. Food Sci. Technol. Int. 2012, 18, 559–568. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Carneiro, L.M.; Silva, J.P.; Roberto, I.C.; Teixeira, J. A study on chemical constituents and sugars extraction from spent coffee grounds. Carbohydr. Polym. 2011, 83, 368–374. [Google Scholar] [CrossRef]
- Kwon, E.E.; Yi, H.; Jeon, Y.J. Sequential co-production of biodiesel and bioethanol with spent coffee grounds. Bioresour. Technol. 2013, 136, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Couto, R.M.; Fernandes, J.; Da Silva, M.G.; Simoes, P.C. Supercritical fluid extraction of lipids from spent coffee grounds. J. Supercrit. Fluids 2009, 51, 159–166. [Google Scholar] [CrossRef]
- Burniol-Figols, A.; Cenian, K.; Skiadas, I.V.; Gavala, H.N. Integration of chlorogenic acid recovery and bioethanol production from spent coffee grounds. Biochem. Eng. J. 2016, 116, 54–64. [Google Scholar] [CrossRef]
- Panusa, A.; Zuorro, A.; Lavecchia, R.; Marrosu, G.; Petrucci, R.; Farmaco, D. Recovery of natural antioxidants from spent coffee grounds. J. Agric. Food Chem. 2013, 61, 4162–4168. [Google Scholar] [CrossRef]
- Stuper-Szablewska, K.; Perkowski, J. Phenolic acids in cereal grain: Occurrence, biosynthesis, metabolism and role in living organisms. Crit. Rev. Food Sci. Nutr. 2019, 59, 664–675. [Google Scholar] [CrossRef]
- Ferri, M.; Happel, A.; Zanaroli, G.; Bertolini, M.; Chiesa, S.; Commisso, M.; Guzzo, F.; Tassoni, A. Advances in combined enzymatic extraction of ferulic acid from wheat bran. New Biotechnol. 2020, 56, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, H.; Arkell, A.; Jönsson, A.S. Impact of prefiltration on membrane performance during isolation of hemicelluloses extracted from wheat bran. Separ. Purif. Technol. 2013, 116, 192–198. [Google Scholar] [CrossRef]
- Merali, Z.; Collins, S.R.; Elliston, A.; Wilson, D.R.; Käsper, A.; Waldron, K.W. Characterization of cell wall components of wheat bran following hydrothermal pretreatment and fractionation. Biotechnol. Biofuels 2015, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Prinsen, P.; Gutiérrez, A.; Faulds, C.B.; Del Río, J.C. Comprehensive study of valuable lipophilic phytochemicals in wheat bran. J. Agric. Food Chem. 2014, 62, 1664–1673. [Google Scholar] [CrossRef]
- Musaalbakri, A.M.; Webb, C. Estimation of growth in solid state fermentation: A review. Malays. J. Microbiol. 2018, 14, 61–69. [Google Scholar]
- Sharma, P.D.; Fisher, P.J.; Webster, J. Critique of the chitin assay technique for the estimation of fungal biomass. Transact. British Mycol. Soc. 1977, 69, 479–483. [Google Scholar] [CrossRef]
- Andersen, M.R.; Lehmann, L.; Nielsen, J. Systemic analysis of the response of Aspergillus niger to ambient pH. Genome Biol. 2009, 10, 1–14. [Google Scholar] [CrossRef]
