Featured Application
This work demonstrates that a trained deep neural network is able to accurately correct the blurring present in PET images when using radionuclides with large positron ranges, such as 68Ga. This approach can be used in any preclinical and clinical PET studies.
Abstract
Positron emission tomography (PET) is a molecular imaging technique that provides a 3D image of functional processes in the body in vivo. Some of the radionuclides proposed for PET imaging emit high-energy positrons, which travel some distance before they annihilate (positron range), creating significant blurring in the reconstructed images. Their large positron range compromises the achievable spatial resolution of the system, which is more significant when using high-resolution scanners designed for the imaging of small animals. In this work, we trained a deep neural network named Deep-PRC to correct PET images for positron range effects. Deep-PRC was trained with modeled cases using a realistic Monte Carlo simulation tool that considers the positron energy distribution and the materials and tissues it propagates into. Quantification of the reconstructed PET images corrected with Deep-PRC showed that it was able to restore the images by up to 95% without any significant noise increase. The proposed method, which is accessible via Github, can provide an accurate positron range correction in a few seconds for a typical PET acquisition.
1. Introduction
Positron emission tomography (PET) is a molecular imaging technique that provides information about biochemical or physiological processes in the body in vivo [1]. PET images are obtained by detecting the radiation emitted by radionuclides bound to molecules (radiotracers) of interest administered to patients or animals under study [2]. PET currently provides very valuable information in clinical fields such as oncology, cardiology, and neurology, as well as in many preclinical studies [3].
PET is based on the detection of the two collinear 511 keV γ rays resulting from the annihilation of the positrons emitted by the unstable radionuclides. After being emitted, a positron will lose its initial kinetic energy through multiple interactions (mostly inelastic collisions [4]) with the electrons of the surrounding tissues. Positron–electron annihilation is most likely to occur when the positron has lost all its kinetic energy [5]. The distance between the emission and annihilation points is known as positron range (PR), and it is one of the main limiting factors of the spatial resolution of PET scanners [6,7,8,9,10,11]. PR makes the spatial distribution of the annihilation points a somewhat blurred version of that of the emission points (see Figure 1b). PR modeling is not straightforward, as it depends on both the kinetic energy of the emitted positrons (Figure 1a) and the electron density (i.e., the density and composition) of the surrounding tissues.
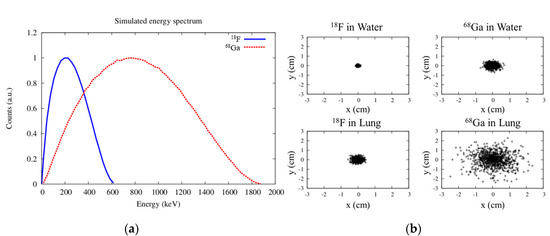
Figure 1.
Simulated energy spectra of positrons emitted by 18F and 68Ga obtained with the code PenNuc [18] (a), and distribution of annihilation points for 18F and 68Ga in water and lung tissue (b).
18F is by far the most widely used PET radionuclide, accounting for about 90% of clinical studies [12]. Nevertheless, many other radionuclides have been proposed for use in PET imaging, and hundreds of PET radiotracers based on them have been developed. A challenge associated with the use of some of these radionuclides, such as such as 68Ga, 82Rb, 124I, 76Br, or 86Y, is that they emit positrons with a large initial kinetic energy (see Figure 1), which results in large PR in tissues, yielding images with reduced spatial resolution and lower contrast in small volumes due to partial volume effects [7,11,13]. This effect is even more important when using high-resolution scanners designed to image small animals, which are able to achieve millimeter resolution [14].
The quantitative accuracy of PET depends on an accurate positron range correction (PRC). As 18F has a relatively small PR in soft tissue, accurate PRC for different radionuclides has been largely neglected, and to the best of our knowledge is not yet performed in standard PET image reconstruction. Nevertheless, the improved resolution of current scanners and the use of radionuclides with large PR such as 68Ga requires this correction to be improved.
1.1. Positron Range Models
PR can be modeled in many ways, which can be grouped into the following categories in order of increasing complexity:
Isotropic and tissue-independent model: A uniform model is used for all voxels, assuming water (or soft tissue) as the medium through which positrons travel and annihilate. This is a fast and easy procedure to implement, and only requires the activity distribution as input. PR distributions for most common radionuclides used in PET in water- or plastic-equivalent materials can be measured [15] or computed using Monte Carlo (MC) simulations based on well-established libraries such as GEANT4, PENELOPE, and FLUKA [10,16,17]. The simulated spatial distributions can be fitted to an equation [9,10]. These models can consider differences in the energy distribution of the positrons emitted by each radionuclide (see Figure 1a), but in general they are not quite realistic and can yield artefacts in heterogeneous media.
Isotropic but tissue-dependent model: The positron range effect is modeled as a blurring kernel that depends on the material of the voxel from which the positron is emitted, irrespective of the surrounding media. In this case, each blurring kernel is homogeneous and isotropic. This correction requires the coregistration of the CT image with the activity distribution and it is expected to work well everywhere except near tissue boundaries. However, a non-negligible number of positrons close to the skin–air boundary can escape and using space-invariant filters can result in severe artefacts [19]. For instance, these artefacts may be observed in clinical 124I PET imaging of thyroid glands close to the trachea [20].
