ATP Bioluminescence for Rapid and Selective Detection of Bacteria and Yeasts in Wine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Wine Samples
2.2. Bacterial Strains and Deliberate Wine Contamination
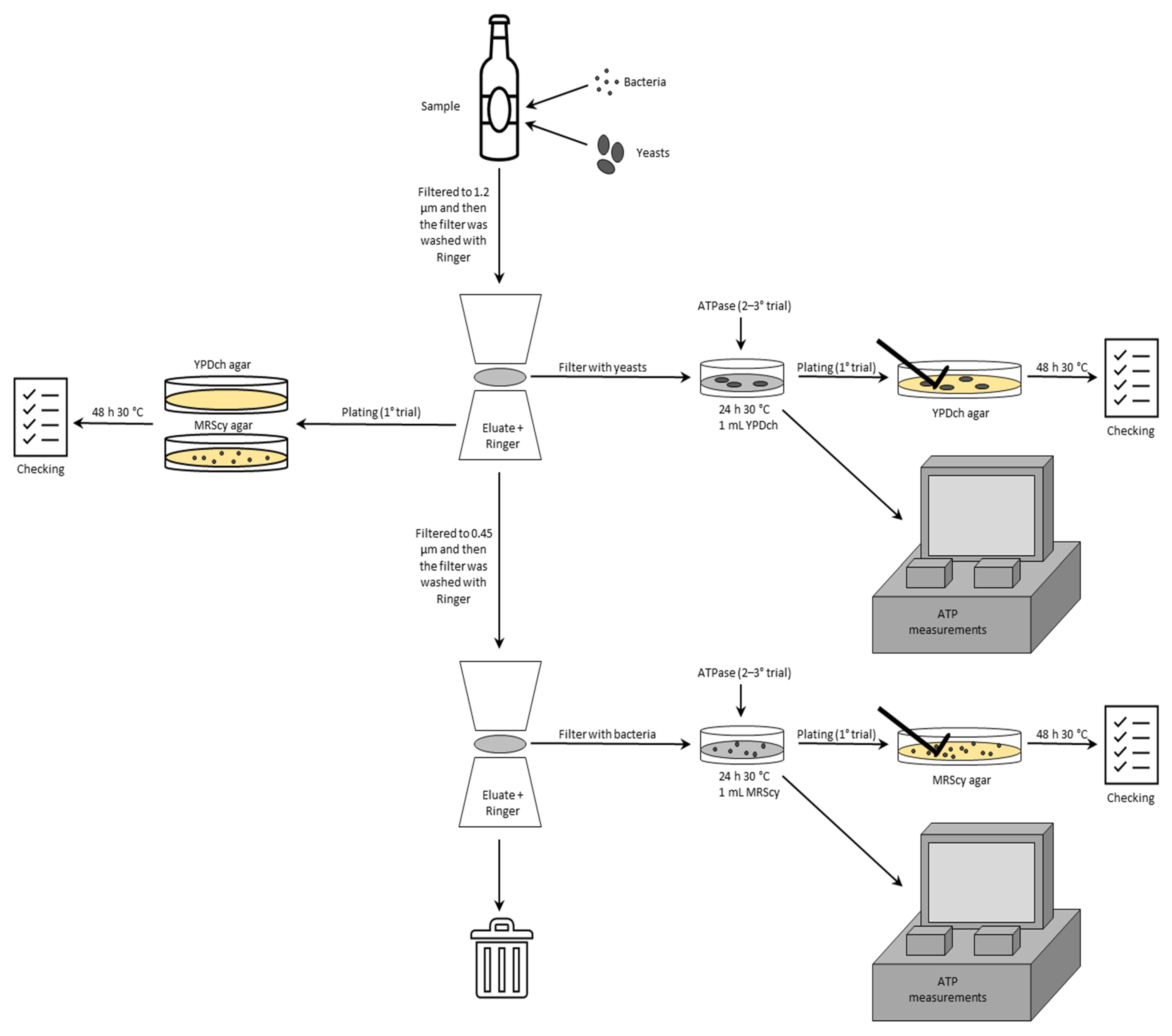
2.3. Bioluminescence Assay
2.4. Plating
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Janagama, H.K.; Mai, T.; Han, S.; Nadala, L.; Nadala, C.; Samadpour, M. Simultaneous Detection of Multiple Wine-Spoilage Organisms Using a PCR-Based DNA Dipstick Assay. J. AOAC Int. 2019, 102, 490–496. [Google Scholar] [CrossRef]
- Fugelsang, K.C.; Edwards, C.G. Wine Microbiology, 2nd ed.; Springer: New York, NY, USA, 2017; ISBN 978-0-387-33341-0. [Google Scholar]
- Okanojo, M.; Miyashita, N.; Tazaki, A.; Tada, H.; Hamazoto, F.; Hisamatsu, M.; Noda, H. Attomol-Level ATP Bioluminometer for Detecting Single Bacterium: ATP Calibration Curve Is Linear from 0 to 5 Amol. Luminescence 2017, 32, 751–756. [Google Scholar] [CrossRef] [PubMed]
- McElroy, W.D.; Green, A. Function of Adenosine Triphosphate in the Activation of Luciferin. Arch. Biochem. Biophys. 1956, 64, 257–271. [Google Scholar] [CrossRef]
- Lonvaud-Funel, A.; Joyeux, A. Application de La Bioluminescence Au Dénombrement Des Microorganismes Vivants Dans Les Vins. OENO One 1982, 16, 241. [Google Scholar] [CrossRef]
- Chollet, R.; Kukuczka, M.; Halter, N.; Romieux, M.; Marc, F.; Meder, H.; Beguin, V.; Ribault, S. Rapid Detection and Enumeration of Contaminants by Atp Bioluminescence Using the Milliflex® Rapid Microbiology Detection and Enumeration System. J. Rapid Methods Autom. Microbiol. 2008, 16, 256–272. [Google Scholar] [CrossRef]
- Bottari, B.; Santarelli, M.; Neviani, E. Determination of Microbial Load for Different Beverages and Foodstuff by Assessment of Intracellular ATP. Trends Food Sci. Technol. 2015, 44, 36–48. [Google Scholar] [CrossRef]
- Littel, K.J.; Rocco, K.A.L. ATP Screening Method for Presumptive Detection of Microbiologically Contaminated Carbonated Beverages. J. Food Sci. 1986, 51, 474–476. [Google Scholar] [CrossRef]
- Osborne, J.P. Advances in microbiological quality control. In Managing Wine Quality; Elsevier: Amsterdam, The Netherlands, 2010; pp. 162–188. ISBN 978-1-84569-484-5. [Google Scholar]