- Mai, H.T.N.; Lee, K.M.; Choi, S.S. Enhanced oxalic acid production from corncob by a methanol-resistant strain of Aspergillus niger using semi solid-sate fermentation. Process Biochem. 2016, 51, 9–15. [Google Scholar] [CrossRef]
- Poulsen, L.; Andersen, M.R.; Lantz, A.E.; Thykaer, J. Identification of a transcription factor controlling pH-dependent organic acid response in Aspergillus niger. PLoS ONE 2012, 7, e50596. [Google Scholar] [CrossRef]
- Mejias, L.; Cerda, A.; Barrena, R.; Gea, T.; Sánchez, A. Microbial strategies for cellulase and xylanase production through solid-state fermentation of digestate from biowaste. Sustainability 2018, 10, 2433. [Google Scholar] [CrossRef]
- Hatakka, A.; Hammel, K.E. Fungal biodegradation of lignocelluloses. In Industrial Applications; Esser, K., Hofrichter, M., Eds.; Springer: Berlin, Germany, 2011; pp. 319–340. [Google Scholar]
- Cardoso Duarte, J.; Costa-Ferreira, M. Aspergilli and lignocellulosics: Enzymology and biotechnological applications. FEMS Microbiol. Rev. 1994, 13, 377–386. [Google Scholar] [CrossRef]
- Santos, J.I.; Fillat, Ú.; Martín-Sampedro, R.; Ballesteros, I.; Manzanares, P.; Ballesteros, M.; Eugenio, M.E.; Ibarra, D. Lignin-enriched fermentation residues from bioethanol production of fast-growing poplar and forage sorghum. Bioresources 2015, 10, 5215–5232. [Google Scholar] [CrossRef][Green Version]
- Hasanin, M.S.; Darwesh, O.M.; Matter, I.A.; El-Saied, H. Isolation and characterization of non-cellulolytic Aspergillus flavus EGYPTA5 exhibiting selective ligninolytic potential. Biocatal. Agric. Biotechnol. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.P.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef]
- Eijsink, V.G.; Petrovic, D.; Forsberg, Z.; Mekasha, S.; Røhr, Å.K.; Várnai, A.; Bissaro, B.; Vaaje-Kolstad, G. On the functional characterization of lytic polysaccharide monooxygenases (LPMOs). Biotechnol. Biofuels 2019, 12, 1–16. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Eveleigh, D.E.; Mandels, M.; Andreotti, R.; Roche, C. Measurement of saccharifying cellulase. Biotechnol. Biofuels 2009, 6, 1–8. [Google Scholar] [CrossRef]
- Crognale, S.; Liuzzi, F.; D’Annibale, A.; de Bari, I.; Petruccioli, M. Cynara cardunculus a novel substrate for solid-state production of Aspergillus tubingensis cellulases and sugar hydrolysates. Biomass Bioenerhy 2019, 127, 105276. [Google Scholar] [CrossRef]
