Evolution of Microbial Composition and Enzymatic Activities during the Composting of Textile Waste
Abstract
1. Introduction
2. Materialsand Methods
2.1. Composting Process
2.2. Physico-Chemical Analysis
2.3. Infrared Spectroscopy Analysis
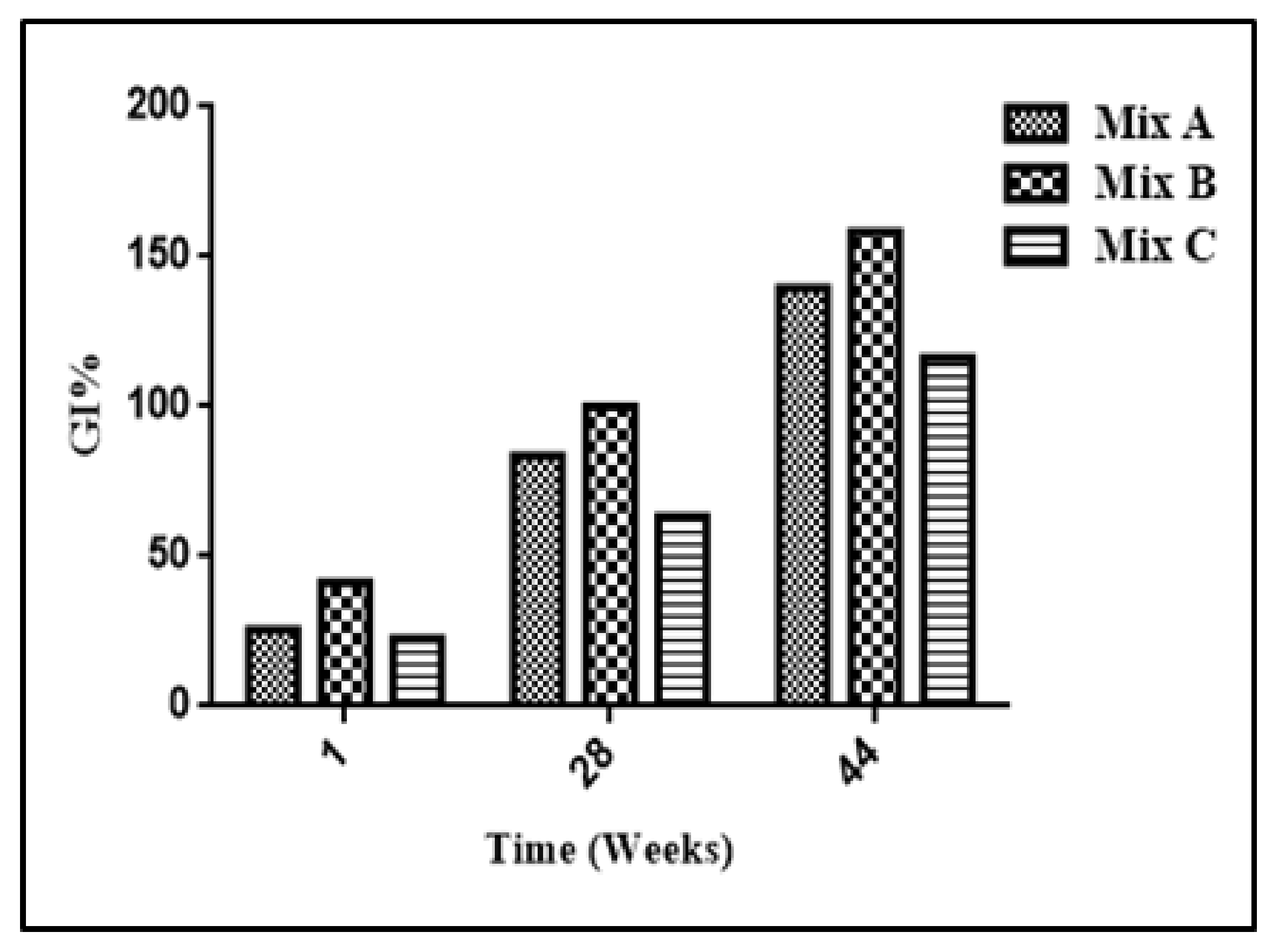
2.4. Germination Testing
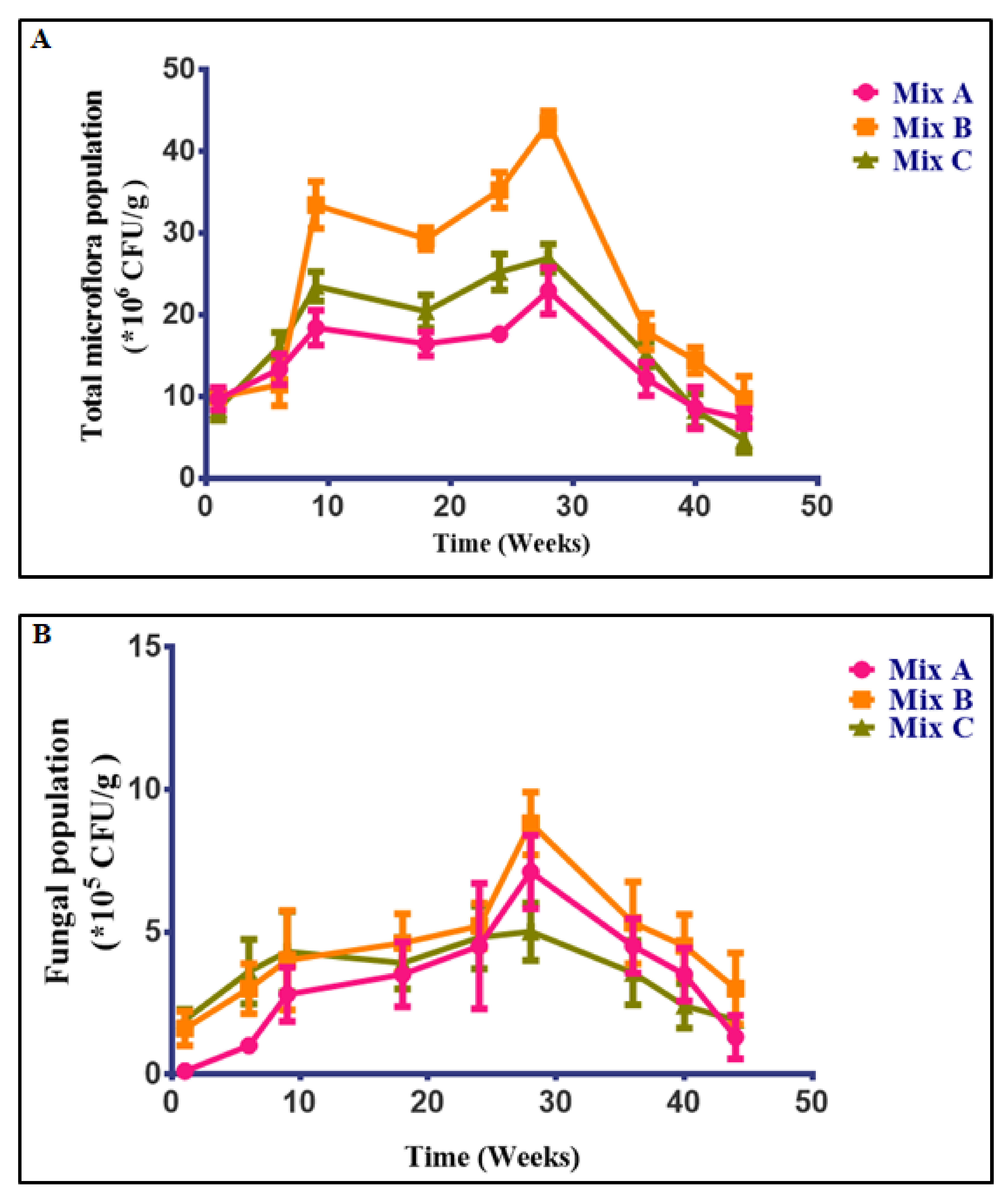
2.5. Microbiological Analysis
2.6. Enzymatic Activity Assays
2.6.1. Cellulase Activity
2.6.2. Phosphatase Activity
2.6.3. Identification of Microorganisms Using NGS
2.6.4. Statistical Analyses
3. Results
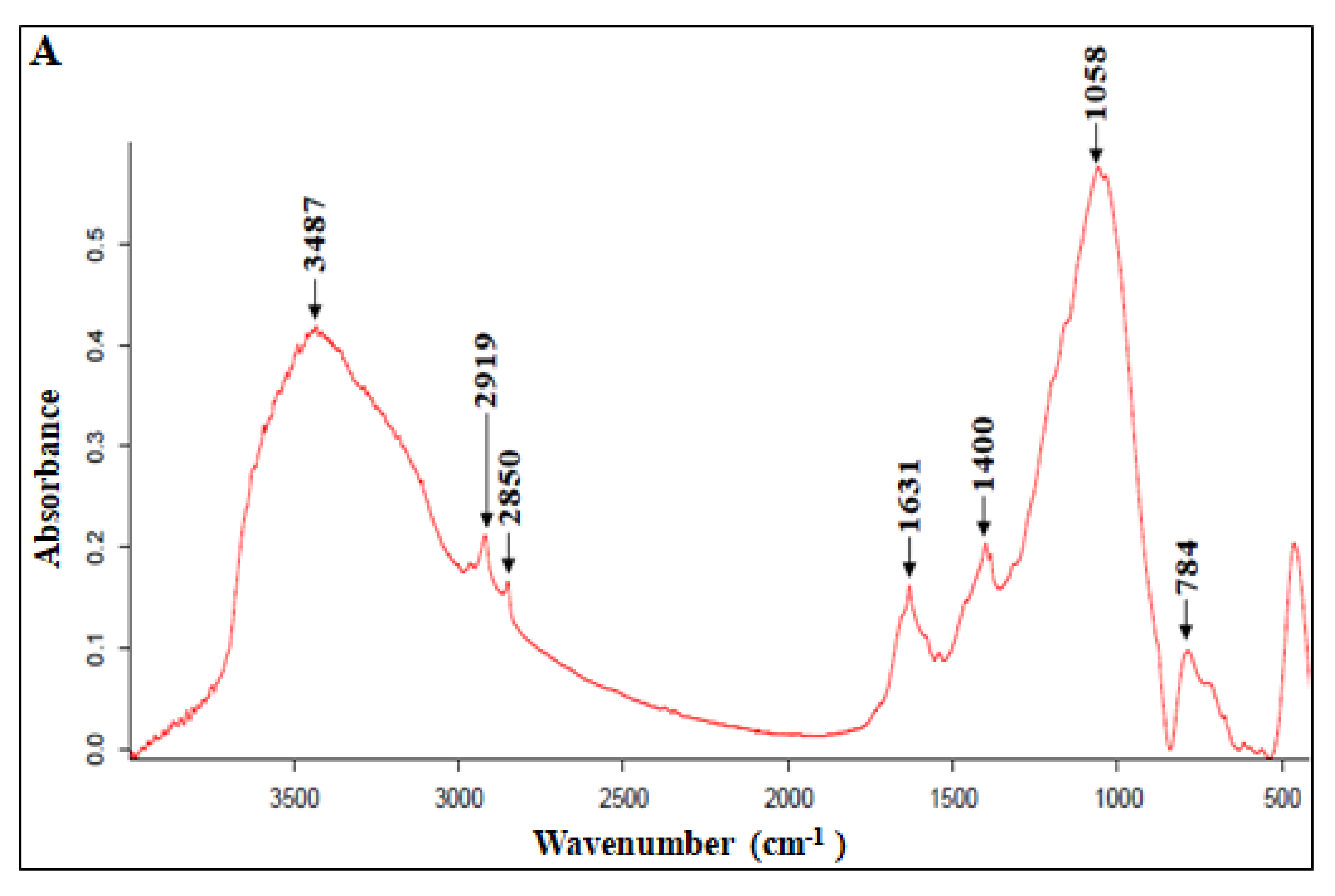
3.1. Infrared Spectroscopy Analysis
3.2. Changes in Total Organic Carbon and Total Nitrogen Concentrations and the C/N Ratio during the Composting Process
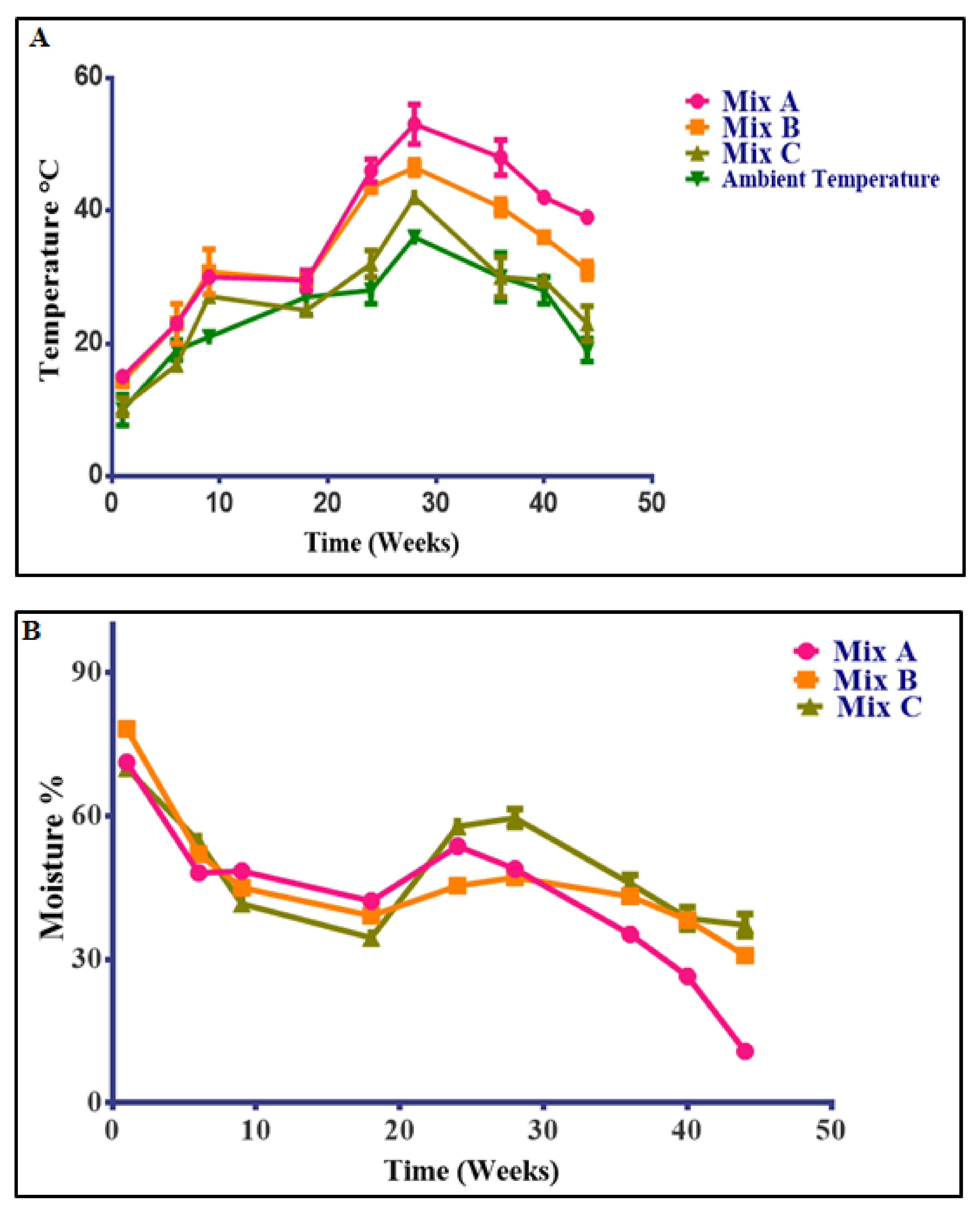
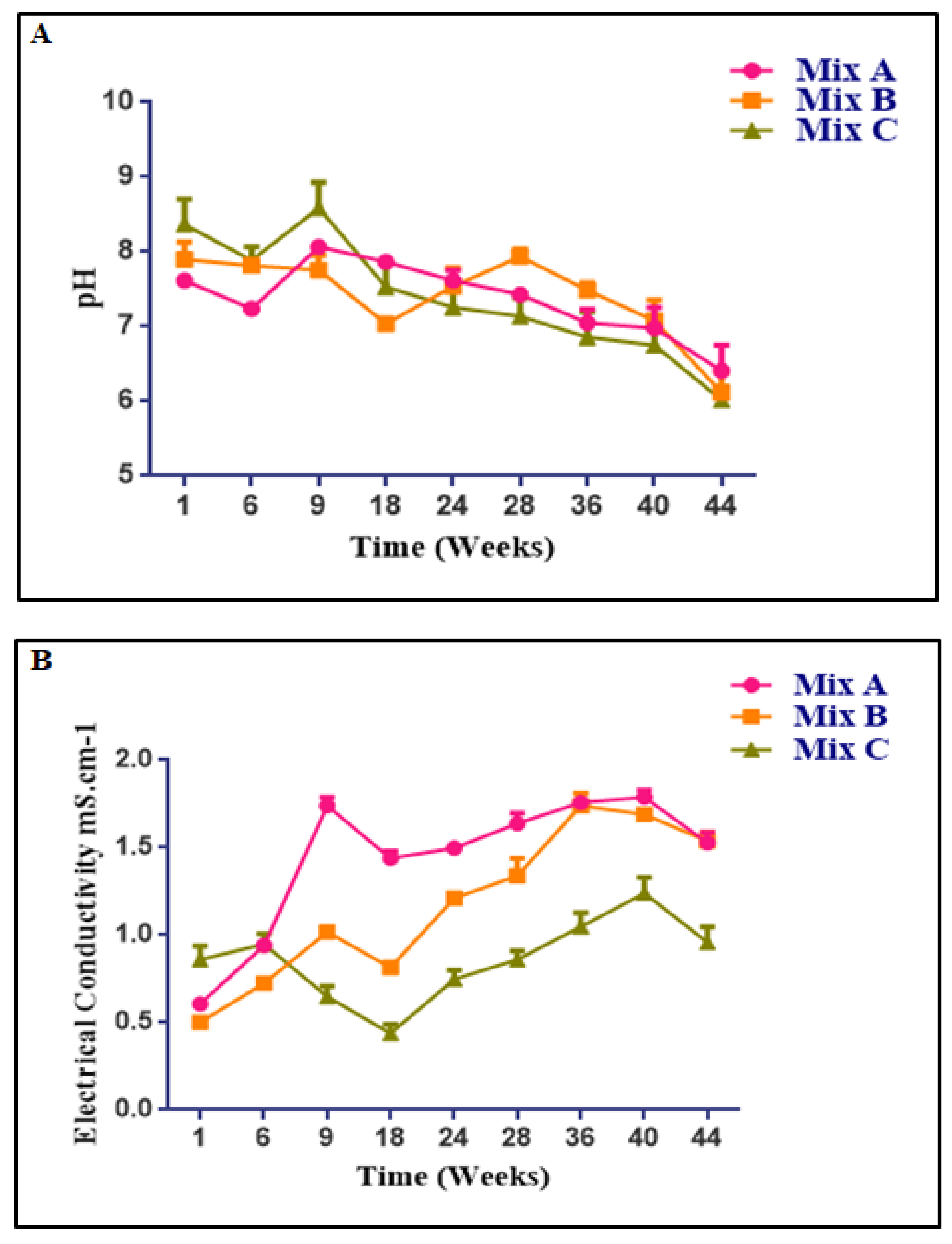
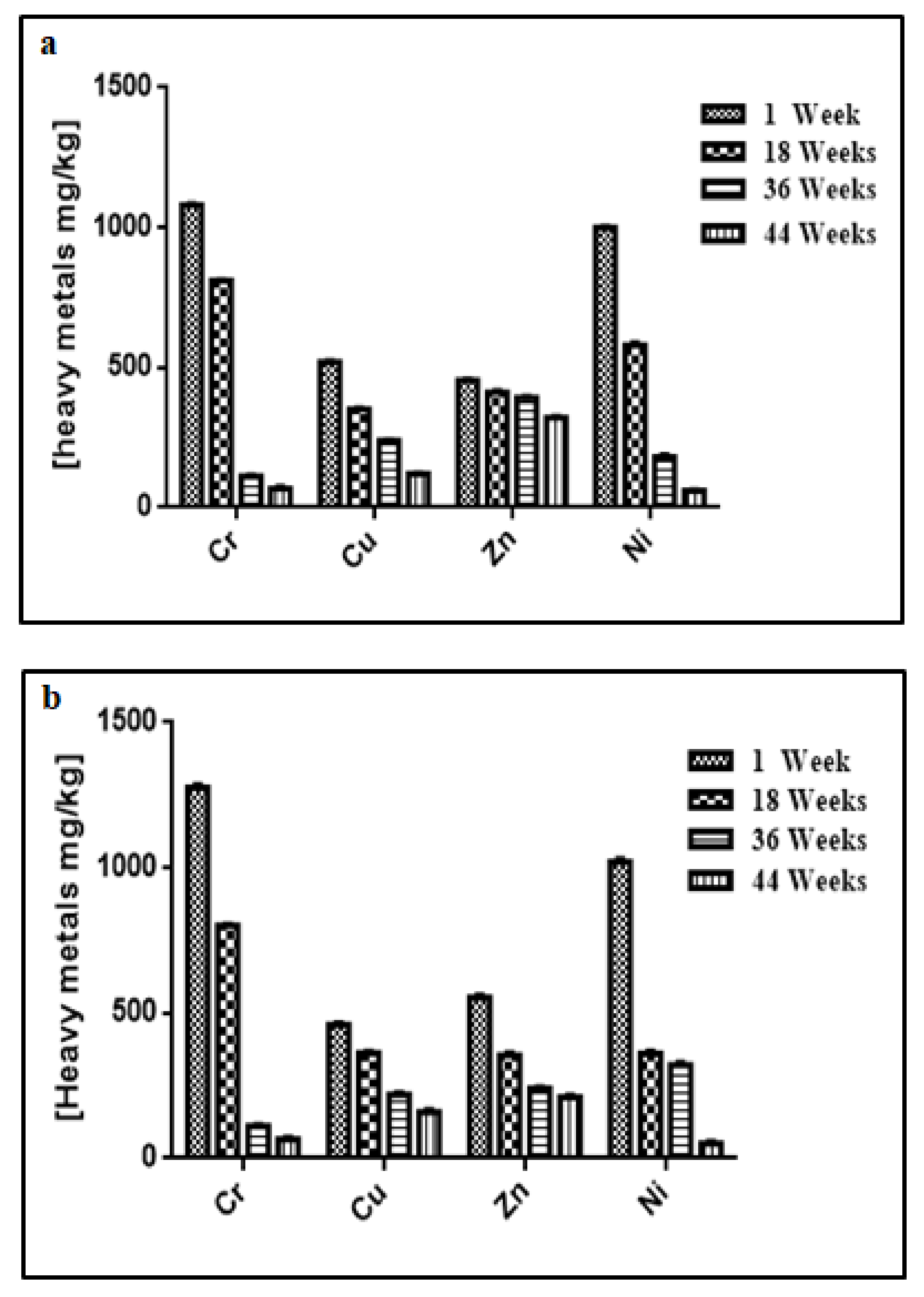
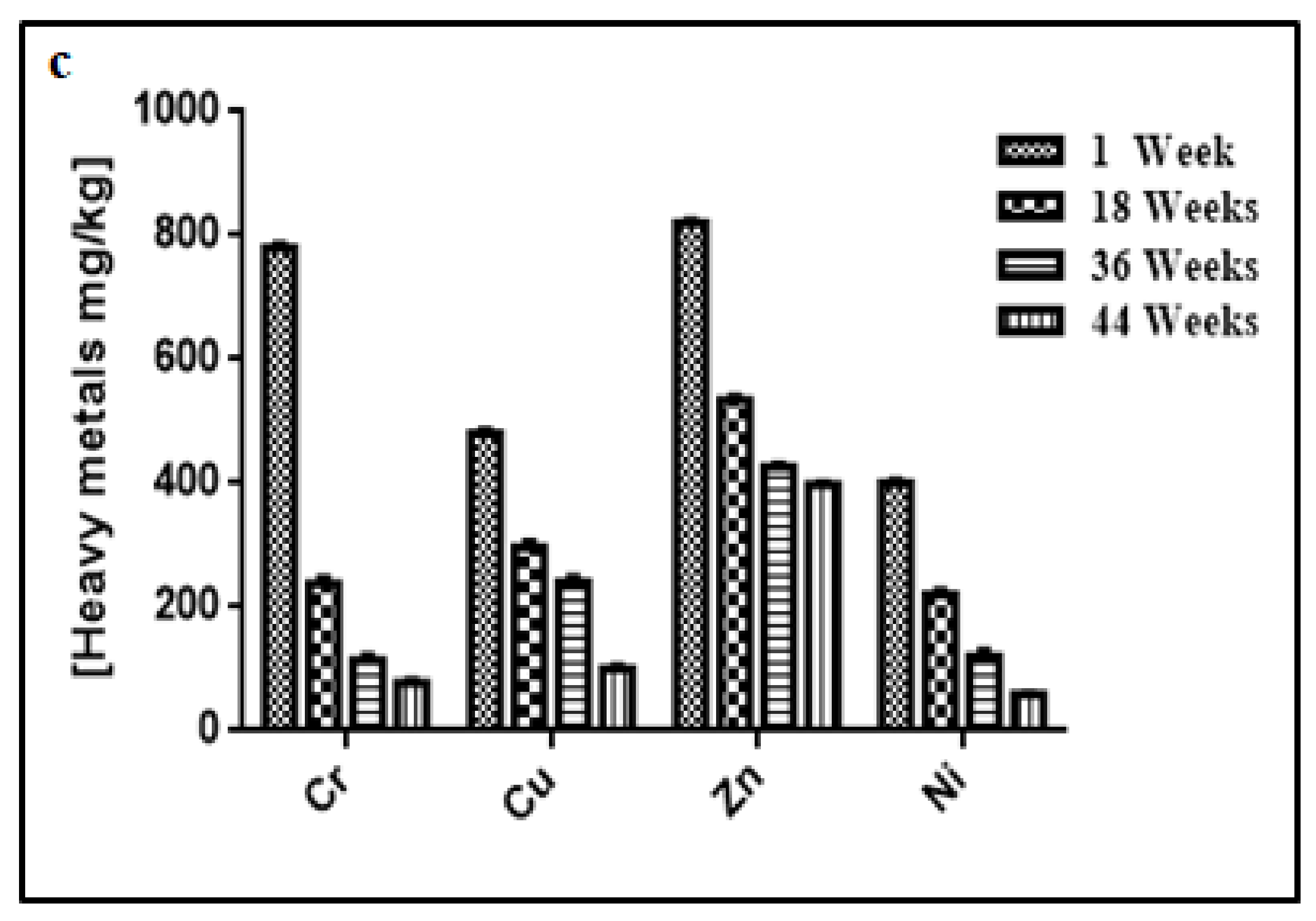
3.3. Changes in Temperature, Moisture, pH, Electrical Conductivity, Heavy Metal Contents, and Microbial Concentration during the Composting Process
3.4. Enzymatic Activity Changes during Composting
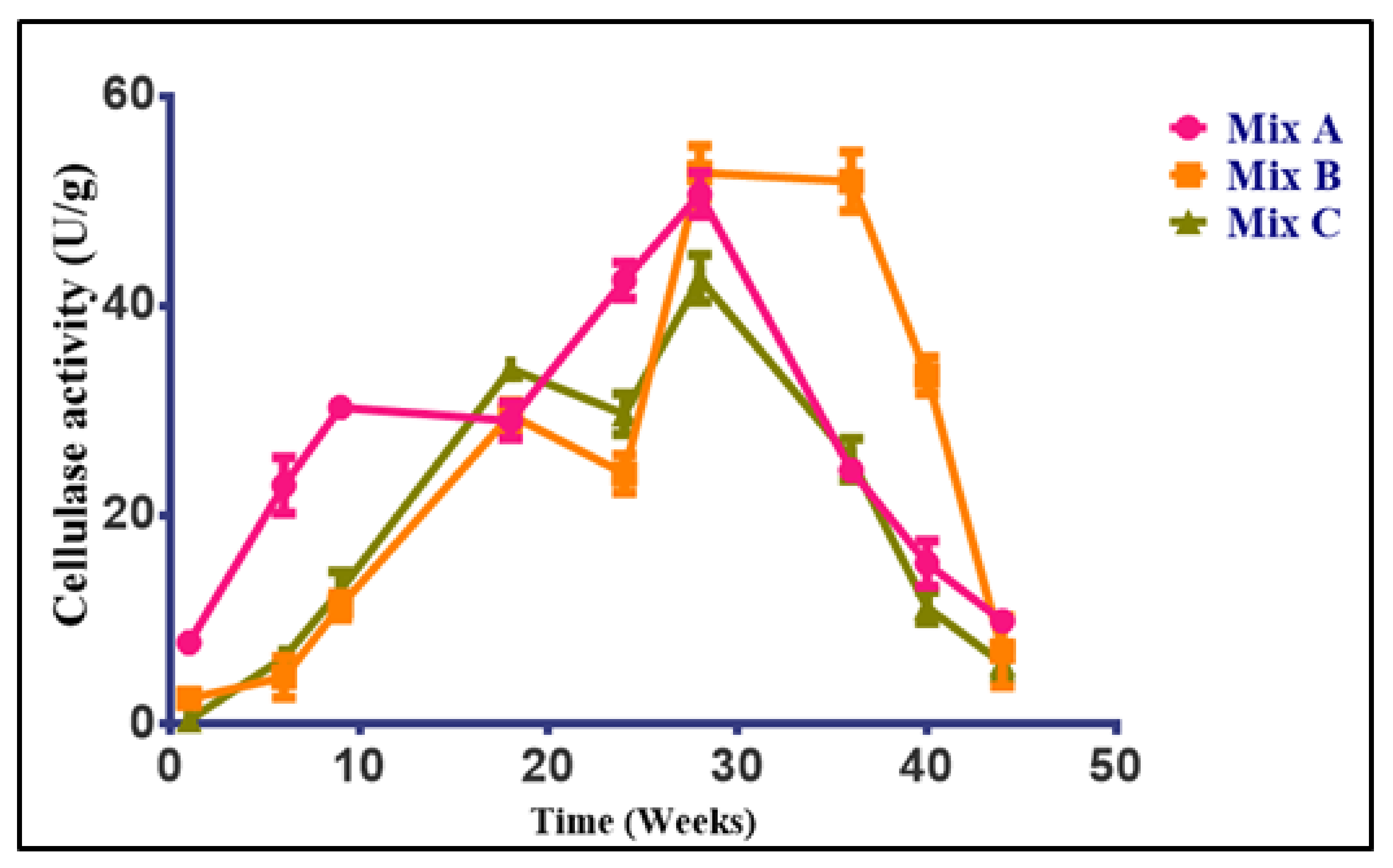
3.4.1. Changes in Cellulase Activity
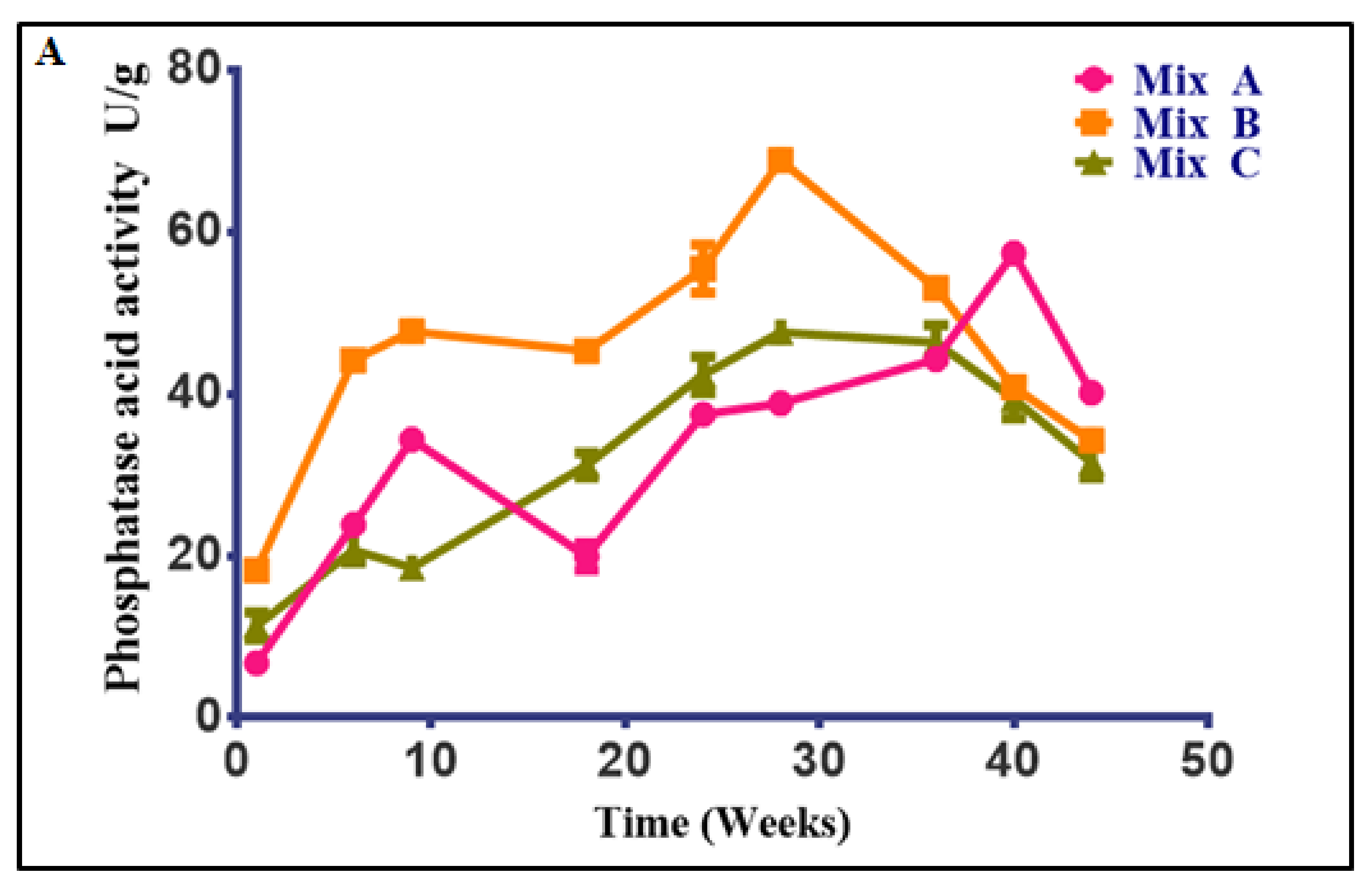
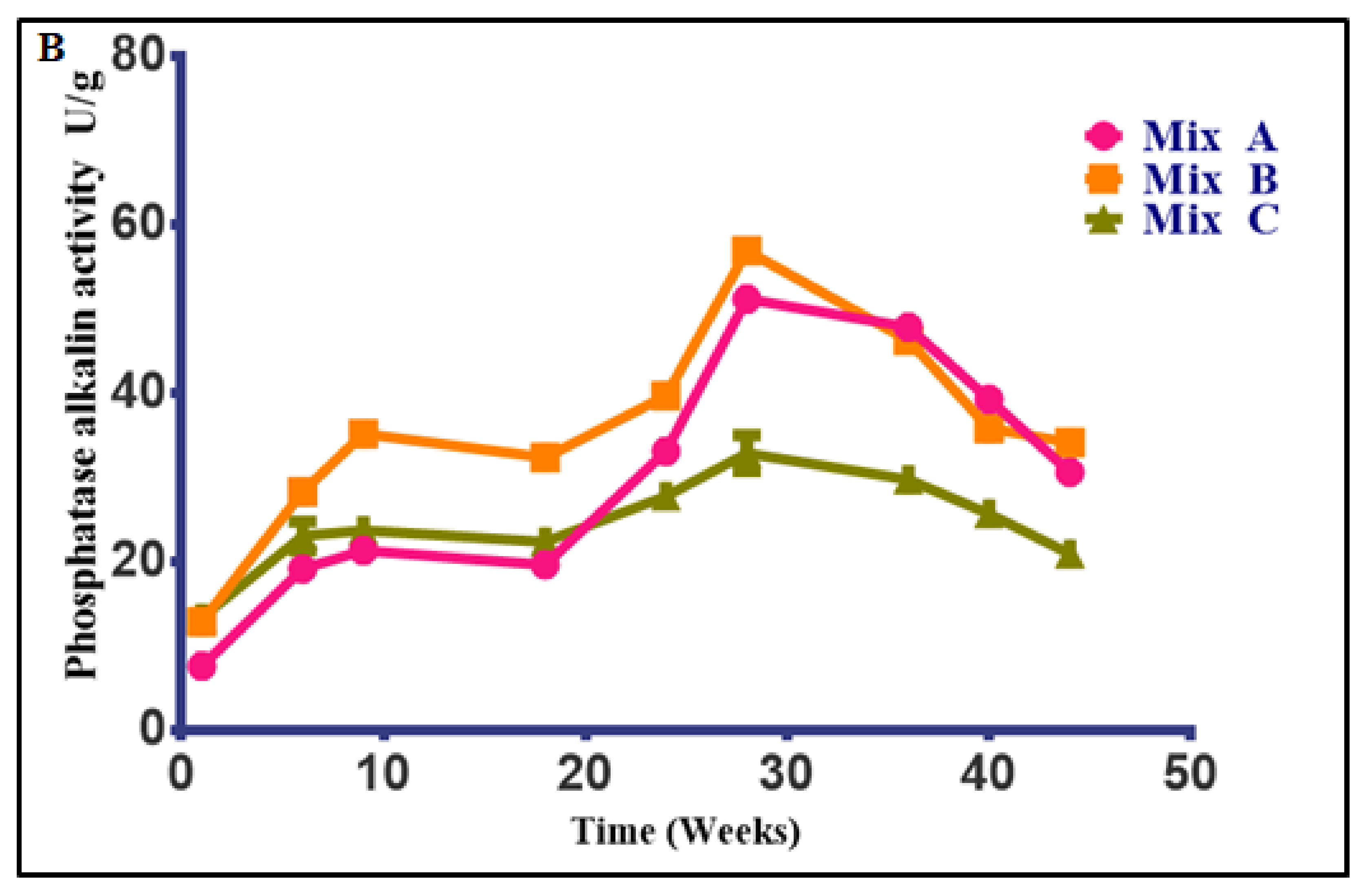
3.4.2. Changes in Phosphatase Activity
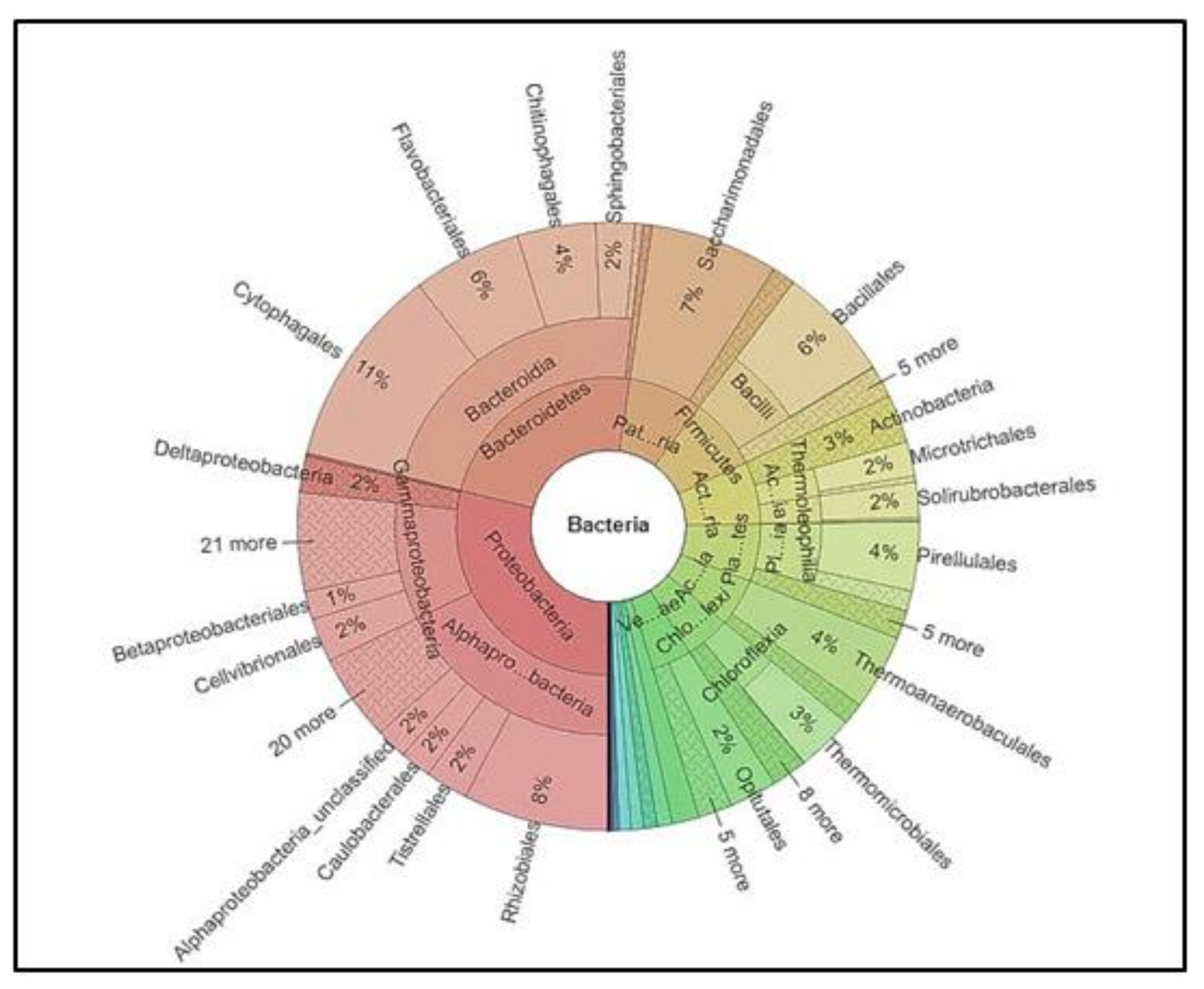
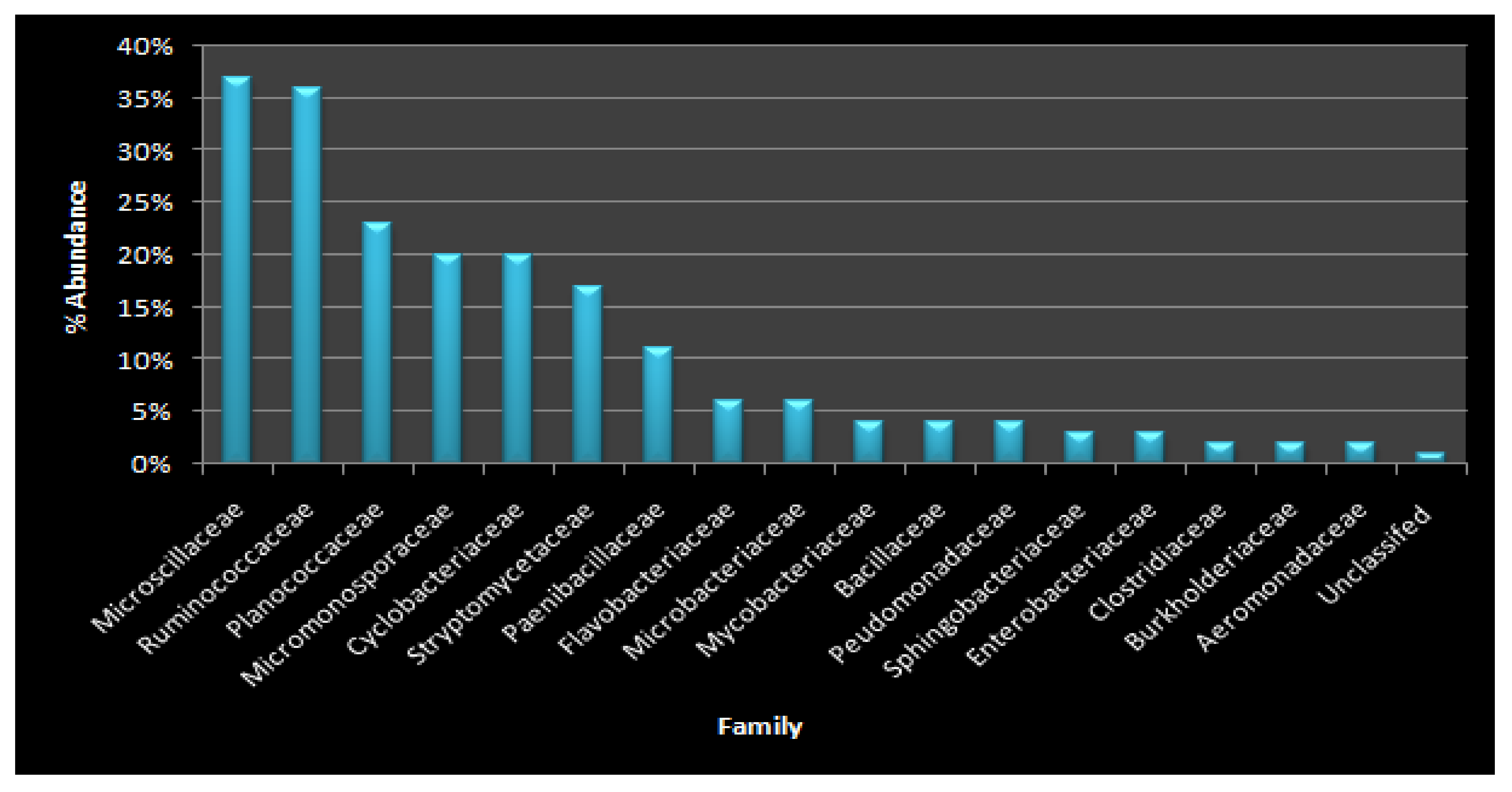
3.5. Identification of Microbial Diversity by Next-Generation Sequencing
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Francou, C. Stabilisation de la Matière Organique au Cours du Compostage de Déchets Urbains: Influence de la Nature des Déchets et du Procédé; Paris-Grignon National Agronomic Institute: Paris, France, 2004. [Google Scholar]
- Talouizte, H.; Merzouki, M.; Benlemlih, M. Treatment of Real textile wastwater using SBR technology: Effect of sludge age and operational parameters. J. Biotechnol. Lett. 2017, 4, 79–83. [Google Scholar]
- Soobhany, N.; Mohee, R.; Kumar, V. Comparative assessment of heavy metals content during the composting and vermicomposting of Municipal Solid Waste employing Eudrilus eugeniae. Waste Manag. 2015, 39, 130–145. [Google Scholar] [CrossRef] [PubMed]
- López-González, J.A.; del Carmen Vargas-García, M.; López, M.J.; Suárez-Estrella, F.; Jurado, M.; Moreno, J. Enzymatic characterization of microbial isolates from lignocellulose waste composting: Chronological evolution pez-Gonz a. J. Environ. Manag. 2014, 145, 137–146. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Nour El-Din, M.; Refaat, B.M.; Abdel-Shakour, E.H.; Ewais, E.E.D.; Alrefaey, H.M.A. Biotechnological Application of Thermotolerant Cellulose-Decomposing Bacteria in Composting of Rice Straw. Ann. Agric. Sci. 2016, 61, 135–143. [Google Scholar] [CrossRef]