Anisotropic and tissue-dependent (approximated) model: To address the presence of inhomogeneous media, space-variant models have been proposed [21,22]. The anisotropic PR has been modeled by anisotropically truncating an isotropic point probability density function based on tissue type, performing successive convolution operations of tissue-dependent PR kernels, or using the average of the fitting parameters of annihilation densities for the originating voxel and the target voxel [23]. These filters provide a fast and robust method for implementing a PR model, but due to the complexity of positron migration at irregular interfaces, it may not be always accurate.
Anisotropic and tissue-dependent (full) model: In this final case, the blurring kernel takes into account the material from which the positron is emitted and all the different materials that the positron travels through until it annihilates. These models are accurate even when the activity is distributed at extreme tissue boundaries. Monte Carlo simulations, despite their large computational cost, can be used to generate these models [24], including when the PR is in the presence of magnetic fields [17,25,26].
1.2. Positron Range Correction Methods
Many correction approaches have been proposed to remove the blurring caused by PR on reconstructed PET images. These methods differ not only for the PR model they are based on, but also for how they are applied. Two main approaches for PRC in PET can be identified.
PRC applied as a postprocessing step to the reconstructed images: In this approach, the reconstructed images before PRC are considered to represent the distribution of the positron annihilations, instead of the distribution of the positron emissions. Therefore, the goal of the postprocessing PRC procedure is to convert the annihilation distribution into the positron emission distribution (the one expected to be seen with PET). Postprocessing PRC has the advantage of being fast, simple, and to some extent independent of the procedures, algorithms, and codes used in the image reconstruction. On the other hand, it has the risk of increasing the noise in the final corrected images [25]. This approach has been applied using Fourier deconvolution techniques [27] with isotropic and tissue-independent kernels, as well as iterative deconvolution methods such as Richardson–Lucy [28], which enables the use of more realistic PR models.
PRC applied within the tomographic iterative image reconstruction process: PR models can be included in a point-spread-function (PSF) or resolution kernel [13,14,25], or within the system response matrix (SRM) used in the iterative reconstruction. This can be argued to be the most common approach used for PRC with 18F as, in fact, many image-reconstruction methods use SRM from point source measurements [29] obtained from 18F in water, 22Na in plastic, or realistic simulations [30] that incorporate PR effects. This approach has some important limitations: On one hand, adapting an existing SRM to other radionuclides could be difficult, unless a SRM is used wherein PR is factored out [31]. Furthermore, fully realistic PR models would require the SRM to be evaluated in each acquisition.
In some works, a mismatched projector/backprojector pair has been proposed, with PR blurring only applied right before the forward projection operation. It has been shown that using a PR blurring kernel in the backprojector has only the effect of reducing the convergence speed of the iterative algorithm [8]. In this case, the SRM used should not incorporate any PR effects. In many cases, it is difficult to separate the PR from other blurring effects considered in the SRM, and including an isotope-dependent PRC in a reconstruction procedure introduces the risk of overcorrecting the PRC, yielding overshooting and Gibbs artefacts [32].
1.3. Neural Networks in Medical Imaging
Machine learning (ML), and more specifically the area of ML known as deep learning, are having a huge impact on many areas, including medical imaging and PET [33]. Deep learning methods are able to create accurate mappings between inputs and outputs by means of artificial neural networks (NNs) with a large (deep) number of layers. The fact that the same framework can connect many different inputs, such as measurements, raw images, outputs such as labels, and reference images, allows its application in a large variety of problems and disciplines. A recent overview on ML and deep learning for PET imaging can be found in Reference [34].
The components of the NN are learned from example training datasets. In the case of supervised learning, the example inputs are paired with their corresponding desired outputs. After the training, the NN can then be used on new input data to predict their outputs.
Various studies based on convolutional neural networks (CNNs) have been proposed for medical image generation, especially for segmentation, many of them using the U-Net structure [35]. However, to the best of our knowledge, such networks have not yet been applied for PRC.
1.4. Proposed Method
In this work, we propose a deep-learning based PRC method (Deep-PRC) applied as a postprocessing step to the reconstructed PET images. Our goal was to develop a fast PRC method for 3D PET imaging that is able to provide PET images for medium- and large-range radionuclides, rivaling in spatial resolution the ones reconstructed with the standard short-range 18F radionuclide. The NN was trained with realistic simulated cases of preclinical studies of reconstructed images of 68Ga and 18F corresponding to the same activity distribution, and it was able to produce accurate and precise 68Ga PR-corrected images similar to the 18F ones.
We assumed that the image reconstruction method used to obtain the images had already incorporated the PRC for 18F, and, therefore, the purpose of this work was to obtain a PRC for 68Ga that made it look similar to the corresponding 18F counterpart, without the risk of double correcting this effect. The source code is available in Github [36].
2. Materials and Methods
Supervised ML requires a set of reference cases to train the NN. In this work, we generated simulated cases with realistic activity, material, and density distribution. The complete workflow is depicted in Figure 2, and each component is described in detail in the sections below.
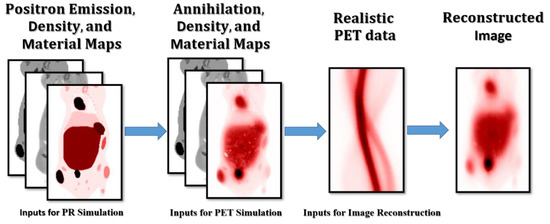
Figure 2.