- Gao, J.; Weng, H.; Zhu, D.; Yuan, M.; Guan, F.; Xi, Y. Production and characterization of cellulolytic enzymes from the thermoacidophilic fungal Aspergillus terreus M11 under solid-state cultivation of corn stover. Biores. Technol. 2008, 99, 7623–7629. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Paul, T.; Halder, S.K.; Jana, A.; Maity, C.; Mohapatra, P.K.D.; Pati, B.R.; Mondal, K.C. Production of cellulolytic enzymes by Aspergillus fumigatus ABK9 in wheat bran-rice straw mixed substrate and use of cocktail enzymes for deinking of waste office paper pulp. Biores. Technol. 2013, 128, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Favaro, C.P.; Baraldi, I.J.; Casciatori, F.P.; Farinas, C.S. β-Mannanase production using coffee industry waste for application in soluble coffee processing. Biomolecules 2020, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Rodrigues, P.; Gurgel, L.V.A.; Pasquini, D.; Badotti, F.; Góes-Neto, A.; Baffi, M.A. Lignocellulose-degrading enzymes production by solid-state fermentation through fungal consortium among Ascomycetes and Basidiomycetes. Renew. Energy 2020, 145, 2683–2693. [Google Scholar] [CrossRef]
- Crognale, S.; Pesciaroli, L.; Petruccioli, M.; D’Annibale, A. Phenoloxidase-producing halotolerant fungi from olive brine wastewater. Process Biochem. 2012, 47, 1433–1437. [Google Scholar] [CrossRef]
- Dos Santos, T.C.; dos Santos Reis, N.; Silva, T.P.; Machado, F.D.P.P.; Bonomo, R.C.F.; Franco, M. Prickly palm cactus husk as a raw material for production of ligninolytic enzymes by Aspergillus niger. Food Sci. Biotechnol. 2016, 25, 205–211. [Google Scholar] [CrossRef] [PubMed]
| Parameter (g kg−1) | RTB | SGC | DSF | Barley Straw | WB † |
|---|---|---|---|---|---|
| Hemicellulose | 189.2 ± 3.2 a | 375.2 ± 16.8 c | 172.2 ± 14.9 a | 279.3 ± 3.0 b | 187.6 ± 12.2 c |
| Cellulose | 405.9 ± 12.1 c | 120.2 ± 3.2 a | 393.4 ± 24.4 c | 420.6 ± 2.2 c | 201.1 ± 24.3 b |
| Klason lignin | 204.2 ± 4.6 c | 201.7 ± 7.2 c | 257.4 ± 14.2 d | 165.2 ± 4.6 b | 73.2 ± 3.9 a |
| Crude lipids | 20.8 ± 1.4 a | 139.8 ± 5.4 c | 19.3 ± 2.7 a | 16.2 ± 1.8 a | 45.4 ± 4.3 b |
| Reducing sugars | 3.6 ± 0.4 a | 4.1 ± 0.2 a | 9.2 ± 0.4 b | 41.8 ± 2.7 c | 49.3 ± 3.2 d |
| Phenols | 17.2 ± 1.5 b | 49.2 ± 2.4 d | 15.3 ± 2.4 b | 21.4 ± 1.6 c | 9.2 ± 1.7 a |
| Ash | 85.3 ± 7.0 d | 20.4 ± 2.5 a | 82.1 ± 3.6 d | 70.1 ± 5.3 c | 43.6 ± 2.1 b |
| Nitrogen | 5.9 ± 0.3 a | 22.8 ± 1.6 c | 19.8 ± 1.1 b | 5.4 ± 0.4 a | 30.1 ± 1 d |
| Carbon | 423.4 ± 8.0 ab | 508.9 ± 11.1 d | 415.3 ± 8.2 a | 467.4 ± 5.4 c | 441.2 ± 4.2 b |
| Hydrogen | 62.1 ± 2.0 b | 75.6 ± 1.8 c | 59.4 ± 2.1 a | 55.2 ± 4.1 a | 69.3 ± 4.2 c |
| Sulphur | 1.7 ± 0 b | 1.0 ± 0 a | 2.2 ± 0.1 b | 0.1 ± 0.0 a | 1.8 ± 0.0 b |
| Solid-State Cultures On: | d-Glucosamine (mg g−1) | EXP (mg g−1) | ΔpH † | OML ‡ (%) | YGLC/S (mg g−1) |
|---|---|---|---|---|---|
| RTB | 0.87 ± 0.05 ab | 2.89 ± 0.24 b | −0.80 (6.93 ± 0.03) | 12.64 ± 0.40 c | 6.88 ± 0.50 b |
| RTB + WB | 1.89 ± 0.07 c | 3.49 ± 0.11 c | −0.56 (6.43 ± 0.04) | 17.01 ± 0.60 e | 11.11 ± 0.41 c |