- Daeschel, M.A.; Jung, D.-S.; Watson, B.T. Controlling Wine Malolactic Fermentation with Nisin and Nisin-Resistant Strains of Leuconostoc Oenos. Appl. Environ. Microbiol. 1991, 57, 601–603. [Google Scholar] [CrossRef] [Green Version]
- Nobile, J. Use of Membrane Filter Technique in the Microbiological Control for the Brewing Industry. Appl. Microbiol. 1966, 15, 736–737. [Google Scholar] [CrossRef]
- Millet, V.; Lonvaud-Funel, A. The Viable but Non-Culturable State of Wine Micro-Organisms during Storage. Lett. Appl. Microbiol. 2000, 30, 136–141. [Google Scholar] [CrossRef]
- Venkateswaran, K.; Hattori, N.; La Duc, M.T.; Kern, R. ATP as a Biomarker of Viable Microorganisms in Clean-Room Facilities. J. Microbiol. Methods 2003, 52, 367–377. [Google Scholar] [CrossRef]
- Lucas, P.M.; Claisse, O.; Lonvaud-Funel, A. High Frequency of Histamine-Producing Bacteria in the Enological Environment and Instability of the Histidine Decarboxylase Production Phenotype. AEM 2008, 74, 811–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loureiro, V.; Malfeito-Ferreira, M. Spoilage Yeasts in the Wine Industry. Int. J. Food Microbiol. 2003, 86, 23–50. [Google Scholar] [CrossRef]
- Ammor, M.S.; Belén Flórez, A.; Mayo, B. Antibiotic Resistance in Non-Enterococcal Lactic Acid Bacteria and Bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Pérez, F.; Regodón, J.A.; Valdés, M.E.; De Miguel, C.; Ramı́rez, M. Cycloheximide Resistance as Marker for Monitoring Yeasts in Wine Fermentations. Food Microbiol. 2000, 17, 119–128. [Google Scholar] [CrossRef]
- Thompson, A. Application in Beverages Microbiology. In ATP Bioluminescence; Academic Press, Scottish Courage Brewing Ltd.: Edinburgh, UK, 1999; pp. 101–109. [Google Scholar]
- Takahashi, T.; Nakakita, Y.; Watari, J.; Shinotsuka, K. A New Rapid Technique for Detection of Microorganisms Using Bioluminescence and Fluorescence Microscope Method. J. Biosci. Bioeng. 2000, 89, 509–513. [Google Scholar] [CrossRef]
- Yan, S.-L.; Miao, S.-N.; Deng, S.-Y.; Zou, M.-J.; Zhong, F.-S.; Huang, W.-B.; Pan, S.-Y.; Wang, Q.-Z. ATP Bioluminescence Rapid Detection of Total Viable Count in Soy Sauce: Bioluminescence for Rapid Detection of Microbes in Soy Sauce. Luminescence 2012, 27, 34–38. [Google Scholar] [CrossRef]