- Tiquia, S.M. Microbiological parameters as indicators of compost maturity. J. Appl. Microbiol. 2005, 99, 816–828. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Zyl, W.H.; Van Pretorius, I.S. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Albrecht, R.; Petit, J.L.; Calvert, V.; Terrom, G.; Périssol, C. Changes in the level of alkaline and acid phosphatase activities during green wastes and sewage sludge co-composting. Bioresour. Technol. 2010, 101, 228–233. [Google Scholar] [CrossRef]
- Yi, H. Next generation sequencing-based metagenomics for monitoring soil microbiota. In Biosafety and The Environmental uses of Microorganisms; OECD: Paris, France, 2015. [Google Scholar]
- Tkachuk, V.L.; Krause, D.O.; Knox, N.C.; Hamm, A.C.; Zvomuya, F.; Ominski, K.H. Targeted 16S rRNA high-throughput sequencing to characterize microbial communities during composting of livestock mortalities. J. Appl. Microbiol. 2014, 116, 1181–1194. [Google Scholar] [CrossRef]
- Petrosino, J.F.; Highlander, S.; Luna, R.A.; Gibbs, R.A. Metagenomic Pyrosequencing and Microbial Identification. Clin. Chem. 2009, 55, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Biyada, S.; Merzouki, M.; Imtara, H.; Benlemlih, M. Assessment of the Maturity of Textile Waste Compost and their Capacity of Fertilization. Eu. J. Sci. Res. 2019, 154, 399–412. [Google Scholar]
- Afnor Amendements du sol et Support de Culture-Préparation des Echantillons Pour les Essais Physiques et Chimiques, Détermination de la Teneur en Matière Sèche, du Taux D’humidité et de la Masse Volumique Compactée en Laboratoire; The French Association for Standardization: Saint-Denis, France, 2000.
- Dias, S.; Welton, N.J.; Caldwell, D.M.; Ades, A.E. Checking consistency in mixed treatment comparison meta-analysis. Stat. Med. 2010, 29, 932–944. [Google Scholar] [CrossRef]
- Mancinelli, E.; Baltrėnaitė, E.; Baltrėnas, P.; Marčiulaitienė, E.; Limane, B.; Bartkevic, V. Dissolved organic carbon content and leachability of biomass waste biochar for trace metal (Cd, Cu and Pb) speciation modelling. J. Environ. Eng. Landsc. Manag. 2017, 25, 354–366. [Google Scholar] [CrossRef]
- Alsac, N. Dosage des métaux lourds (As, Cd, Cr, Cu, Ni, Pb, Zn et Hg) dans les sols par ICP-MS. Ann. Toxicol. Anal. 2007, XIX, 37–41. [Google Scholar] [CrossRef]
- Acharya, S.; Hu, Y.; Moussa, H.; Abidi, N. Preparation and characterization of transparent cellulose films using an improved cellulose dissolution process. J. Appl. Polym. Sci. 2017, 44871, 1–12. [Google Scholar] [CrossRef]