| SCG | 0.94 ± 0.06 b | 2.00 ± 0.09 a | −1.75 (5.77 ± 0.04) | 10.77 ± 1.21 b | 8.73 ± 0.46 b |
| SCG + WB | 2.47 ± 0.08 d | 1.89 ± 0.12 a | −0.73 (5.34 ± 0.02) | 14.70 ± 0.81 d | 16.80 ± 0.54 d |
| BS | 0.70 ± 0.06 a | 5.46 ± 0.33 d | n.s. * (6.50 ± 0.04) | 8.43 ± 0.34 a | 8.30 ± 1.30 b |
| BS + WB | 2.99 ± 0.21 e | 5.04 ± 0.18 d | −0.17 (6.83 ± 0.02) | 12.24 ± 0.78 bc | 24.44 ± 1.63 e |
| DSF | 0.79 ± 0.04 ab | 3.68 ± 0.07 c | +0.24 (6.60 ± 0.05) | 17.73 ± 0.51 e | 4.46 ± 0.49 a |
| DSF + WB | 0.87 ± 0.03 ab | 3.70 ± 0.38 c | +0.64 (6.92 ± 0.08) | 17.03 ± 0.46 e | 5.11 ± 0.7 ab |
| Culture | Cellulose (g kg−1) | Hemicellulose (g kg−1) | Klason Lignin (g kg−1) | Reducing Sugars (g kg−1) | Crude Lipids (g kg−1) | Total Phenols (g kg−1) |
|---|---|---|---|---|---|---|
| RTB (IC) | 405.6 ± 12.1 c | 187.0 ± 3.3 c | 201.1 ± 8.0 a | 3.3 ± 0.6 a | 20.9 ± 2.0 b | 17.3 ± 2.3 a |
| RTB (SSC) | 354.2 ± 4.0 b | 121.9 ± 4.4 a | 217.5 ± 2.2 a | 17.7 ± 2.2 b | 14.5 ± 1.3 a | 38.3 ± 4.2 c |
| RTB + WB (IC) | 365.6 ± 13.4 b | 167.7 ± 6.2 b | 181.2 ± 14.4 a | 3.7 ± 0.5 a | 26.5 ± 2.1 c | 15.6 ± 3.4 a |
| RTB + WB (SSC) | 279.6 ± 16.3 a | 123.4 ± 12.1 a | 204.8 ± 19.5 a | 16.4 ± 0.6 b | 15.3 ± 3.2 a | 23.8 ± 2.8 b |
| SCG (IC) | 119.2 ± 3.0 b | 370.2 ± 16.3 c | 199.4 ± 18.5 b | 3.2 ± 0.0 a | 139.0 ± 6.4 c | 48.7 ± 3.2 b |
| SCG (SSC) | 105.4 ± 4.4 a | 326.3 ± 18.3 b | 240.0 ± 21.3 c | 7.6 ± 1.1 b | 109.2 ± 4.1 b | 56.0 ± 2.3 c |
| SCG + WB (IC) | 136.2 ± 3.2 c | 337.1 ± 8.6 bc | 171.4 ± 5.2 a | 3.1 ± 0.5 a | 121.2 ± 8.2 b | 40.8 ± 2.1 a |
| SCG + CG (SSC) | 103.5 ± 3.1 a | 255.4 ± 21.6 a | 176.5 ± 5.2 ab | 9.6 ± 0.4 c | 75.8 ± 10.3 a | 51.5 ± 2.6 bc |
| BS (IC) | 420.1 ± 3.0 d | 279.5 ± 11.2 c | 165.0 ± 14.3 a | 41.3 ± 2.8 a | 15.6 ± 1.1 a | 20.7 ± 2.1 ab |
| BS (SSC) | 393.9 ± 3.2 c | 237.0 ± 9.3 b | 172.6 ± 8.2 a | 51.3 ± 2.0 b | 12.0 ± 1.9 a | 27.1 ± 4.3 b |
| BS + WB (IC) | 376.1 ± 8.3 b | 262.7 ± 8.3 c | 159.0 ± 14.8 a | 43.4 ± 4.6 a | 21.4 ± 2.4 b | 18.4 ± 4.2 a |
| BS + WB (SSC) | 339.7 ± 5.0 a | 210.4 ± 7.6 a | 170.6 ± 13.3 a | 40.8 ± 5.4 a | 20.3 ± 3.0 b | 25.4 ± 3.0 b |
| DSF (IC) | 393.2 ± 4.6 c | 174.1 ± 12.3 c | 257.2 ± 11.6 bc | 10.9 ± 0.5 b | 19.2 ± 3.1 b | 15.6 ± 1.6 b |
| DSF (SSC) | 363.0 ± 7.1 b | 70.2 ± 9.1 a | 272.0 ± 8.3 c | 4.6 ± 0.3 a | 21.4 ± 0.2 b | 11.9 ± 2.0 a |
| DSF + WB (IC) | 354.2 ± 4.1 b | 177.3 ± 8.6 c | 220.3 ± 4.8 a | 17.4 ± 3.1 c | 22.4 ± 3.1 b | 14.3 ± 1.3 ab |
| DSF + WB (SSC) | 321.3 ± 12.0 a | 92.7 ± 8.3 b | 236.1 ± 5.2 b | 4.9 ± 0.1 a | 4.4 ± 0.2 a | 12.9 ± 1.1 a |
| Substrate | Enzyme Activity | CAZyme Families | Number of Protein-Coding Predicted Genes | |||||
|---|---|---|---|---|---|---|---|---|
| Aol † | Afa | Anda | Anga | Aora | ||||
| Cellulose | β-1,4-Endoglucanase | GH3, -5, -6, -7, -9, -12, -45 | 35 (22) | 41 | 42 | 36 | 41 | |
