Abstract
Advances in genome sequencing technology and computing power have brought about the explosive growth of sequenced genomes in public repositories with a concomitant increase in annotation errors. Many protein sequences are annotated using computational analysis rather than experimental verification, leading to inaccuracies in annotation. Confirmation of existing protein annotations is urgently needed before misannotation becomes even more prevalent due to error propagation. In this work we present a novel approach for automatically confirming the existence of manually curated information with experimental evidence of protein annotation. Our ensemble learning method uses a combination of recurrent convolutional neural network, logistic regression, and support vector machine models. Natural language processing in the form of word embeddings is used with journal publication titles retrieved from the UniProtKB database. Importantly, we use recall as our most significant metric to ensure the maximum number of verifications possible; results are reported to a human curator for confirmation. Our ensemble model achieves 91.25% recall, 71.26% accuracy, 65.19% precision, and an F1 score of 76.05% and outperforms the Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT) model with fine-tuning using the same data.
1. Introduction
The development of high-throughput sequencing technologies and computational capabilities has revolutionized genetics and genomics, resulting in a tremendous reduction in the cost of genome sequencing and the exponential growth of genomes in public repositories. Unfortunately, the annotation of both genes and proteins has not kept pace with genome sequencing, and this has led to a problem with misannotations. Recent research has shown that not only are protein annotations incorrect or confusing, but many protein sequences are missing annotations [1,2,3,4,5]. For example, in [5], protein sequences downloaded in 2016 using the National Center for Biotechnology Information (NCBI) GenBank ftp service [6] were clustered, and it was found that of 2826 proteobacterial protein sequences that clustered into a single homologous group, 82% included GroEL or GroL in their annotation while the remaining sequences had a variety of annotations including “not yet annotated”, mopA (obsolete gene name), thermosome, and 60 kDa chaperonin (11.78%). Moreover, 44 non-GroEL protein sequences were incorrectly annotated as chaperonin GroEL by the original annotators. The preferred annotation for this sequence listed in the UniProtKB/Swiss-Prot database is 60 kDa chaperonin while NCBI RefSeq annotates it as molecular chaperone GroEL [5]. With a lack of consensus between these two mainstream databases for such a universally conserved protein, it is not surprising that major issues exist with protein annotation.
How can we reduce the problems with protein annotation? Manual curation would be ideal, but it demands substantial time and effort and, thus, the exponential growth in sequences has only been matched by a linear increase, with a relatively small slope, in the number of annotated entries with experimental validation [7]. In the following paragraphs, we discuss our approach to this problem.
Unlike gene misannotation, which often occurs because of sequence discrepancies, protein misannotation arises from confusion whereby different names are used by the original annotators, and these names are propagated, sometimes appropriately and sometimes not; from lack of domain knowledge; and by error propagation. To improve protein annotation, we decided to use peer-reviewed biomedical papers that provide experimental evidence for manually curated protein function for a given protein annotation. Unfortunately, classifying whether a biomedical paper contains such evidence poses numerous challenges when determining the correct feature representation for the necessary relevant biological relationships. In [8], protein features (protein lexicons) and assertion features (sentences expressing the main point of a paper) are extracted from publications, and a document classification corpus is constructed using evidence from the Function category of UniProtKB/Swiss-Prot. However, the set of target lexicons they use to indicate experimental confirmation, for example, “requir(e)”, “function”, “promot(e)”, and “control”, is very limited.
Compared with traditional techniques for language modeling such as hidden Markov models and handcrafted features as used in [8], the simple yet useful bag-of-n-grams representation [9] relies on a predetermined vocabulary of known words and measurement of the occurrence of each word. For n-grams (sequences of n successive words), the probability of a word is computed from the n-1 previous words using maximum likelihood estimation based on word frequency in a corpus and count normalization. Although a bag-of-n-grams contains some information on word order within a context size of n, it is subject to the problem of sparsity because it underestimates the probability of words rarely occurring in a training corpus, and a zero or near-zero probability of the existence of such words makes it impossible to compute the probability of neighboring words in the test set [10]. Most importantly, it is impractical to manually design a comprehensive vocabulary that contains the essential words from biomedical papers given the vast number of biomedical terms that exist.
More complex features such as part-of-speech (POS) tags, noun phrases [11], and tree kernels [12] are also widely used in feature representation. However, it is difficult to evaluate when POS tags and noun phrases are discriminatory features for a specific classification task [13]. Tree kernels are able to generate many useful and relevant syntactic features, but their computational time complexity is superlinear with the number of tree nodes (syntactic features), and the accuracy using features generated by tree kernels is lower than that of linear models using manually crafted features [14].