Workflow used to produce the realistic simulated cases with different radionuclides.
2.1. Positron Range Simulator
The MC tool PenEasy (v2020) [37] based on PENELOPE [37] was used to simulate the PR for different radionuclides in several heterogeneous biological tissues. PENELOPE is a code for the MC simulation of coupled transport of electrons, positrons, and photons. It is suitable for the range of energies between 100 eV and 1 GeV and allows for definition of complex materials and geometries. In this work, PenEasy was adapted to generate 3D images of the spatial distribution of the positron annihilation points, using as an input the 3D images of the positron emissions and CT (see Figure 2). PenEasy considers the path traveled by each positron until its annihilation, taking into account its energy distribution and all the materials in the field of view.
The energy distributions of the positrons emitted by 68Ga and 18F were obtained with the code PenNuc [18], which considers all possible decay branches and nuclear properties for a large set of tabulated radionuclides (see Figure 1a).
2.2. Simulated Cases
We used numerical models of mice from a repository [38] to simulate the different cases needed for training, testing, and validation of the NN. The material composition and density of each segmented tissue in the models were directly obtained from the repository, while different activities were assigned to each tissue type, such as heart, liver, kidneys, and tumors, using a range of typical values found in 18F-Fluorodeoxyglucose (18F-FDG) acquisitions. The numerical models consisted of 154 × 154 × 242 cubic voxels of 0.28 × 0.28 × 0.28 mm, which covered a major part of the bodies of the mice (except for the brain, which was not included in the segmentation of the repository).
A total of eight different whole-body mouse models were used with PenEasy to generate the positron annihilation distributions from the initial positron emission, material, and density distributions (see Figure 3). Each model was simulated twice, once for 18F and once for 68Ga. Each simulation consisted of around 3 × 108 positron emissions, simulated at a rate of 3.4 × 104 histories per second for the 18F simulations and 2.2 × 104 histories per second for the 68Ga simulations in an Intel(R) Xeon(R) CPU @ 2.30GHz computer.
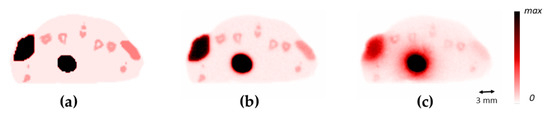
Figure 3.
Comparison of cross-sections of the mouse between the emission image (a), the 18F positron annihilation image (b), and the 68Ga positron annihilation image (c).
2.3. PET Acquisition Simulation and Reconstruction
In order to generate images more similar to actual PET reconstructed images, the positron annihilation distributions obtained from PenEasy were used to simulate a realistic PET acquisition in a generic preclinical scanner similar to an Inveon PET/CT scanner [39] using the MC simulator MCGPU-PET [40]. MCGPU-PET is a version of the MC-GPU software adapted for PET, which was developed for X-ray imaging [41]. MCGPU-PET allows very fast and realistic simulation of PET acquisitions from voxelized activity, material, and density distribution. MCGPU-PET can generate data which can be histogrammed into 3D sinograms, in our case with 147 × 168 × 1293 bins considering a maximum ring difference of 79, an axial compression factor of 11 and a radial bin size of 0.795 mm. MCGPU-PET simulations in a computer with a GeForce GTX 1080 8 Gb GPU contain around 1.2 × 109 coincidences in a minute (2 × 107 coincidences/second), including scatter and non-scattered true coincidences.
For reconstruction of the sinograms, we used GFIRST [42], which is the GPU-accelerated version of FIRST [30], a 3D-OSEM algorithm which allows a physical model to be incorporated into the SRM. In this case, the SRM used was the standard one created based on 18F in water. We used one subset and 40 iterations. The final images consisted of 154 ×154 × 80 voxels with a size of 0.28 × 0.28 × 0.795 mm, which is the typical size of the images reconstructed in the Inveon scanner [39]. The total reconstruction time was 50 s in a GTX 1080 8 Gb GPU. The values of the reconstructed images were converted into standardized uptake value units (SUV) to make it easier to evaluate the performance of the method. Using SUV units, a radiotracer with uniform distribution in the body would have a SUV of 1.
At the end of the whole simulation workflow (see Figure 2), eight mice were simulated with 18F and 68Ga, for a total of 640 slices. In order to make the size of the slices more tractable to the neural network, each image of 154 × 154 pixels was padded with zeros to reach a size of 160 × 160 pixels.
2.4. Neural Network
The CNN was implemented in Python within the Tensorflow framework [43] (v 2.3.0) with Keras [44]. It was based on the U-Net network [35] which has been demonstrated to be useful in many medical imaging applications [45,46]. We directly used the U-Net model available in Keras with four levels, 64 filters, and a dropout factor of 0.2 (see Figure 4). The source code can be found in Reference [36].
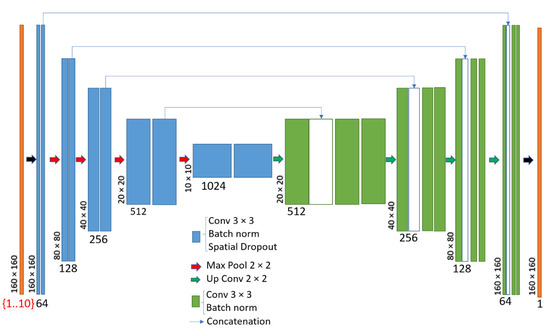
Figure 4.