| Cellobiohydrolase | GH7 | 3 (3) | 4 | 3 | 2 | 3 | ||
| β-1,4-Glucosidase | GH1, -3 | 17 (11) | 23 | 23 | 22 | 26 | ||
| Cellobiose dehydrogenase | AA3, AA8 | 2 (2) | 2 | 2 | 2 | 2 | ||
| Lytic polysaccharide monooxygenase | AA9 | 6 (6) | 7 | 9 | 8 | 8 | ||
| Hemicellulose | Xylan | β-1,4-Endoxylanase | GH10, -11, -30 | 6 (5) | 8 | 5 | 6 | 8 |
| β-1,4-Xylosidase | GH3, -43 | 30 (15) | 36 | 35 | 30 | 43 | ||
| Galactomannan | β-1,4-Endomannanase | GH5 | 15 (5) | 13 | 15 | 10 | 13 | |
| β-1,4-Mannosidase | GH2 | 6 (2) | 6 | 10 | 6 | 7 | ||
| β-1,4-Galactosidase | GH2, -35 | 9 (3) | 11 | 14 | 11 | 14 | ||
| α-1,4-Galactosidase | GH27, -36 | 3 (2) | 3 | 4 | 2 | 3 | ||
| α-Arabinofuranosidase | GH51, -54 | 3 (3) | 3 | 3 | 5 | 4 | ||
| Xyloglucan | β-1,4-endoglucanase | GH12, -74 | 3 (3) | 6 | 3 | 4 | 4 | |
| α-, β-Arabinofuranosidase | GH51, -54, -127 | 2 (3) | 3 | 4 | 2 | 3 | ||
| α-Xylosidase | GH31 | 10 (1) | n.a. | 10 b | 7 b | 10 b | ||
| α-Fucosidase | GH29, -95 | 2 (0) | n.a. | 3 b | 3 b | 3 b | ||
| α-1,4-Galactosidase | GH27, -36 | 3 (2) | 3 | 4 | 2 | 3 | ||
| β -1,4-Galactosidase | GH2, -35 | 9 (3) | 11 | 14 | 11 | 14 | ||
| Arabinoxylan | Arabinofuranohydrolase | GH62 | 2 (2) | 2 | 2 | 1 | 2 | |
| α-, β-Glucuronidase | GH67, -115, -154 | 4 (2) | 2 | 2 | 1 | 5 | ||
| α-1,4-Galactosidase | GH27, -36 | 3 (2) | 3 | 4 | 2 | 3 | ||
| β-1,4-Galactosidase | GH2, -35 | 9 (3) | 11 | 14 | 11 | 14 | ||
| Acetyl xylan esterase | CE1, -5 | 3 (4) | 10 | 7 | 8 | 10 | ||
| Feruloyl esterase | CE1 | 1 (1) | 5 | 3 | 3 | 5 | ||
| Pectin | Endo-polygalacturonases | GH28 | 3 (1) | 14 | 9 | 22 | 21 | |
| α-Rhamnosidase | GH78, 106 | 7 (0) | 6 | 8 | 8 | 9 | ||
| α-Arabinofuranosidase, Arabinofuranohydrolase | GH51, -54, -62 | 4 (5) | 5 | 5 | 6 | 6 | ||
| Endoarabinanase | GH43 | 14 (5) | 18 | 15 | 11 | 20 | ||
| Exoarabinanase | GH93 | 3 (2) | 3 | 2 | 1 | 3 | ||
| β -1,4-Endogalactanase | GH53 | 1 (1) | 1 | 1 | 1 | 1 | ||
| Unsaturated glucuronyl hydrolase | GH88 | 1 (0) | n.a. | 2 b | 1 b | 3 b | ||
| Unsaturated rhamnogalacturonan hydrolase | GH105 | 2 (0) | n.a. | 3 b | 2 b | 4 b | ||
| β-1,4-Xylosidase | GH3, -43 | 30 (15) | 36 | 35 | 30 | 43 | ||
| β-1,4-Galactosidase | GH2, -35 | 9 (3) | 11 | 14 | 11 | 14 | ||
| Pectate lyase | PL1 | 2 (0) | 6 | 8 | 7 | 12 | ||
| Rhamnogalacturonan lyase | PL4 | 1 (0) | 3 | 4 | 2 | 4 | ||
| Pectin methyl esterase PME | CE8 | 1 (0) | 6 | 3 | 3 | 5 | ||
| Rhamnogalacturonan acetyl esterase | CE12 | 2 (0) | 3 | 2 | 2 | 4 | ||
| Feruloyl esterase CE1 | CE1 | 1 (3) | 5 | 3 | 3 | 5 | ||
| Lignin | Laccase/p-diphenol:oxygen oxidoreductase | AA1 | 3 (2) | 1 c | 2 b | 2 c | 4 b | |
| Lignin-modifying peroxidases | AA2 | 2 (0) | 3 c | 0 b | 2 c | 0 b | ||