With recent improvements in deep learning, embedding methods such as word2vec [15] and doc2vec [10], which transform a word or a paragraph into a numeric vector representing its semantics, are widely used in feature representation. By using the densely distributed feature representation of word embeddings for each word as learned from its usage, words used in a similar context have similar representations to denote their meaning. Although word embedding uses a distinct numeric vector to represent a word, it overlooks the internal structure of words and is poor at learning words that do not appear in the training data.
In our work, we use BioWordVec [16], a word embedding method that captures subword information, i.e., each word is further represented as a bag of n-gram characters [17], and exploits the internal structure of words to enrich feature representation in the numeric vector. Once such an effective feature representation of a text input is available, it can be widely used for classification in deep learning models such as recurrent neural networks (RNNs) [18] and convolutional neural networks (CNNs) [19]. In RNNs, a sequence is broken into multiple words and the output is passed from the previous layer of the network to the current layer; in CNNs, a matrix is formed using word embeddings, and then convolutions using filters are performed on the matrix to generate feature maps. For our work, we chose to use a recurrent convolutional neural network (RCNN) to better capture contextual information and discriminative phrases using max pooling (a discretization process) and filtering [20]. Given that ensemble learning works well when errors for the individual models are uncorrelated, we also use two discriminative classifiers, logistic regression and a support vector machine (SVM) with a linear kernel. Logistic regression is efficient for high-dimensional inputs, and regularization can be used to avoid overfitting. Similarly, regularization can be used with the SVM model. For the logistic regression and SVM models, we use BioSentVec [21] rather than BioWordVec. The former is trained on large-scale biomedical texts to generate sentence embeddings and is more efficient for the traditional models.
To demonstrate the effectiveness of our ensemble learning method, we compare its performance with the state-of-the-art Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT) fine-tuned using the same data for the same task. The Bidirectional Encoder Representations from Transformers (BERT) model is a contextualized word representation model based on a masked language model, which predicts masked words randomly in a sequence [22]. It learns information from both the left and right contexts of an input token during training and has been proven to achieve state-of-the-art performance for most NLP tasks. BioBERT [23] is a biomedical-domain language representation model pre-trained on large-scale biomedical corpora (PubMed abstracts and PMC full-text articles), which extends BERT to the biomedical domain.
In an effort to ameliorate protein misannotation, we developed a smart software program based on natural language processing (NLP), which periodically queries the UniProtKB database for peer-reviewed articles on a given protein annotation. Word embedding using BioWordVec or BioSentVec is used on all pertinent publication titles retrieved, and an ensemble model is used to determine whether the publications demonstrate laboratory validation of protein function for a given protein annotation. Each of the three models used for ensemble learning—RCNN, logistic regression, and SVM—is individually trained using positive and negative data, and voting is used to classify whether a publication provides the necessary experimental confirmation. Annotations of proteins of which functions have been confirmed experimentally in peer reviewed publications are considered validated. Importantly, because we want to ensure the maximum number of verifications and because the results are reported to a human curator for confirmation, we use recall as our foremost metric.
In this paper, we describe a smart program used to verify protein annotation based on publications in the UniProtKB database and, in particular, classifying publications without any evidence tag for experimental validation, the majority of which are in the uncurated UniProtKB/TrEMBL database. While this smart program is the key to the success of our project, it is only a part of it. In the near future, we will present a system for individual researchers to use to assist them with annotating protein sequences of interest to them. More details on this system are presented in the Conclusion and Future Work section.
2. Methods
2.1. Smart Classification Program
We propose a smart classification program that takes protein annotations as input and outputs whether an annotation is correct together with the publications confirming the annotation. Our smart program uses an ensemble learning approach [24], which combines the predictions of logistic regression, SVM with a linear kernel, and RCNN models individually trained on literature from the UniProtKB/Swiss-Prot database.
2.1.1. Problem Formulation
We formulate the verification of protein annotation as a binary classification problem: whether or not peer-reviewed biomedical publications indicate experimental evidence of the target protein. If such publications exist, we classify the annotation as verified; otherwise, we classify the annotation as to-be-verified.
2.1.2. Smart Program Architecture
The complete system architecture for our smart program is shown in Figure 1 and Figure 2, which depict the training workflow and classification workflow, respectively.