Deep convolutional neural network architecture (U-Net) used in this work.
The inputs to the model were slices of the 68Ga (PET) and μ-maps (CT) volumes. In this work, we wanted to evaluate the amount of input information needed to perform an accurate PRC with a neural network. Therefore, we considered six different cases (see Table 1). In three of them, only the 68Ga PET images were considered for the input, and in three of them both the 68Ga and μ-maps (PET/CT) were considered. In each of them, we evaluated the performance of the method when using one, three, and five input slices. The different slices were used as channels in the input layer (see Figure 4). In all cases, the output was the corresponding central slice from the 18F-PET image with 160 × 160 × (1 channel). The sizes of the different input layers are shown in Table 1. In all cases, the total number of parameters was 31.7 million and the size of the model (hdf5 file) was 485 Mb.

Table 1.
Sizes of the input layers for the different evaluated cases of the Deep-PRC network.
In this work, the Swish activation function [47] was used instead of ReLU (except for the final output layer). Swish is a new, self-gated activation function which performs better than ReLU with a similar level of computational efficiency.
2.5. Model Training
The simulated cases were separated as follows: One volume was set aside and not used in the training/validation process, while the other seven volumes were processed and data augmentation was used with flip and shifts in horizontal and vertical directions, and in-plane rotations. This enabled the network to be trained with a much larger variety of cases than the initially simulated ones. Note that zoom could not be used in this case for data augmentation, as it would have distorted the PR effect.
Model training was performed with the recently proposed Rectified Adam optimizer [48] (learning rate of 1 × 10−3) and with the Lookahead technique [49]. The combination of these techniques provided a much faster convergence of the training process compared to the more commonly used Adam optimizer. The loss function used was the L1-norm between each slice of the ground truth of the 18F image, and the output slice of the Deep-PRC network.
For training, we used an NVIDIA T4 (NVIDIA Corporation, Santa Clara, CA, USA) graphics processing unit (GPU) with 16 GB of memory from Google Cloud (AI notebook running Jupyter Lab, with CUDA 10.1). The models were trained for 50 epochs with 100 iterations each. It took around 1 h to train each considered case.
2.6. Application and Quantitative Analysis
The trained models were saved as Keras models in hdf5 format. The models were then loaded and applied to a simulated study not included in either the training or the validation datasets.
The input was adapted to the specific characteristics of each trained network:
- Selecting the corresponding input slices—in the case of the models with three and five input slices, the slices closer to the edges of the axial FOV were extended to avoid truncation.
- Zero-padding the images to obtain 160 × 160 pixels in each slice.
- Normalizing the values to be between −1 and 1. This normalization was restored in the output, so that the PRC preserved the appropriate units.
A quantitative analysis of the resulting image was performed to obtain the mean (µ) and standard deviation (σ) in different organs. The noise was defined as the σ:µ ratio in uniform regions away from any boundaries and edges. The recovery coefficients were obtained by defining regions over the whole organs, and their values were then normalized respective to the reference reconstruction with 18F. The differences between the 68Ga images before and after the proposed PRC could be easily evaluated from the obtained coefficients.
3. Results
3.1. Model Training
Figure 5 shows the evolution of loss function during the training step in the different cases (depending on the amount and type of input information provided). It can be seen that the L1 was significantly minimized in all cases, although the convergence was more monotonic and reached a lower L1 loss function in the case of using five PET slices (Figure 5c). Some significant spikes were noticed in the validation cases. This reflected some possible directions in the training which may have yielded neural networks that produced images with artefacts. Fortunately, these unwanted solutions did not last long, and the convergence process continued without problems.
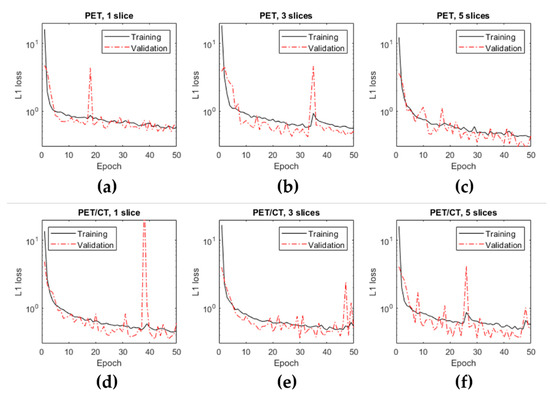
Figure 5.
Evolution of the loss function over the epochs during the training using the training and the validation datasets for the different cases evaluated.
Figure 6 shows a coronal view of a mouse with the μ-map obtained from the CT, and the reconstructed images of 18F, 68Ga, and 68Ga after the PRC (using the model with five slices and PET-only input). It can be easily seen that the proposed method was able to recover the resolution loss in 68Ga images with respect to 18F, and that this increased the values in some areas with higher uptake.
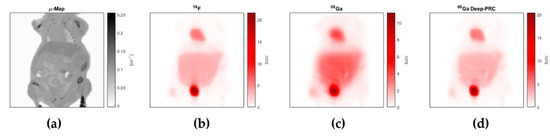
Figure 6.