- El Rayess, Y.; Albasi, C.; Bacchin, P.; Taillandier, P.; Raynal, J.; Mietton-Peuchot, M.; Devatine, A. Cross-Flow Microfiltration Applied to Oenology: A Review. J. Membr. Sci. 2011, 382, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Horinouchi, T.; Maeda, T.; Furusawa, C. Understanding and Engineering Alcohol-Tolerant Bacteria Using OMICS Technology. World J. Microbiol. Biotechnol. 2018, 34, 157. [Google Scholar] [CrossRef] [Green Version]
- Panesar, P.S.; Kumar, N.; Marwaha, S.S.; Joshi, V.K. Vermouth Production Technology–An Overview; CSIR: Delhi, India, 2009; Volume 8, p. 11. [Google Scholar]
- Ding, J.; Huang, X.; Zhang, L.; Zhao, N.; Yang, D.; Zhang, K. Tolerance and Stress Response to Ethanol in the Yeast Saccharomyces Cerevisiae. Appl. Microbiol. Biotechnol. 2009, 85, 253–263. [Google Scholar] [CrossRef]
- Ebani, V.V.; Nardoni, S.; Bertelloni, F.; Giovanelli, S.; Rocchigiani, G.; Pistelli, L.; Mancianti, F. Antibacterial and Antifungal Activity of Essential Oils against Some Pathogenic Bacteria and Yeasts Shed from Poultry: Antimicrobial Activity of Essential Oils against Pathogens of Poultry. Flavour Fragr. J. 2016, 31, 302–309. [Google Scholar] [CrossRef]
| Wine Samples | Sparkling | Alcohol (%) | Sugar (g/L) |
|---|---|---|---|
| R1 | No | 13.00 | 3.7 |
| R2 | No | 13.00 | 1 |
| R3 | No | 13.00 | 3.1 |
| R4 | No | 6.00 | 125 |
| R5 | Yes | 6.00 | 110 |
| R6 | Yes | 12.00 | 15 |
| W1 | No | 12.00 | 3.5 |
| W2 | No | 13.00 | 23.9 |
| W3 | Yes | 5.00 | 125 |
| W4 | Yes | 11.00 | 13 |
| W5 | Yes | 6.00 | 110 |
| W6 | Yes | 12.00 | 5 |
| W7 | Yes | 8.00 | 80 |
| W8 | Yes | 12.00 | 10 |
| R7 | Yes | 12.00 | 10 |
| V1 | No | 15.00 | 152 |
| V2 | No | 15.00 | 168 |
| Pass/Fail Limits | RLU Value |
|---|---|
| Sterile | ≤2 times blank |
| Re-test | 2 times blank < RLU value < 3 times blank |
| Non-sterile | ≥3 times blank |
| Inoculum (CFU/mL) | Yeast (104) + Bacteria (107) | Yeast (104) + Bacteria (103) | ||||||
|---|---|---|---|---|---|---|---|---|
| Filter | 1.2 µm | 0.45 µm | 1.2 µm | 0.45 µm | ||||
| Analysis | RLU YPDch | SD | RLU MRScy | SD | RLU YPDch | SD | RLU MRScy | SD |
| R1 | 7.04 × 106 | 2.24 × 104 | 1.61 × 106 | 6.03 × 104 | 9.23 × 105 | 8.84 × 104 | 1.01 × 103 | 2.91 × 10 |