- Aguelmous, A.; Lahsaini, S.; El Fels, L.; Souabi, S.; Zamama, M.; Hafidi, M. Biodegradation assessment of biological oil sludge from a petroleum refinery. J. Mater. Environ. Sci. 2016, 7, 3421–3430. [Google Scholar]
- Albrecht, R.; Périssol, C.; Ruaudel, F.; Petit, J.L.e.; Terrom, G. Functional changes in culturable microbial communities during a co-composting process: Carbon source utilization and co-metabolism. Waste Manag. 2010, 30, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, R. Co-Compostage de Boues de Station D’épuration de Déchets Verts: Nouvelle Méthodologie du Suivi des Transformations de la Matiere Organique; University Paul Cezanne Aix-Marseille III: Marseille, Paris, 2007. [Google Scholar]
- Miller, G.; Blum, R.; Glennon, W.; Burton, A.L. Measurement of carboxymethylcellulase activity. Anal. Biochem. 1960, 132, 127–132. [Google Scholar] [CrossRef]
- Eivazi, F.; Tabatabai, M.A. Phosphatases in Soils. Soil Biol. Biochem. 1977, 9, 167–172. [Google Scholar] [CrossRef]
- Albrecht, R.; Petit, J.L.e.; Terrom, G.; Périssol, C. Comparison between UV spectroscopy and nirs to assess humification process during sewage sludge and green wastes co-composting. Bioresour. Technol. 2011, 102, 4495–4500. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Ryan, A.; Oakley, B.B.; Parks, D.H.; Courtney, J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Soobhany, N.; Gunasee, S.; Pooja, Y.; Joyram, H.; Raghoo, P. Spectroscopic, thermogravimetric and structural characterization analyses for comparing Municipal Solid Waste composts and vermicomposts stability and maturity. Bioresour. Technol. 2017, 236, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.F.; Wu, Q.T.; Wong, J.W.C.; Nagar, B.B. Transformation of organic matter during co-composting of pig manure with sawdust. Bioresour. Technol. 2006, 97, 1834–1842. [Google Scholar] [CrossRef] [PubMed]
- Hajjouji, H.; El Fakharedine, N.; Baddi, G.A.; Winterton, P.; Bailly, J.R.; Revel, J.C.; Hafidi, M. Treatment of olive mill waste-water by aerobic biodegradation: An analytical study using gel permeation chromatography, ultraviolet – visible and Fourier transform infrared spectroscopy. Bioresour. Technol. 2007, 98, 3513–3520. [Google Scholar] [CrossRef]
- Hachicha, R.; Hachicha, S.; Trabelsi, I.; Woodward, S.; Mechichi, T. Evolution of the fatty fraction during co-composting of olive oil industry wastes with animal manure: Maturity assessment of the end product. Chemosphere 2009, 75, 1382–1386. [Google Scholar] [CrossRef]
- Cunningham, C.J.; Philp, J.C. Comparison of bioaugmentation and biostimulation in ex situ treatment of diesel contaminated soil. Land Contam. Reclam. 2000, 8, 261–269. [Google Scholar]