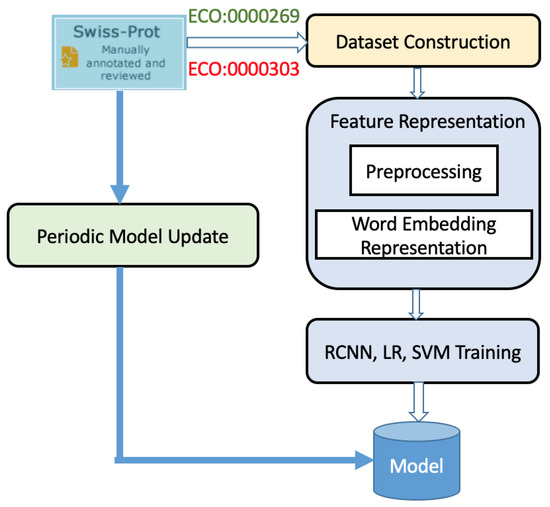
Figure 1.
Overview of the training workflow for ensemble learning. Publication titles from the UniProtKB/Swiss-Prot literature database associated with ECO (Evidence and Conclusion Ontology) codes were used to construct the training data. For positive examples, PubMed IDs with the ECO code ECO:0000269 were used; this code is used for manually curated information for which published experimental evidence exists [25,26]. For negative examples, PubMed IDs with the ECO code ECO:0000303 were used; this code is used for manually curated information with no experimental evidence of statements presented in scientific articles.
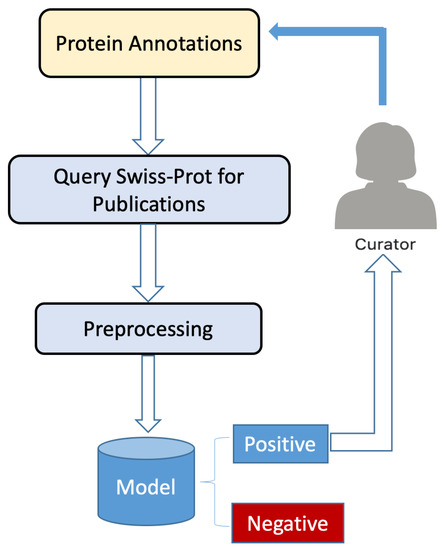
Figure 2.
Overview of the classification workflow for protein annotation.
As shown in Figure 1, we extract publications from the Function category of the UniProtKB/Swiss-Prot database, the manually annotated and reviewed section of the UniProt Knowledgebase, during training and then represent these publications using biomedical word embedding of their titles. Afterward, we separately train the logistic regression, SVM with linear kernel, and RCNN models and combine the results to obtain our ensemble learning model. Because new publications are added to the UniProtKB/Swiss-Prot database as they become available, periodic updating of the model can be performed. In Section 2.2, we present the details of our training workflow.
In Figure 2, we see that protein annotations are used as the initial input to our classification workflow. Each annotation is used to query the UniProtKB database for publications. Publication titles are extracted, word embedded, and the resulting vectors of numbers are used in our trained ensemble model, which uses them to classify the query protein sequence as either having experimental confirmation of protein function or not. We note, however, that publications for sequences in UniProtKB/Swiss-Prot tagged in the Function category have already been confirmed as having experimental evidence, and there is no need to run our smart program for these. Our smart program is only needed for unreviewed publications in UniProtKB, although these are the vast majority. For all sequences classified positively, the publication titles together with the protein accession number associated with the publications are submitted to a human curator for manual inspection. After confirmation that the publications have identified protein annotation, the protein sequence is marked by the curator as having been verified. Details of the classification workflow are presented in Section 2.3.
2.2. Training Workflow Phase
2.2.1. Training Corpus Construction and Pre-Processing
To train our individual models, we needed to acquire both positive and negative training data. We used publications from the UniProtKB/Swiss-Prot literature database, which are associated with ECO (Evidence and Conclusion Ontology) codes to represent the evidence type. Initially, we considered the use of publication abstracts, but publication titles resulted in greater accuracy with faster training and classification. We believe this is because both negative and positive abstracts share many words in common, which introduce considerable noise, thereby making classification more difficult. In general, publication titles available in the UniProtKB/Swiss-Prot database, which have been manually curated, seem to best summarize the substance of the publications. They often include significant words denoting experimental validation of protein function. For example, in the title “Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens”, “Identification”, “isolation”, and “cloning” infer experimentation and “toxin-binding” infers function. Such word usage in titles often distinguishes positive examples (titles) from negative examples (titles). In contrast, although negative examples may contain words such as “characterization”, the titles do not indicate experimental validation of protein function. Some examples are “Structure and transcription of the Drosophila melanogaster vermilion gene and several mutant alleles” and “Variation in fumonisin and ochratoxin production associated with differences in biosynthetic gene content in Aspergillus niger and A. welwitschiae isolates from multiple crop and geographic origins”. For positive examples, PubMed IDs with the ECO code ECO:0000269 were used; this code is used for manually curated information for which published experimental evidence exists [25,26]. For negative examples, PubMed IDs with the ECO code ECO:0000303 were used; this code is used for manually curated information with no experimental evidence of statements presented in scientific articles. Both the positive and negative publication titles were collected by means of mapping from PubMed IDs to publication titles using BioPython programming tools [27]. The data were obtained from the UniProtKB/Swiss-Prot database on or before 10 April 2019, and we processed 14,101 positive examples and 14,101 negative examples to use for our training data.