| R2 | 3.86 × 106 | 3.32 × 104 | 1.14 × 106 | 2.24 × 104 | 7.66 × 106 | 2.14 × 104 | 1.01 × 105 | 4.28 × 103 |
| R3 | 4.28 × 106 | 2.96 × 104 | 1.43 × 106 | 6.24 × 104 | 9.40 × 106 | 4.05 × 104 | 1.11 × 103 | 2.36 × 102 |
| R4 | 3.02 × 106 | 2.13 × 104 | 9.23 × 105 | 1.90 × 104 | 7.38 × 106 | 4.11 × 104 | 1.14 × 103 | 8.55 × 10 |
| R5 | 1.29 × 106 | 1.89 × 103 | 4.11 × 105 | 4.83 × 103 | 3.75 × 106 | 1.87 × 104 | 1.37 × 106 | 9.89 × 103 |
| R6 | 1.45 × 106 | 4.88 × 104 | 9.58 × 105 | 5.23 × 104 | 4.84 × 106 | 3.76 × 104 | 5.70 × 105 | 1.57 × 104 |
| W1 | 5.25 × 106 | 5.91 × 104 | 1.32 × 106 | 7.58 × 104 | 6.80 × 106 | 1.97 × 104 | 1.57 × 104 | 1.45 × 102 |
| W2 | 3.96 × 106 | 1.20 × 104 | 4.91 × 105 | 7.99 × 103 | 7.35 × 106 | 5.69 × 104 | 1.02 × 103 | 8.59 × 10 |
| W3 | 4.34 × 106 | 4.05 × 104 | 1.47 × 106 | 4.48 × 104 | 8.07 × 106 | 6.91 × 104 | 1.24 × 104 | 1.41 × 102 |
| W4 | 9.69 × 106 | 3.19 × 104 | 1.33 × 106 | 6.91 × 104 | 3.85 × 106 | 3.01 × 104 | 1.31 × 106 | 2.82 × 104 |
| W5 | 3.91 × 106 | 1.63 × 104 | 2.19 × 104 | 5.05 × 103 | 7.75 × 106 | 9.16 × 104 | 1.80 × 105 | 2.48 × 103 |
| W6 | 1.74 × 106 | 1.44 × 104 | 4.93 × 105 | 1.04 × 104 | 8.04 × 106 | 5.14 × 104 | 5.39 × 105 | 2.28 × 103 |
| Blank | 4.24 × 103 | 6.55 × 102 | 3.65 × 103 | 3.16 × 102 | 4.04 × 103 | 1.10 × 103 | 2.03 × 103 | 7.12 × 102 |
| Inoculum (CFU/mL) | Filter | Analysis | R1 | R2 | R3 | R4 | R5 | R6 | W1 | W2 | W3 | W4 | W5 | W6 | TtD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yeast (104) + Bacteria (107) | 1.2 µm | RLU YPDch 1 | + | + | + | + | + | + | + | + | + | + | + | + | 24 h |
| plating RLU-YPDch on YPDch agar 2 | + | + | + | + | + | + | + | + | + | + | + | + | 48 h | ||
| Eluate on YPDch agar | − | − | − | − | − | − | − | − | − | − | − | − | |||
| Eluate on MRScy agar | + | + | + | + | + | + | + | + | + | + | + | + | |||
| 0.45 µm | RLU MRScy 1 | + | + | + | + | + | + | + | + | + | + | + | + | 24 h | |
| Plating RLU-MRScy on MRScy agar 2 | + | + | + | + | + | + | + | + | + | + | + | + | 48 h | ||
| Yeast (104) + Bacteria (103) | 1.2 µm | RLU YPDch | + | + | + | + | + | + | + | + | + | + | + | + | 24 h |
| plating RLU-YPDch on YPDch agar | + | + | + | + | + | + | + | + | + | + | + | + | 48 h | ||