- Mondini, C.; Fornasier, F.; Sinicco, T. Enzymatic activity as a parameter for the characterization of the composting process. Soil Biol. Biochem. 2004, 36, 1587–1594. [Google Scholar] [CrossRef]
- El Fels, L.; El Ouaqoudi, F.Z.; Barje, F.; Hafidi, M.; Ouhdouch, Y. Two culture approaches used to determine the co-composting stages by assess of the total microflora changes during sewage sludge and date palm waste co-composting. J. Environ. Health Sci. Eng. 2014, 12, 1–5. [Google Scholar] [CrossRef]
- Herna, T.; Masciandaro, G.; Moreno, J.I.; Garcia, C. Changes in organic matter composition during composting of two digested sewage sludges. Waste Manag. 2006, 26, 1370–1376. [Google Scholar]
- Baddi, G.A.; Alburquerque, J.A.; Gonzálvez, J.; Cegarra, J.; Hafidi, M. Chemical and spectroscopic analyses of organic matter transformations during composting of olive mill wastes. Int. Biodeterior. Biodegrad. 2004, 54, 39–44. [Google Scholar] [CrossRef]
- Goyal, S.; Dhull, S.K.; Kapoor, K.K. Chemical and biological changes during composting of different organic wastes and assessment of compost maturity. Bioresour. Technol. 2005, 96, 1584–1591. [Google Scholar] [CrossRef] [PubMed]
- Queda, A.C.C.; Vallini, G.; Agnolucci, M.; Coelho, C.A.; Camposl, L. Microbiological and Chemical Characterisation of Composts at Different Levels of Maturity, with Evaluation of Phytotoxicity and Enzymatic activities. In Microbiology of Composting; Springer: Berlin/Heidelberg Germany, 2002. [Google Scholar]
- Huang, G.F.; Wong, J.W.C.; Wu, Q.T.; Nagar, B.B. Effect of C/N on composting of pig manure with sawdust. Waste Manag. 2004, 24, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Barje, F.; El Fels, L.; El Hajjouji, H.; Winterton, P.; Hafidi, M. Biodegradation of organic compounds during co-composting of olive oil mill waste and municipal solid waste with added rock phosphate. Environ. Technol. 2013, 34, 2965–2975. [Google Scholar] [CrossRef]
- Fels, L. El Suivi Physico-Chiique, Microbiologique et Ecotoxicologique du Compostage de Boues de step Mélangées à des Déchets de Palmier: Validation de Nouveaux Indices de Maturation; Université de Toulouse: Toulouse, France, 2014. [Google Scholar]
- García, C.; Hernandez, T.; Costa, F. The influence of composting on the fertilizing value of an aerobic sewage sludge. Plant Soil. 1991, 136, 269–272. [Google Scholar] [CrossRef]
- Amir, S. Contribution à la Valorisation de Boues de Stations D’épuration par Compostage: Devenir des Micropolluants Métalliques et Organiques et Bilan Humique du Compost; National Polytechnical Institute of Toulouse: Toulouse, France, 2005. [Google Scholar]