Example of a coronal view of one of the evaluated cases, corresponding to the μ-map obtained from the CT (a), and the reconstructed images of 18F (b), 68Ga (c), and 68Ga after the PRC (d).
3.2. Model Deployment and Quantitative Analysis
In order to test the model, the trained Deep-PRC network (PET, five slices) was applied to a simulated case considered in neither the training nor the validation process. The time required to obtain the PRC on the 80 slices of the whole volume was 5.14 s in a T4 GPU and 2.85 s in a V100 GPU. Although these results could be sped up by using multiple GPUs, they are already fast enough to be used in preclinical and clinical applications.
Profiles along some organs of interest, such as the bladder, heart and tumor in the 18F, 68Ga, and 68Ga with Deep-PRC images are shown in Figure 7. The significant impact of the PR in these cases is quite clear, as well as the capacity of the proposed method to correct for this effect. The differences in SUV units between the estimated images (68Ga with Deep-PRC) and the reference one (18F) are shown in Figure 8. Areas with high uptake, such as the bladder, still had some residual error (as it can be also seen in Figure 7a), but it was a small deviation in relative terms.
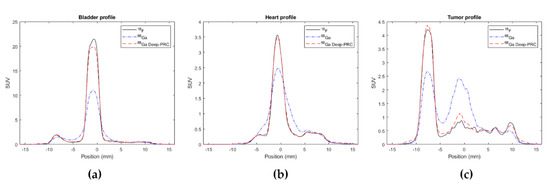
Figure 7.
Profiles across different regions in the 18F, 68Ga, and 68Ga with Deep-PRC images: bladder (a), heart (b), and tumor (c).
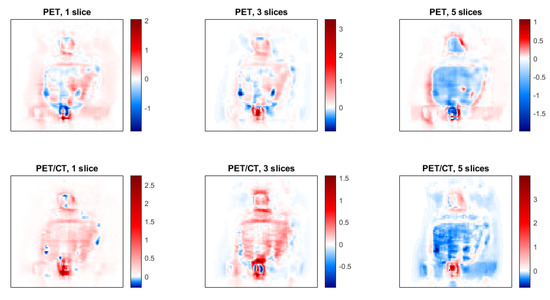
Figure 8.
Images of the difference between the estimated images (68Ga with Deep-PRC) and the reference ones (18F) for each of the cases considered.
The quantitative analysis of the results is shown in Table 2. From the table, it is clear that the 68Ga images corrected by PR images were very similar to the 18F images (with recoveries greater than 95% of the reference values). Additionally, the noise level of the estimated images was comparable to that of the reference ones, which indicates that the proposed method did not trade noise for resolution, as is the case in many deconvolution-based approaches for PRC.

Table 2.
Quantitative analysis of the results over different regions.
4. Discussion
This paper presents the use of a deep convolutional neural network to provide an accurate PRC in PET. The method was evaluated in simulations in preclinical studies and its performance characterized. To the best of our knowledge, this is the first work to successfully combine deep learning and PRC in a coherent framework.
Our results indicate that overall, the image quality produced by the learned model is comparable to that of the reference images, with recoveries going from around 60% to more than 95% while maintaining low noise levels.
One interesting question that we wanted to address in this work is the amount of input data required for these types of algorithms. With PR being a three-dimensional effect (i.e., it affects not only a particular single slice) and being dependent on both the PET activity and the material distribution (CT), it may be reasonable to expect that many PET and CT input slices will be needed to generate an accurate output slice. On the other hand, many works have shown that the information in PET and CT images is not independent, and that a deep neural network may be able to estimate to some extent images of one modality from the other. This fact may indicate that using only the PET image as input may be enough.
The results obtained with all methods considered were good, as shown by the L1 loss function in the validation cases. Nevertheless, it seems that the training with just the PET images as input, and with large enough axial slices was the best option. It is important to note that in our case, each slice was two times larger than the pixel size in the transverse direction. Therefore, considering five slices corresponded to using 4 mm in the axial direction, which seemed enough to consider the effects of surrounding slices for a specific slice. In this work the number of slices were limited by the amount of memory available in the GPU. Nevertheless, this is something that will be easily solved with new GPU models.
The training was based on minimization of the L1 norm between the reference images and the estimated ones. The L1 norm is known to be more stable and robust to the presence of noise than the L2 norm [50]. In any case, other loss functions could be explored in this context, including a loss term from an adversarial network (GAN).
A detailed comparison of the performance of the proposed method with previously proposed ones (described in Section 1.2) is outside the scope of this work, but we plan to perform this detailed comparison in a future work. In any case, the fact that the proposed Deep-PRC had no significant impact on the noise level of the images is a clear advantage compared to previous approaches [25].
It is important to note that although we have proposed the method as a postprocessing step, the same neural network architecture could be used to generate a PR model that could be applied in the forward projection within an image reconstruction (simply by inverting the input and outputs of the NN). This line of research will be explored in future works.
In this work, the axial FOV of the scanner was large enough for mice, so no significant axial truncation of the PET activity was present. If there is significant truncation (as it usually happens in whole-body PET acquisitions in which only a section of the body is examined in each bed axial position), it may be advisable to perform this PRC on the final 3D whole-body volume to avoid possible truncation artefacts (from activity out of the axial FOV in each particular bed position). This can be argued to be an advantage of the postprocessing PRC approach, as it can be easily applied to multibed studies in which the activity from other bed positions might have a non-negligible effect.