Our pre-processing includes segmenting the text into component words and symbols, converting uppercase characters and words to lowercase, removing all the punctuation and non-alphabetic symbols, and stop-word filtering. Specifically, our filtering uses a dictionary of stop words from the Natural Language ToolKit [28]. Stop words such as “the” and “an” convey no experimental evidence; thus, we eliminate them to reduce the dimensionality of our term space, that is, the number of words we need to vectorize. We also added word stemming, which truncates words that are phonologically related to their root word. While this reduced the total number of words, performance was degraded because the pre-trained word embedding models are based on complete words. Hence, we dispensed with word stemming.
2.2.2. Feature Representation
In order to include semantic relationships and to maximize biomedical domain specificity, we used BioWordVec word embeddings [21] for feature representation. BioWordVec, trained on corpora obtained using the PubMed search engine as well as clinical notes from the MIMIC-III clinical database [16,29], is a set of biomedical word embeddings that incorporates subword information (each word is further represented as a bag of n-gram characters) from unlabeled biomedical publications with Medical Subject Headings (MeSH), a hierarchically-organized biomedical vocabulary [16].
BioWordVec is trained using the skip-gram model, an unsupervised learning method which uses probability to determine words most related to a given word. It generates 200-dimensional word embeddings with a window size of 30 (the 30 words preceding and following the target word and considered as context), a sampling threshold of 0.001, which controls how much subsampling occurs, and ten negative examples that are randomly selected as negative words, which should not be predicted as context and which are used to update the weights. These values were used by the creator of BioWordVec [16]. We used both the domain-specific BioWordVec word embeddings and word2vec from Google News and found that BioWordVec significantly outperformed word2vec for our application.
For our RCNN model, each input publication title was converted into a numeric feature vector for classification using BioWordVec embeddings [16]. For the logistic regression and SVM models, we use BioSentVec [21], which converts a sentence into a biomedical sentence embedding more efficiently than using BioWordVec embeddings. BioSentVec [21] is generated by applying sent2vec [30] to large-scale biological and clinical texts from corpora obtained using the PubMed search engine and clinical notes from the MIMIC-III clinical database [29]; therefore, the sentence embeddings are domain specific.
2.2.3. Model Selection and Training
Our ensemble learning method is a combination of traditional machine learning models and a deep learning (neural network) model, the latter of which has become very popular in recent years. The training data for each model are publication titles; the output of each model is a binary class (positive or negative). Titles classified as positive are considered to contain experimental evidence of protein function for a given protein annotation.
For our traditional machine learning models, we use the discriminative classifiers, logistic regression and SVM with a linear kernel. Logistic regression can be used for high-dimensional inputs, and it is also fast and easy to fit the data. We improve the model generalization and reduce overfitting by adding regularization to penalize the sum of the magnitudes of the model weights. We used both positive and negative validation data (9398 publications, half positive and half negative) for parameter selection. We trained the models using different values of the parameter for regularization in order to find the value with the best performance for our validation set. After validation was performed, we selected 100 as the parameter for regularization based on its validation performance. Because our data are high-dimensional and features are individually informative, the decision boundary can likely be represented as a linear combination of the original features. Therefore, we adopted the linear kernel for the SVM model. Based on validation, we set the parameter for regularization to 10 for our SVM model to make the loss greater for data violating the margin.
Prior to determining that a single model was insufficient and that an ensemble model was needed to obtain the level of accuracy (recall) we desired, we considered using only an RCNN model for our NLP problem. This was because deep learning models have enjoyed some success for such problems, particularly when used in conjunction with word embeddings [20,31]. With word embedding, to more precisely capture the meaning of a word it is represented by the concatenation of its left context vector, its word embedding, and its right context vector using a bidirectional recurrent structure. The left context is obtained using a forward scan and the right context using a backward scan. A linear transformation of the resulting numeric vector is passed on to the next layer. Once word representations have been calculated, we apply max pooling to convert texts of different lengths into numeric vectors of fixed length. This approach allows extraction of the words that capture the most meaning for different input sequences. Finally, a softmax function, which normalizes the possible outcomes into a probability distribution in the final layer, calculates the probability needed to determine the classification, i.e., whether or not a publication provides experimental confirmation of protein function.