- Singh, K.P.; Mohan, D.; Sinha, S.; Dalwani, R. Impact assessment of treated / untreated wastewater toxicants discharged by sewage treatment plants on health, agricultural, and environmental quality in the wastewater disposal area. Chemosphere 2004, 55, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Iravani, S.; Varma, R.S. Bacteria in Heavy Metal Remediation and Nanoparticle Biosynthesis Bacteria in Heavy Metal Remediation and Nanoparticle Biosynthesis. ACSSustain. Chem. Eng. 2020, 8, 5395–5409. [Google Scholar]
- Sharma, D.; Kanchi, S.; Bisetty, K. Biogenic synthesis of nanoparticles: A review. Arab. J. Chem. 2019, 12, 3576–3600. [Google Scholar] [CrossRef]
- Amendements Organiques Dénominations, Spécifications et Marquage; Afnor: Paris, France, 2018.
- FAO. Méthodes de Compostage au Niveau de L’exploitation Agricole; FAO: Rome, Italy, 2005. [Google Scholar]
- Raut, M.P.; William, S.P.M.P.; Bhattacharyya, J.K.; Chakrabarti, T.; Devotta, S. Microbial dynamics and enzyme activities during rapid composting of municipal solid waste – A compost maturity analysis perspective. Bioresour. Technol. 2008, 99, 6512–6519. [Google Scholar] [CrossRef]
- Tuomela, M.; Vikman, M.; Hatakka, A.; Itävaara, M. Biodegradation of lignin in a compost environment: A review. Bioresour. Technol. 2000, 72, 169–183. [Google Scholar] [CrossRef]
- Vikman, M.; Karjomaa, S.; Kapanen, A.; Wallenius, K.; Itavaara, M. The influence of lignin content and temperature on the biodegradation of lignocellulose in composting conditions. Appl. Microbiol. Biotechnol. 2002, 59, 591–598. [Google Scholar]
- Zhou, X.; Ren, L.; Meng, Q.; Li, Y.; Yu, Y.; Yu, J. The next-generation sequencing technology and application. Protein Cell. 2010, 1, 520–536. [Google Scholar] [CrossRef]
- Bandounas, L.; Wierckx, N.J.P.; Winde, J.H.; De Ruijssenaars, H.J. Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol. 2011, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Boisset, C.; Chanzy, H.; Henrissat, B. Optimized Mixtures of Recombinant Humicola Insolens Cellulases for the Biodegradation of Crystalline Cellulose; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Adsul, M.G.; Bastawde, K.B.; Varma, A.J.; Gokhale, D.V. Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresour. Technol. 2007, 98, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Khan, M.; Fagan, B.; Mullins, E.; Doohan, F.M. Exploiting the inter-strain divergence of Fusarium oxysporum for microbial bioprocessing of lignocellulose to bioethanol. AMB Express. 2012, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chen, S. The white-rot fungus Phanerochaete chrysosporium: Conditions for the production of lignin-degrading enzymes. Appl. Microbiol. Biotechnol. 2008, 81, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xiao, Y.; Yu, H.Q. Degradation of lignin in pulp mill wastewaters by white-rot fungi on biofilm. Bioresour. Technol. 2005, 96, 1357–1363. [Google Scholar] [CrossRef]