In this work, the effect of positronium formation in the positron range was not considered [9,51]. This can occur when, after losing its kinetic energy, the positron reaches thermal velocities (a few eV) and instead of annihilating directly with an electron into two gammas, forms an intermediate state called positronium, which may extend its lifetime. Its effect on PR is not well established, but it may have a non-negligible effect in porous materials or low-density ones such as the lungs.
The cases developed in this work are not exclusive of any scanner or radionuclide in particular. In this work, we used the preclinical scanner Inveon and 68Ga as a reference, but the proposed approach is flexible and suitable for any preclinical and clinical PET systems and with any radionuclide.
5. Conclusions
We developed and evaluated a deep convolutional neural network (Deep-PRC) that provides a fast and accurate PRC method to recover the resolution loss present in PET studies using radionuclides that emit positrons with large PR. We demonstrated its quantitative accuracy in realistic simulations of preclinical PET/CT studies with 68Ga.
Our results suggest that it is sufficient to use PET images as input for the neural network (i.e., without the corresponding CT or the μ-map extracted from it), but it is important to include not only the reference slice (i.e., 2D case), but also some additional neighbor slices.
The correction of PR effects in PET image reconstruction is becoming mandatory in light of the increasing use of high-energy positron emitters in preclinical and clinical PET imaging and their improved spatial resolution. Convolutional neural networks seem to be very well suited for this type of correction.
Author Contributions
Study concept and design, J.L.H.; data simulation, A.L.-M.; validation, A.B.; software, all authors; obtained funding, J.L.H.; writing—original draft preparation, J.L.H.; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge support from the Spanish Government (RTI2018-095800-A-I00), from Comunidad de Madrid (B2017/BMD-3888 PRONTO-CM). and the NIH R01 CA215700-2 grant. JLH also acknowledges support from a Google Cloud Academic Grant.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The source code and some training data are available in Github (https://github.com/jlherraiz/deepPRC).
Acknowledgments
This is a contribution to the Moncloa Campus of International Excellence.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vaquero, J.J.; Kinahan, P. Positron Emission Tomography: Current Challenges and Opportunities for Technological Advances in Clinical and Preclinical Imaging Systems. Annu. Rev. Biomed. Eng. 2015, 17, 385–414. [Google Scholar] [CrossRef]
- Bailey, D.L.; Townsend, D.W.; Valk, P.E.; Maisey, M.N. (Eds.) Positron Emission Tomography: Basic Sciences; Springer: London, UK, 2005; ISBN 978-1-85233-485-7. [Google Scholar]
- Mettler, F.A.; Guiberteau, M.J. Essentials of Nuclear Medicine and Molecular Imaging; Elsevier: New York, NY, USA, 2019; ISBN 978-0-323-48319-3. [Google Scholar]
- PENELOPE-2018: A Code System for Monte Carlo Simulation of Electron and Photon Transport. Available online: https://www.oecd-nea.org/jcms/pl_46441/penelope-2018-a-code-system-for-monte-carlo-simulation-of-electron-and-photon-transport?details=true (accessed on 26 November 2020).
- Progress in Positron Annihilation |Book| Scientific.Net. Available online: https://www.scientific.net/book/progress-in-positron-annihilation/978-3-03813-451-0 (accessed on 26 November 2020).
- Moses, W.W. Fundamental Limits of Spatial Resolution in PET. Nucl. Instrum. Methods Phys. Res. Sect. Accel. Spectrometers Detect. Assoc. Equip. 2011, 648 (Suppl. S1), S236–S240. [Google Scholar] [CrossRef]
- Levin, C.S.; Hoffman, E.J. Calculation of positron range and its effect on the fundamental limit of positron emission tomography system spatial resolution. Phys. Med. Biol. 1999, 44, 781–799. [Google Scholar] [CrossRef]
- Cal-González, J.; Herraiz, J.L.; España, S.; Vicente, E.; Herranz, E.; Desco, M.; Vaquero, J.J.; Udías, J.M. Study of CT-based positron range correction in high resolution 3D PET imaging. Nucl. Instrum. Methods Phys. Res. Sect. Accel. Spectrometers Detect. Assoc. Equip. 2011, 648, S172–S175. [Google Scholar] [CrossRef]
- Jødal, L.; Le Loirec, C.; Champion, C. Positron range in PET imaging: An alternative approach for assessing and correcting the blurring. Phys. Med. Biol. 2012, 57, 3931–3943. [Google Scholar] [CrossRef]