| Eluate on YPDch agar | − | − | − | − | − | − | − | − | − | − | − | − | |||
| Eluate on MRScy agar | − | − | − | − | + | − | − | − | + | − | + | − | |||
| 0.45 µm | RLU MRScy | − | + | − | − | + | + | + | − | + | + | + | + | 24 h | |
| Plating RLU-MRScy on MRScy agar | − | − | − | − | + | + | + | − | + | + | + | + | 48 h |
| Inoculum (CFU/L) | Yeast (50) + LAB (1000) | |||||||
|---|---|---|---|---|---|---|---|---|
| Filter | 1.2 µm | 0.45 µm | ||||||
| Res.Medium | YPDch | SD | RESch | SD | MRScy | SD | REScy | SD |
| W7 | 1.93 × 106 | 2.08 × 102 | 4.35 × 105 | 2.74 × 104 | 2.89 × 103 | 4.08 × 102 | 1.22 × 104 | 4.83 × 103 |
| W8 | 1.93 × 104 | 1.37 × 103 | 5.02 × 106 | 5.19 × 104 | 8.72 × 104 | 1.88 × 103 | 5.35 × 104 | 1.04 × 103 |
| R7 | 1.92 × 105 | 3.29 × 104 | 6.81 × 106 | 4.09 × 103 | 1.73 × 106 | 1.75 × 104 | 7.20 × 104 | 2.13 × 103 |
| V1 | 1.77 × 106 | 9.21 × 105 | 6.81 × 106 | 1.24 × 106 | 2.15 × 102 | 1.02 × 105 | 7.28 × 102 | 6.02 × 102 |
| V2 | 3.47 × 102 | 2.86 × 102 | 5.59 × 106 | 2.75 × 105 | 7.07 × 102 | 7.62 × 102 | 5.78 × 103 | 6.84 × 102 |
| Blank | 2.37 × 102 | 3.46 × 10 | 1.35 × 102 | 2.43 × 10 | 1.73 × 102 | 2.26 × 10 | 1.35 × 102 | 1.20 × 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monica, S.; Bancalari, E.; Castellone, V.; Rijkx, J.; Wirth, S.; Jahns, A.; Bottari, B. ATP Bioluminescence for Rapid and Selective Detection of Bacteria and Yeasts in Wine. Appl. Sci. 2021, 11, 4953. https://doi.org/10.3390/app11114953
Monica S, Bancalari E, Castellone V, Rijkx J, Wirth S, Jahns A, Bottari B. ATP Bioluminescence for Rapid and Selective Detection of Bacteria and Yeasts in Wine. Applied Sciences. 2021; 11(11):4953. https://doi.org/10.3390/app11114953
Chicago/Turabian StyleMonica, Saverio, Elena Bancalari, Vincenzo Castellone, Jos Rijkx, Sarah Wirth, Anika Jahns, and Benedetta Bottari. 2021. "ATP Bioluminescence for Rapid and Selective Detection of Bacteria and Yeasts in Wine" Applied Sciences 11, no. 11: 4953. https://doi.org/10.3390/app11114953
APA StyleMonica, S., Bancalari, E., Castellone, V., Rijkx, J., Wirth, S., Jahns, A., & Bottari, B. (2021). ATP Bioluminescence for Rapid and Selective Detection of Bacteria and Yeasts in Wine. Applied Sciences, 11(11), 4953. https://doi.org/10.3390/app11114953