The vanishing gradient problem occurs when the gradients, i.e., the values used to update an RNN’s or RCNN’s weights, become too small because of backpropagation through the layers, and learning of longer text does not occur. To avoid this problem, we apply a long short-term memory (LSTM) unit with a gate mechanism, which essentially learns to keep relevant data and to forget non-relevant data. During training and after parameter tuning using our validation data, we set one hidden layer to a size of 150, the learning rate of the stochastic gradient descent to 0.01, the vector size of word embeddings to 200, and the size of the context vector to 150. In addition, we set the regularization parameter to 0.5, the batch size (the amount of training data used in one iteration) to 32, and the dropout-keep probability (probability of keeping units in a layer as well as their connections in the RCNN) to 0.7 to avoid overfitting as suggested by [20] and by related literature on neural networks.
2.3. Classification Workflow Phase
For our project, we use the literature available in the entire UniProtKB database to determine whether experimental evidence of protein function exists. The UniProtKB/Swiss-Prot database currently lists about 560,000 manually curated protein sequences. Many of these sequences have publications tagged in the Function category, which ensures that experimental evidence of their function exists, and these do not require use of our smart program. On the other hand, the UniProtKB/TrEMBL database has more than 200 million unreviewed entries. This unreviewed database clearly contains the majority of sequences with publications to use with our smart program. In addition, while it is not the objective of our project, it also seems clear that our smart program could be of use in the curation of unreviewed sequences in the UnitProtKB database.
We periodically query the UniProtKB database for protein sequence accession numbers using the UniProt Java application program interface. For each query, we consider only entries with two or more associated journal publications; this is a proxy indicating that sufficient research exists for a protein. We then extract the publication titles for the queried sequences and perform the same pre-processing steps used during the training phase, including tokenization, conversion of uppercase characters to lowercase, removal of punctuation and non-alphabetic characters, and stop-word filtering. For the logistic regression and SVM with linear kernel models, we convert the titles into 700-dimensional biomedical sentence embeddings using BioSentVec [21]. For the RCNN, we use BioWordVec embeddings [16] to convert each word in the title into a numeric feature vector and incorporate context information into the bidirectional recurrent structure. Again, as with our training phase, after linear transformation and max pooling operations have been performed, titles of different lengths are converted into a numeric vector of a fixed length, and this vector is used for classification by the softmax layer.
During ensemble learning, each trained model classifies the input title independently, and the final decision on whether the query protein sequence has experimental confirmation or not is determined by a majority vote.
If at least one publication associated with a protein entry in UniProtKB is classified as positive, the publication title and the protein accession number are submitted to a human curator for manual review. All publications classified as negative are ignored. After manual confirmation, the protein annotation is marked by the curator as having been verified.
3. Results and Discussion
All data, both positive and negative examples, discussed in this paper were procured from the UniProtKB/Swiss-Prot database as of 10 April 2019. The dataset was randomly shuffled and divided into training (60%), validation (20%), and test (20%) sets. These consisted of 28202, 9398, and 9398 publications, respectively, with positive and negative examples equal in number. As mentioned previously, we tried to include word stemming in our pre-processing step, but non-stemmed words performed better with our pre-trained word embedding model. Thus, all results are based on the use of non-stemmed words. To test the effectiveness of our approach, we compared its performance with the state-of-the-art BioBERT model fine-tuned appropriately.
To evaluate the performance of our model, we used four metrics: accuracy, precision, recall, and the F1 score, as calculated using the following formulas:
where TP (True Positive) correctly indicates publications have experimental evidence of protein function, TN (True Negative) correctly indicates publications have no experimental evidence of protein function, FP (False Positive) incorrectly indicates publications have experimental evidence of protein function, and FN (False Negative) incorrectly indicates publications have no experimental confirmation of protein function.
Of the four evaluation metrics, recall is the most significant for our application. This is because our goal is to identify as many protein annotations as possible with experimental evidence. Thus, we would rather check publications that may not have experimental verification than to miss any possibilities of those that do. Because all results are sent to a human curator for manual review and curation after each periodic query has been performed, 100% precision can be guaranteed in the end. However, we did not ignore the other metrics. We wanted to achieve an overall balance, and in particular, we ensured a good F1 score, which is the harmonic mean of precision and recall. We did not want to completely sacrifice precision for recall. We could have, in fact, striven for a recall of one, but this would result in wasted time on the part of the curator.