- Cal-González, J.; Herraiz, J.L.; España, S.; Corzo, P.M.G.; Vaquero, J.J.; Desco, M.; Udias, J.M. Positron range estimations with PeneloPET. Phys. Med. Biol. 2013, 58, 5127–5152. [Google Scholar] [CrossRef]
- Carter, L.M.; Leon Kesner, A.; Pratt, E.C.; Sanders, V.A.; Massicano, A.V.F.; Cutler, C.S.; Lapi, S.E.; Lewis, J.S. The Impact of Positron Range on PET Resolution, Evaluated with Phantoms and PHITS Monte Carlo Simulations for Conventional and Non-conventional Radionuclides. Mol. Imaging Biol. 2020, 22, 73–84. [Google Scholar] [CrossRef]
- Dash, A.; Chakravarty, R. Radionuclide generators: The prospect of availing PET radiotracers to meet current clinical needs and future research demands. Am. J. Nucl. Med. Mol. Imaging 2019, 9, 30–66. [Google Scholar]
- Cal-Gonzalez, J.; Perez-Liva, M.; Herraiz, J.L.; Vaquero, J.J.; Desco, M.; Udias, J.M. Tissue-Dependent and Spatially-Variant Positron Range Correction in 3D PET. IEEE Trans. Med. Imaging 2015, 34, 2394–2403. [Google Scholar] [CrossRef]
- Cal-Gonzalez, J.; Vaquero, J.J.; Herraiz, J.L.; Pérez-Liva, M.; Soto-Montenegro, M.L.; Peña-Zalbidea, S.; Desco, M.; Udías, J.M. Improving PET Quantification of Small Animal [68Ga]DOTA-Labeled PET/CT Studies by Using a CT-Based Positron Range Correction. Mol. Imaging Biol. 2018, 20, 584–593. [Google Scholar] [CrossRef]
- Alva-Sánchez, H.; Quintana-Bautista, C.; Martínez-Dávalos, A.; Ávila-Rodríguez, M.A.; Rodríguez-Villafuerte, M. Positron range in tissue-equivalent materials: Experimental microPET studies. Phys. Med. Biol. 2016, 61, 6307–6321. [Google Scholar] [CrossRef] [PubMed]
- Augusto, R.S.; Bauer, J.; Bouhali, O.; Cuccagna, C.; Gianoli, C.; Kozłowska, W.S.; Ortega, P.G.; Tessonnier, T.; Toufique, Y.; Vlachoudis, V.; et al. An overview of recent developments in FLUKA PET tools. Phys. Med. 2018, 54, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Caribé, P.R.R.V.; Vandenberghe, S.; Diogo, A.; Pérez-Benito, D.; Efthimiou, N.; Thyssen, C.; D’Asseler, Y.; Koole, M. Monte Carlo Simulations of the GE Signa PET/MR for Different Radioisotopes. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- García-Toraño, E.; Peyres, V.; Salvat, F. PenNuc: Monte Carlo simulation of the decay of radionuclides. Comput. Phys. Commun. 2019, 245, 106849. [Google Scholar] [CrossRef]
- Bai, B.; Ruangma, A.; Laforest, R.; Tai, Y.-; Leahy, R.M. Positron range modeling for statistical PET image reconstruction. In Proceedings of the 2003 IEEE Nuclear Science Symposium. Conference Record (IEEE Cat. No.03CH37515), Portland, OR, USA, 19–25 October 2003; Volume 4, pp. 2501–2505. [Google Scholar]
- Abdul-Fatah, S.B.; Zamburlini, M.; Halders, S.G.E.A.; Brans, B.; Teule, G.J.J.; Kemerink, G.J. Identification of a Shine-Through Artifact in the Trachea with 124I PET/CT. J. Nucl. Med. 2009, 50, 909–911. [Google Scholar] [CrossRef]
- Bai, B.; Laforest, R.; Smith, A.M.; Leahy, R.M. Evaluation of MAP image reconstruction with positron range modeling for 3D PET. In Proceedings of the IEEE Nuclear Science Symposium Conference Record, Fajardo, PR, USA, 23–29 October 2005; Volume 5, pp. 2686–2689. [Google Scholar]
- Alessio, A.M.; Kinahan, P.E.; Lewellen, T.K. Modeling and incorporation of system response functions in 3-D whole body PET. IEEE Trans. Med. Imaging 2006, 25, 828–837. [Google Scholar] [CrossRef]
- Alessio, A.; MacDonald, L. Spatially Variant Positron Range Modeling Derived from CT for PET Image Reconstruction. In Proceedings of the 2008 IEEE Nuclear Science Symposium Conference Record, Dresden, Germany, 19–25 October 2008; pp. 3637–3640. [Google Scholar] [CrossRef]
- Fu, L.; Qi, J. A residual correction method for high-resolution PET reconstruction with application to on-the-fly Monte Carlo based model of positron range. Med. Phys. 2010, 37, 704–713. [Google Scholar] [CrossRef]
- Bertolli, O.; Eleftheriou, A.; Cecchetti, M.; Camarlinghi, N.; Belcari, N.; Tsoumpas, C. PET iterative reconstruction incorporating an efficient positron range correction method. Phys. Med. 2016, 32, 323–330. [Google Scholar] [CrossRef]
- Li, C.; Cao, X.; Liu, F.; Tang, H.; Zhang, Z.; Wang, B.; Wei, L. Compressive effect of the magnetic field on the positron range in commonly used positron emitters simulated using Geant4. Eur. Phys. J. Plus 2017, 132, 484. [Google Scholar] [CrossRef]
- Derenzo, S.E. Mathematical Removal of Positron Range Blurring in High Resolution Tomography. IEEE Trans. Nucl. Sci. 1986, 33, 565–569. [Google Scholar] [CrossRef]
- Rukiah, A.L.; Meikle, S.R.; Gillam, J.E.; Kench, P.L. An investigation of 68Ga positron range correction through de-blurring: A simulation study. In Proceedings of the 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), Sydney, Australia, 10–17 November 2018; pp. 1–2. [Google Scholar]