- Danon, M.; Franke-whittle, I.H.; Insam, H.; Chen, Y.; Hadar, Y. Molecular analysis of bacterial community succession during prolonged compost curing. Fed. Eur. Microbiol. Soc. 2008, 65, 133–144. [Google Scholar] [CrossRef]
- Łaba, W.; Rodziewicz, A.; Choin, A. Enhancement of pig bristles waste bioconversion by inoculum of keratinolytic bacteria during composting. Waste Manag. 2019, 84, 269–276. [Google Scholar]
- Kukolya, J. Thermobifida cellulolytica sp. nov., a novel lignocellulose-decomposing actinomycete. Int. J. Syst. Evolut. Microbiol. 2002, 52, 1193–1199. [Google Scholar]
- Tripathi, B.M.; Kaushik, R.; Kumari, P.; Saxena, A.K.; Arora, D.K. Genetic and metabolic diversity of streptomycetes in pulp and paper mill effluent treated crop fields. World J. Microbiol. Biotechnol. 2011, 27, 1603–1613. [Google Scholar] [CrossRef]
- Sashidhar, B.; Podile, A.R. Mineral phosphate solubilization by rhizosphere bacteria and scope for manipulation of the direct oxidation pathway involving glucose dehydrogenase. J. Appl. Microbiol. 2010, 109, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, J.P.; Orozco, M.O.; Gianinazzi-pearson, V.; Gianinazzi, S. Influence of phosphate fertilization on fungal alkaline phosphatase and succinate dehydrogenase activities in arbuscular mycorrhiza of soybean and pineapple. Agric. Ecosyst. Environ. 1995, 53, 63–69. [Google Scholar] [CrossRef]
- Braud, A. Effects of organic and mineral amendments on available P and phosphatase activities in a degraded Mediterranean soil under short-term incubation experiment. Soil Tillage Res. 2008, 98, 164–174. [Google Scholar]
- Cunha-Queda, A.C.; Ribeiro, H.M.; Ramos, A.; Cabral, F. Study of biochemical and microbiological parameters during composting of pine and eucalyptus bark. Bioresour. Technol. 2007, 98, 3213–3220. [Google Scholar] [CrossRef] [PubMed]
- Rodrı, H.; Fraga, R.; Gonzalez, T.; Bashan, Y. Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil. 2006, 287, 15–21. [Google Scholar]
- Ferreira, A.S.; Leitão, J.H.; Sousa, S.A.; Cosme, A.M.; Sá-correia, I.; Moreira, L.M. Functional Analysis of Burkholderia cepacia Genes bceD and bceF, Encoding a Phosphotyrosine Phosphatase and a Tyrosine Autokinase, Respectively: Role in Exopolysaccharide Biosynthesis and Biofilm Formation. Appl. Environ. Microbiol. 2007, 73, 524–534. [Google Scholar] [CrossRef]