As shown in Table 1, the logistic regression (threshold = 0.5) and SVM models give similar results. This might be because the two classification categories are not well separated. The RCNN model outperforms both the logistic regression (threshold = 0.5) and SVM models by more than 10% for recall. This is most likely because the RCNN model is better at including contextual information and keywords. Recall that feature representation for the traditional logistic regression and SVM models differs from feature representation for the deep-learning RCNN model. For the traditional methods, conversion of a complete sentence into a numeric vector is accomplished using the BioSentVec model while for the deep learning model, key words are better captured by max pooling after we apply a non-linear activation function and a linear transformation on the word representation.

Table 1.
Performance of logistic regression (threshold = 0.5), logistic regression (threshold = 0.25), support vector machine (SVM), recurrent convolutional neural network (RCNN), ensemble learning, and BioBERT models using test data (9398 peer-reviewed journal publications).
While the RCNN model achieves higher recall than the logistic regression (threshold = 0.5) and SVM models, the accuracy and precision metrics are not as good. Because recall is our most important metric, we further relaxed our logistic regression model with the use of a threshold of 0.25 and after cross-validation achieved an F1 score of 70% and a recall of 94.64%. With ensemble learning and for our objective, it is reasonable to use a weaker classifier with relatively low precision but high recall. In fact, when the three models are combined to create our ensemble learning model, the metrics generally improve. Importantly, our model achieves a recall of 91.25%. In addition, the F1 score of 76.05% is a marked improvement. Ensemble learning models work well when the error for the individual models is uncorrelated. Given that logistic regression, SVM, and RCNN models differ substantially, it is not surprising that combining them provides considerable improvement.
For comparison, we fine-tuned BioBERT on the same training set for the same classification task. Based on the performance of the validation set, we stopped at six epochs. We found that BioBERT performs better in terms of accuracy and precision mainly because of its deep bidirectionality, which masks random input words and conditions each word bidirectionally to predict the masked words. However, its recall value is much lower than that of our ensemble learning model, and its F1 score is lower as well. In addition, fine-tuning and inference were very slow due to BERT’s huge parameter size.
Table 2 presents results obtained using our ensemble learning model for a number of candidate protein annotations. The entry identifiers are unique and stable accession numbers in the UniProtKB database, which can be easily cross-referenced with accession numbers in the NCBI database. With the first two examples, we demonstrate that our ensemble learning model can identify peer-reviewed biomedical publications indicating experimental evidence of functional annotation for a target protein. We used our smart program with the UniProtKB/Swiss-Prot publications associated with glucarate dehydratase and methylaspartate ammonia-lyase, which are among a “gold standard” set of known enzyme superfamilies rigorously studied by domain experts [32]. Our model successfully identified glucarate dehydratase (entry identifier P0AES2) and methylaspartate ammonia-lyase (entry identifier Q05514) as correct annotations based on the available publications. More specifically, of the six publications associated with glucarate dehydratase, our model identified “Evolution of enzymatic activities in the enolase superfamily: characterization of the (D)-glucarate/galactarate catabolic pathway in Escherichia coli” and “Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli” as positive. Our classification results match exactly with UniProtKB/Swiss-Prot, which classifies only these two publications in the Function category. Of the eight publications associated with methylaspartate ammonia-lyase in UniProtKB/Swiss-Prot, four of them, “The purification and properties of beta-methylaspartase”, “Alteration of the diastereoselectivity of 3-methylaspartate ammonia lyase by using structure-based mutagenesis”, “The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step”, and “Engineering methylaspartate ammonia lyase for the asymmetric synthesis of unnatural amino acids”, were correctly classified as positive by our program. Our model only misclassified “Cloning, sequencing, and expression in Escherichia coli of the Clostridium tetanomorphum gene encoding beta-methylaspartase and characterization of the recombinant protein” as negative. Overall, however, our smart program identified this protein sequence as having the correct functional annotation.

Table 2.
Results using our ensemble learning model for several different protein annotations.