- Panin, V.Y.; Kehren, F.; Rothfuss, H.; Hu, D.; Michel, C.; Casey, M.E. PET reconstruction with system matrix derived from point source measurements. IEEE Trans. Nucl. Sci. 2006, 53, 152–159. [Google Scholar] [CrossRef]
- Herraiz, J.L.; España, S.; Vaquero, J.J.; Desco, M.; Udías, J.M. FIRST: Fast Iterative Reconstruction Software for (PET) tomography. Phys. Med. Biol. 2006, 51, 4547–4565. [Google Scholar] [CrossRef]
- Zhou, J.; Qi, J. Fast and efficient fully 3D PET image reconstruction using sparse system matrix factorization with GPU acceleration. Phys. Med. Biol. 2011, 56, 6739–6757. [Google Scholar] [CrossRef]
- Rahmim, A.; Qi, J.; Sossi, V. Resolution modeling in PET imaging: Theory, practice, benefits, and pitfalls. Med. Phys. 2013, 40. [Google Scholar] [CrossRef]
- Reader, A.J.; Corda, G.; Mehranian, A.; da Costa-Luis, C.; Ellis, S.; Schnabel, J.A. Deep Learning for PET Image Reconstruction. IEEE Trans. Radiat. Plasma Med. Sci. 2020. [Google Scholar] [CrossRef]
- Gong, K.; Berg, E.; Cherry, S.R.; Qi, J. Machine Learning in PET: From Photon Detection to Quantitative Image Reconstruction. Proc. IEEE 2020, 108, 51–68. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:150504597. [Google Scholar]
- Herraiz, J.L. Deep PRC (2020)—Github Repository. Available online: https://github.com/jlherraiz/deepPRC (accessed on 27 December 2020).
- Sempau, J.; Badal, A.; Brualla, L. A PENELOPE-based system for the automated Monte Carlo simulation of clinacs and voxelized geometries—Application to far-from-axis fields. Med. Phys. 2011, 38, 5887–5895. [Google Scholar] [CrossRef]
- Rosenhain, S.; Magnuska, Z.A.; Yamoah, G.G.; Rawashdeh, W.A.; Kiessling, F.; Gremse, F. A preclinical micro-computed tomography database including 3D whole body organ segmentations. Sci. Data 2018, 5, 180294. [Google Scholar] [CrossRef]
- Constantinescu, C.C.; Mukherjee, J. Performance evaluation of an Inveon PET preclinical scanner. Phys. Med. Biol. 2009, 54, 2885–2899. [Google Scholar] [CrossRef]
- Badal, A.; Domarco, J.; Udias, J.M.; Herraiz, J.L. MCGPU-PET: A Real-Time Monte Carlo PET Simulator; International Symposium on Biomedical Imaging: Washington, DC, USA, 2018. [Google Scholar]
- Badal, A.; Badano, A. Accelerating Monte Carlo simulations of photon transport in a voxelized geometry using a massively parallel graphics processing unit. Med. Phys. 2009, 36, 4878–4880. [Google Scholar] [CrossRef] [PubMed]
- Herraiz, J.L.; España, S.; Cabido, R.; Montemayor, A.S.; Desco, M.; Vaquero, J.J.; Udias, J.M. GPU-Based Fast Iterative Reconstruction of Fully 3-D PET Sinograms. IEEE Trans. Nucl. Sci. 2011, 58, 2257–2263. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M. TensorFlow: A System for Large-Scale Machine Learning. In Proceedings of the OSDI’16: Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Chollet, F. Others Keras. 2015. Available online: https://keras.io (accessed on 25 November 2020).
- Berker, Y.; Maier, J.; Kachelrieß, M. Deep Scatter Estimation in PET: Fast Scatter Correction Using a Convolutional Neural Network. In Proceedings of the 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), Sydney, Australia, 10–17 November 2018. [Google Scholar]
- Leung, K.H.; Marashdeh, W.; Wray, R.; Ashrafinia, S.; Rahmim, A.; Jha, A.K. A Physics-Guided Modular Deep-Learning Based Automated Framework for Tumor Segmentation in PET Images. arXiv 2020, arXiv:2002.07969. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Swish: A Self-Gated Activation Function. arXiv 2017, arXiv:171005941. [Google Scholar]
- Liu, L.; Jiang, H.; He, P.; Chen, W.; Liu, X.; Gao, J.; Han, J. On the Variance of the Adaptive Learning Rate and Beyond. arXiv 2020, arXiv:190803265. [Google Scholar]
- Zhang, M.R.; Lucas, J.; Hinton, G.; Ba, J. Lookahead Optimizer: K steps forward, 1 step back. arXiv 2019, arXiv:190708610. [Google Scholar]
- Traonmilin, Y.; Ladjal, S.; Almansa, A. Outlier Removal Power of the L1-Norm Super-Resolution. In Lecture Notes in Computer Science; Scale Space and Variational Methods in Computer Vision; SSVM 2013; Kuijper, A., Bredies, K., Pock, T., Bischof, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 7893. [Google Scholar] [CrossRef]
- Moskal, P.; Kisielewska, D.; Curceanu, C.; Czerwiński, E.; Dulski, K.; Gajos, A.; Gorgol, M.; Hiesmayr, B.; Jasińska, B.; Kacprzak, K.; et al. Feasibility study of the positronium imaging with the J-PET tomograph. Phys. Med. Biol. 2019, 64, 055017. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).