- Tarafdar, J.C.; Panwar, J.; Kathju, S. Efficiency of some phosphatase-producing soil fungi A rapid method for assessment of plant residue quality. J. Plant Nutr. Soil Sci. 2003, 166, 662–666. [Google Scholar]
- Tarafdar, J.C.; Rao, A.V. Role of phosphatase-producing fungi on the growth and nutrition of clusterbean (Cyamopsis tetragonoloba (L) Taub). J. Arid Environ. 1995, 29, 331–337. [Google Scholar] [CrossRef]
- Aleksieva, P.; Spasova, D.; Radoevska, S. Acid Phosphatase Distribution and Localization in the Fungus Humicola lutea. Zeitschrift für Naturforschung C. 2003, 58, 239–243. [Google Scholar] [CrossRef]
- Yadav, R.S.; Tarafdar, J.C. Phytase and phosphatase producing fungi in arid and semi-arid soils and their efficiency in hydrolyzing different organic P compounds. Soil Biol. Biochem. 2003, 35, 1–7. [Google Scholar] [CrossRef]
- Nahas, E. Control of Acid Phosphatases Expression from Aspergillus niger by Soil Characteristics. Braz. Arch. Biol. Technol. 2015, 58, 658–666. [Google Scholar] [CrossRef]
- Reyes, F.; VillaNnueve, P.; Alfonso, C. Comparative study of acid and alkaline phosphatase during the autolysis of filamentous fungi. Lett. Appl. Microbiol. 1990, 10, 175–177. [Google Scholar] [CrossRef]



| Physico-Chemical Parameter | Textile Waste | Green Waste | Paper and Cardboard Waste |
|---|---|---|---|
| Moisture % | 51.28 ± 1.03 | 61.49 ± 1.41 | 11.28 ± 1.07 |
| pH | 7.47 ± 0.15 | 6.61 ± 0.53 | 7.23 ± 0.35 |
| Total OrganicCarbon (TOC %) | 31.63 ± 1.48 | 45.67 ± 1.37 | 59.35 ± 0.90 |
| Total Nitrogen (TN %) | 0.57 ± 0.04 | 1.23 ± 0.04 | 1.05 ± 0.05 |
| C/N ratio | 55.16 | 37.23 | 56.71 |
| Wavenumber (cm−1) | Band Assignments |
|---|---|
| 3700–3000 | H-OH groups of alcohols, phenols, and organicacids, as well as N–H groups of amides and amines |
| 3000–2850 | C–H stretching in aliphatic structures |
| 1650–1630 | C=C stretching in aromatics, C=O stretching in amides (I), C=Ostretching in ketone and quinone groups |
| 1400 | C–H stretching in aliphatic structures |
| 1121 | Amide III, aliphaticalcohols, or acids, C–O and O–H deformation of carboxylic groups of carbohydrates, C–O stretch of arylethers, esters, and organo-sulphur compounds |
| 1060 | C–H vibration, OH deformation in carboxyls, C–O of ethers on an aromatic ring N–H amides II |
| <1000 | Vibrations of aromaticethers, polysaccharides (C–H vibration) |
| Time (Weeks) | Total Organic Carbon (%) | Total Nitrogen (%) | C/N Ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mix A | Mix B | Mix C | Mix A | Mix B | Mix C | Mix A | Mix B | Mix C | |
| 1 | 43.96 ± 0.90 | 40.72 ± 0.62 | 32.64 ± 1.50 | 1.35 ± 0.09 | 1.04 ± 0.06 | 0.56 ± 0.10 | 32.56 | 39.15 | 58.29 |