With the remaining examples, we show that our smart program can also be applied to protein sequences from UniProtKB/Swiss-Prot without publications tagged in the Function category as well as to protein sequences in the unreviewed UniProtKB/TrEMBL database. For the UniProtKB/Swiss-Prot protein sequences 60S ribosomal protein L5-2 (entry identifier Q8L4L4) and Protein PsiE homolog (entry identifier Q81XB6), our model correctly classified both as negative and all their associated publications negative as well. For example, proteins from UniProtKB/TrEMBL, our model identified MEOX2 (entry identifier Q6FHY5) as positive by predicting 18 of 20 publications as positive. The exceptions were “Cloning of human full open reading frames in Gateway(TM) system entry vector (pDONR201)” and “Two candidate tumor suppressor genes, MEOX2 and SOSTDC1, identified in a 7p21 homozygous deletion region in a Wilms tumor”. Some publications in UniProtKB/TrEMBL are assigned to categories, but they are not manually reviewed; only the two publications listed above were assigned to the Sequences category, and our program did not think they contain function-related experimental evidence. Our model identified Beta catenin 1 (entry identifier B1MV73) as positive because it predicted both publications “Equine CTNNB1 and PECAM1 nucleotide structure and expression analyses in an experimental model of normal and pathological wound repair” and “Genome sequence, comparative analysis, and population genetics of the domestic horse” as positive although both these publications are tagged in the Sequences category. However, our human curator determined this to be a false positive case because the latter paper is a genome paper and the former paper looks at the expression of the gene and how it is regulated during wound repair rather than focusing on the target protein. The involvement of a human curator guarantees the accuracy of our final result after prediction by our smart program. Our model identified Maillard deglycase (entry identifier K7G4K8) as negative because it predicted both publications “The complete mitochondrial genome of the Korean soft-shelled turtle Pelodiscus sinensis (Testudines, Trionychidae)” and “The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan” as negative. Our model identified Catenin (Cadherin-associated protein), beta 1, 88 kDa (entry identifier H2R2U1) as negative because it predicted both publications “Initial sequence of the chimpanzee genome and comparison with the human genome” and “De novo assembly of the reference chimpanzee transcriptome from NextGen mRNA sequences” as negative. Overall, our model correctly identified publications containing experimental evidence of protein function regardless of the publication category. As such, it can be used to identify sequences with experimental evidence of protein function. Publications associated with any annotations that have been predicted as positive are sent to a human curator for final review.
Our ensemble learning approach is able to effectively identify publications that are likely to provide experimental evidence of correct protein annotation and a partial solution to the protein annotation problem.
4. Conclusions and Future Work
In this paper, we presented a novel smart program to confirm protein annotations. Our smart program uses ensemble learning together with word embedding to effectively classify whether the title of a journal publication provides the information needed to show that experimental evidence of protein function for a given protein annotation is presented in the publication. Our ensemble learning method achieves 71.26% accuracy, 91.25% recall, 65.19% precision, and an F1 score of 76.05%. When a publication is classified as positive, its title together with the accession number for the protein entry will be sent to a human curator for manual inspection. This ensures a precision of 100%. After the curator has confirmed that the publication or publications do verify protein function experimentally, they indicate that the protein annotation has been confirmed.
We have described our smart program for verifying protein annotation. It will be part of a system we are currently developing, which will have a query-able website as a front end to a query-able database. The database will consist of homologous clusters of protein sequences obtained using the pClust software [5]. pClust will be used to simultaneously cluster proteomes deduced from the genomes of bacteria, archaea, eukarya, and viruses. The website will show the cluster most closely matching the protein sequence query, and each sequence in the cluster will include the FASTA header for the sequence, which gives the accession number, annotation, and organism. Protein sequences that have been marked by the human curator as having been verified will be identified by colored FASTA text. As discussed previously, the smart program will automatically query the UniProtKB database periodically so that annotations are updated on a regular basis. Because homology does not guarantee the same molecular function, users are free to choose an annotation based on their understanding of the relationship of their organism to those in the cluster or else to use a program we will provide that will create a sequence similarity network (SSN) of the protein sequences they have chosen (including any that have been verified). The SSN will indicate the sequences most closely related to their query sequence. A sequence that has been confirmed experimentally will appear in color in the SSN, and this will allow a user to make an informed decision about whether or not to use the annotation for their sequence.