| 1,4-benzoquinone reductase | AA6 | 1 (1) | 0 c | 0 b | 2 c | 1 b | ||
| Culture | PMG (IU g−1 Substrate) | PE (IU g−1 Substrate) | PG (IU g−1 Substrate) | Pectin Lyase (IU g−1 Substrate) | Pectate Lyase (IU g−1 Substrate) |
|---|---|---|---|---|---|
| RTB | 2.30 ± 0.18 c | 1.24 ± 0.20 c | 1.27 ± 0.21 c | 0.33 ± 0.08 bc | 0.11 ± 0.03 a |
| RTB + WB | 2.99 ± 0.27 d | 2.16 ± 0.33 d | 1.93 ± 0.09 d | 0.31 ± 0.01 b | 0.08 ± 0.01 a |
| SCG | 0.66 ± 0.05 a | 0.39 ± 0.03 ab | 0.54 ± 0.06 a | 0.17 ± 0.05 a | 0.12 ± 0.06 ab |
| SCG + WB | 0.73 ± 0.08 a | 0.53 ± 0.08 b | 0.56 ± 0.11 a | 0.15 ± 0.01 a | 0.10 ± 0.02 a |
| BS | 0.99 ± 0.13 a | 0.10 ± 0.01 a | 0.74 ± 0.11 ab | 0.19 ± 0.04 a | 0.20 ± 0.04 ab |
| BS + WB | 1.49 ± 0.13 b | 0.06 ± 0.02 a | 0.91 ± 0.11 b | 0.31 ± 0.02 b | 0.25 ± 0.04 b |
| DSF | 0.49 ± 0.12 a | 0.14 ± 0.05 ab | 0.52 ± 0.07 a | 0.19 ± 0.04 a | 0.22 ± 0.02 b |
| DSF + WB | 0.71 ± 0.09 a | 0.09 ± 0.02 a | 0.73 ± 0.11 a | 0.46 ± 0.05 c | 0.26 ± 0.04 b |
| Culture | Laccase | Peroxidase | ||
|---|---|---|---|---|
| (IU g−1 Substrate) (×10−3) | (IU mg−1 Protein) (×10−3) | (IU g−1 Substrate) (×10−3) | (IU mg−1 Protein) (×10−3) | |
| RTB | 32.8 ± 0.4 c | 11.3 ± 0.1 | 30.0 ± 1.6 b | 10.4 ± 1.3 |
| RTB + WB | 49.2 ± 3.2 d | 14.1 ± 2.5 | 47.4 ± 6.9 c | 13.6 ± 1.0 |
| SCG | 32.9 ± 4.1 c | 16.5 ± 0.4 | 23.6 ± 3.3 b | 11.8 ± 0,6 |
| SCG + WB | 41.3 ± 3.0 d | 21.9 ± 1.9 | 27.6 ± 3.8 b | 14.6 ± 1.8 |
| BS | 20.0 ± 1.6 b | 3.6 ± 0.1 | 13.8 ± 2.5 a | 2.5 ± 0.8 |
| BS + WB | 18.6 ± 1.1 b | 3.7 ± 0.2 | 15.6 ± 3.2 ab | 3.1 ± 1.0 |
| DSF | 2.7 ± 0.6 a | 0.7 ± 0.1 | n. d. ‡ | n. d. ‡ |
| DSF + WB | 3.6 ± 0.8 a | 1.0 ± 0.2 | n. d. ‡ | n. d. ‡ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carota, E.; Crognale, S.; Russo, C.; Petruccioli, M.; D’Annibale, A. Lignocellulolytic Potential of the Recently Described Species Aspergillus olivimuriae on Different Solid Wastes. Appl. Sci. 2021, 11, 5349. https://doi.org/10.3390/app11125349
Carota E, Crognale S, Russo C, Petruccioli M, D’Annibale A. Lignocellulolytic Potential of the Recently Described Species Aspergillus olivimuriae on Different Solid Wastes. Applied Sciences. 2021; 11(12):5349. https://doi.org/10.3390/app11125349
Chicago/Turabian StyleCarota, Eleonora, Silvia Crognale, Cristina Russo, Maurizio Petruccioli, and Alessandro D’Annibale. 2021. "Lignocellulolytic Potential of the Recently Described Species Aspergillus olivimuriae on Different Solid Wastes" Applied Sciences 11, no. 12: 5349. https://doi.org/10.3390/app11125349
APA StyleCarota, E., Crognale, S., Russo, C., Petruccioli, M., & D’Annibale, A. (2021). Lignocellulolytic Potential of the Recently Described Species Aspergillus olivimuriae on Different Solid Wastes. Applied Sciences, 11(12), 5349. https://doi.org/10.3390/app11125349