| 6 | 42.6 ± 0.88 | 36.61 ± 0.59 | 31.32 ± 2.21 | 1.63 ± 0.06 | 1.19 ± 0.20 | 0.66 ± 0.21 | 26.13 | 30.76 | 47.45 |
| 9 | 36.94 ± 0.67 | 35.05 ± 0.84 | 29.81 ± 1.70 | 1.77 ± 0.09 | 1.40 ± 0.08 | 0.98 ± 0.18 | 20.87 | 25.04 | 30.42 |
| 18 | 28.3 ± 0.57 | 29.42 ± 2.59 | 28.26 ± 2.11 | 0.75 ± 0.12 | 0.74 ± 0.19 | 0.76 ± 0.12 | 37.73 | 39.76 | 37.18 |
| 24 | 27.7 ± 0.56 | 27.15 ± 2.53 | 26.17 ± 2.55 | 1.20 ± 0.16 | 1.15 ± 0.13 | 1.11 ± 0.13 | 23.08 | 23.61 | 23.58 |
| 28 | 26.6 ± 0.55 | 26.52 ± 2.05 | 24.92 ± 2.26 | 1.29 ± 0.08 | 1.23 ± 0.15 | 1.14 ± 0.14 | 20.67 | 21.56 | 21.86 |
| 36 | 24.63 ± 1.13 | 25.63 ± 1.75 | 23.91 ± 2.57 | 1.35 ± 0.12 | 1.29 ± 0.07 | 1.17 ± 0.07 | 18.24 | 19.87 | 20.44 |
| 40 | 22.35 ± 1.03 | 23.89 ± 1.55 | 23.62 ± 2.01 | 1.42 ± 0.13 | 1.32 ± 0.09 | 1.19 ± 0.06 | 15.74 | 18.10 | 19.85 |
| 44 | 22.23 ± 0.92 | 23.57 ± 1,39 | 23.53 ± 2.39 | 1.37 ± 0.07 | 1.39 ± 0.06 | 1.10 ± 0.14 | 16.30 | 16.96 | 21.39 |
| Cellulase | Acid Phosphatase | Alkaline Phosphatase | |
|---|---|---|---|
| C/N ratio | −0.366 ** | −0.833 ** | −0.786 ** |
| Temperature | 0.843 | 0.969 | 0.963 |
| Moisture | −0.095 | −0.502 | −0.396 |
| pH | −0.53 | −0.040 | −0.345 |
| Electricalconductivity | 0.417 | 0.851 ** | 0.792 * |
| Bacteria | 0.788 * | 0.459 | 0.495 |
| Actinomycetes | 0.925 ** | 0.853 ** | 0.941 ** |
| Fungi | 0.975 ** | 0.821 ** | 0.864 ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biyada, S.; Merzouki, M.; Dėmčėnko, T.; Vasiliauskienė, D.; Urbonavičius, J.; Marčiulaitienė, E.; Vasarevičius, S.; Benlemlih, M. Evolution of Microbial Composition and Enzymatic Activities during the Composting of Textile Waste. Appl. Sci. 2020, 10, 3758. https://doi.org/10.3390/app10113758
Biyada S, Merzouki M, Dėmčėnko T, Vasiliauskienė D, Urbonavičius J, Marčiulaitienė E, Vasarevičius S, Benlemlih M. Evolution of Microbial Composition and Enzymatic Activities during the Composting of Textile Waste. Applied Sciences. 2020; 10(11):3758. https://doi.org/10.3390/app10113758
Chicago/Turabian StyleBiyada, Saloua, Mohammed Merzouki, Taisija Dėmčėnko, Dovilė Vasiliauskienė, Jaunius Urbonavičius, Eglė Marčiulaitienė, Saulius Vasarevičius, and Mohamed Benlemlih. 2020. "Evolution of Microbial Composition and Enzymatic Activities during the Composting of Textile Waste" Applied Sciences 10, no. 11: 3758. https://doi.org/10.3390/app10113758
APA StyleBiyada, S., Merzouki, M., Dėmčėnko, T., Vasiliauskienė, D., Urbonavičius, J., Marčiulaitienė, E., Vasarevičius, S., & Benlemlih, M. (2020). Evolution of Microbial Composition and Enzymatic Activities during the Composting of Textile Waste. Applied Sciences, 10(11), 3758. https://doi.org/10.3390/app10113758