Author Contributions
J.T. collected the data, pre-processed them, trained the models, designed and performed the experiments, analyzed the results, and prepared the initial manuscript. K.A.B. and S.L.B. analyzed the collected data, approved the method, guided the experiments, edited the manuscript, and further interpreted the experimental results. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Carl M. Hansen Foundation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Our training data, validation data, test data, and trained models as well as code presented in this study are openly available at https://github.com/taojin1992/Automated-confirmation-of-protein-function-annotation-using-NLP.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Galperin, M.Y.; Koonin, E.V. Sources of systematic error in functional annotation of genomes: Domain rearrangement, non-orthologous gene displacement and operon disruption. Silico Biol. 1998, 1, 55–67. [Google Scholar]
- Gilks, W.R.; Audit, B.; de Angelis, D.; Tsoka, S.; Ouzounis, C.A. Percolation of annotation errors through hierarchically structured protein sequence databases. Math. Biosci. 2005, 193, 223–234. [Google Scholar] [CrossRef]
- Schnoes, A.M.; Brown, S.D.; Dodevski, I.; Babbitt, P.C. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009, 5, e1000605. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, S.L. Next-generation genome annotation: We still struggle to get it right. Genome Biol. 2019, 20, 92. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, S.; Brayton, K.A.; Daily, J.A.; Broschat, S.L. Whole Proteome Clustering of 2,307 Proteobacterial Genomes Reveals Conserved Proteins and Significant Annotation Issues. Front. Microbiol. 2019, 10, 383. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2005, 33, D34–D38. [Google Scholar] [CrossRef] [PubMed]
- Cozzetto, D.; Jones, D.T. Computational methods for annotation transfers from sequence. In The Gene Ontology Handbook; Humana Press: New York, NY, USA, 2017; pp. 55–67. [Google Scholar]
- Lim, J.H.; Lee, K.C. Classifying Biomedical Literature Providing Protein Function Evidence. ETRI J. 2015, 37, 813–823. [Google Scholar] [CrossRef]
- Harris, Z.S. Distributional structure. Word 1954, 10, 146–162. [Google Scholar] [CrossRef]
- Le, Q.; Mikolov, T. Distributed representations of sentences and documents. In International Conference on Machine Learning; JMLR W&CP: Beijing, China, 2014; pp. 1188–1196. [Google Scholar]
- Nastase, V.; Sayyad-Shirabad, J.; Sokolova, M.; Szpakowicz, S. Learning noun-modifier semantic relations with corpus-based and WordNet-based features. In Proceedings of the AAAI, Boston, MA, USA, 16–20 July 2006; pp. 781–787. [Google Scholar]
- Plank, B.; Moschitti, A. Embedding semantic similarity in tree kernels for domain adaptation of relation extraction. In Proceedings of the 51st Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Sofia, Bulgaria, 4–9 August 2013; Volume 1, pp. 1498–1507. [Google Scholar]
- Fang, A.C.; Cao, J. Enhanced genre classification through linguistically fine-grained pos tags. In Proceedings of the 24th Pacific Asia Conference on Language, Information and Computation, Miyagi, Japan, 4–7 November 2010; pp. 85–94. [Google Scholar]
- Moschitti, A. Making tree kernels practical for natural language learning. In Proceedings of the 11th Conference of the European Chapter of the Association for Computational Linguistics, Trento, Italy, 3–7 April 2006. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. Adv. Neural Inf. Process. Syst. 2013, 26, 3111–3119. [Google Scholar]
- Zhang, Y.; Chen, Q.; Yang, Z.; Lin, H.; Lu, Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci. Data 2019, 6, 1–9. [Google Scholar] [CrossRef]
- Bojanowski, P.; Grave, E.; Joulin, A.; Mikolov, T. Enriching word vectors with subword information. Trans. Assoc. Comput. Linguist. 2017, 5, 135–146. [Google Scholar] [CrossRef]
- Mikolov, T.; Karafiát, M.; Burget, L.; Černockỳ, J.; Khudanpur, S. Recurrent neural network based language model. In Proceedings of the Eleventh Annual Conference of the International Speech Communication Association, Chiba, Japan, 26–30 September 2010. [Google Scholar]
- Kim, Y. Convolutional neural networks for sentence classification. arXiv 2014, arXiv:1408.5882. [Google Scholar]
- Lai, S.; Xu, L.; Liu, K.; Zhao, J. Recurrent convolutional neural networks for text classification. In Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015. [Google Scholar]
- Chen, Q.; Peng, Y.; Lu, Z. BioSentVec: Creating sentence embeddings for biomedical texts. In Proceedings of the 2019 IEEE International Conference on Healthcare Informatics (ICHI), Xi’an, China, 10–13 June 2019; pp. 1–5. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019. [Google Scholar] [CrossRef]
- Dietterich, T.G. Ensemble learning. Handb. Brain Theory Neural Netw. 2002, 2, 110–125. [Google Scholar]
- Consortium, U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Loper, E.; Bird, S. NLTK: The natural language toolkit. arXiv 2002, arXiv:cs/0205028. [Google Scholar]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Li-wei, H.L.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Pagliardini, M.; Gupta, P.; Jaggi, M. Unsupervised learning of sentence embeddings using compositional n-gram features. arXiv 2017, arXiv:1703.02507. [Google Scholar]
- Young, T.; Hazarika, D.; Poria, S.; Cambria, E. Recent trends in deep learning based natural language processing. IEEE Comput. Intell. Mag. 2018, 13, 55–75. [Google Scholar] [CrossRef]
- Brown, S.D.; Gerlt, J.A.; Seffernick, J.L.; Babbitt, P.C. A gold standard set of mechanistically diverse enzyme superfamilies. Genome Biol. 2006, 7, R8. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).