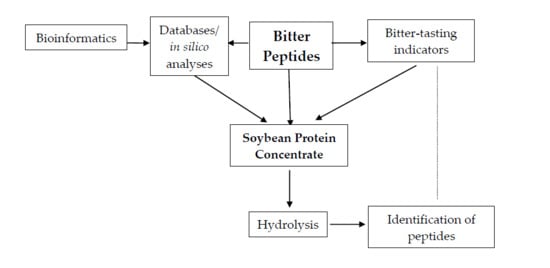
Soybean (Glycine max) Protein Hydrolysates as Sources of Peptide Bitter-Tasting Indicators: An Analysis Based on Hybrid and Fragmentomic Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Soybean Protein Sequences and Computer Simulation of Their Hydrolysis
2.2. Bitter-Tasting Peptide Indicators
2.3. Materials and Reagents
2.4. Hydrolysis of SPC
2.5. RP-HPLC for Monitoring the Process of SPC Hydrolysis and RP-HPLC-MS/MS for the Identification of SPC-Originating Peptides
3. Results
3.1. In Silico Analysis
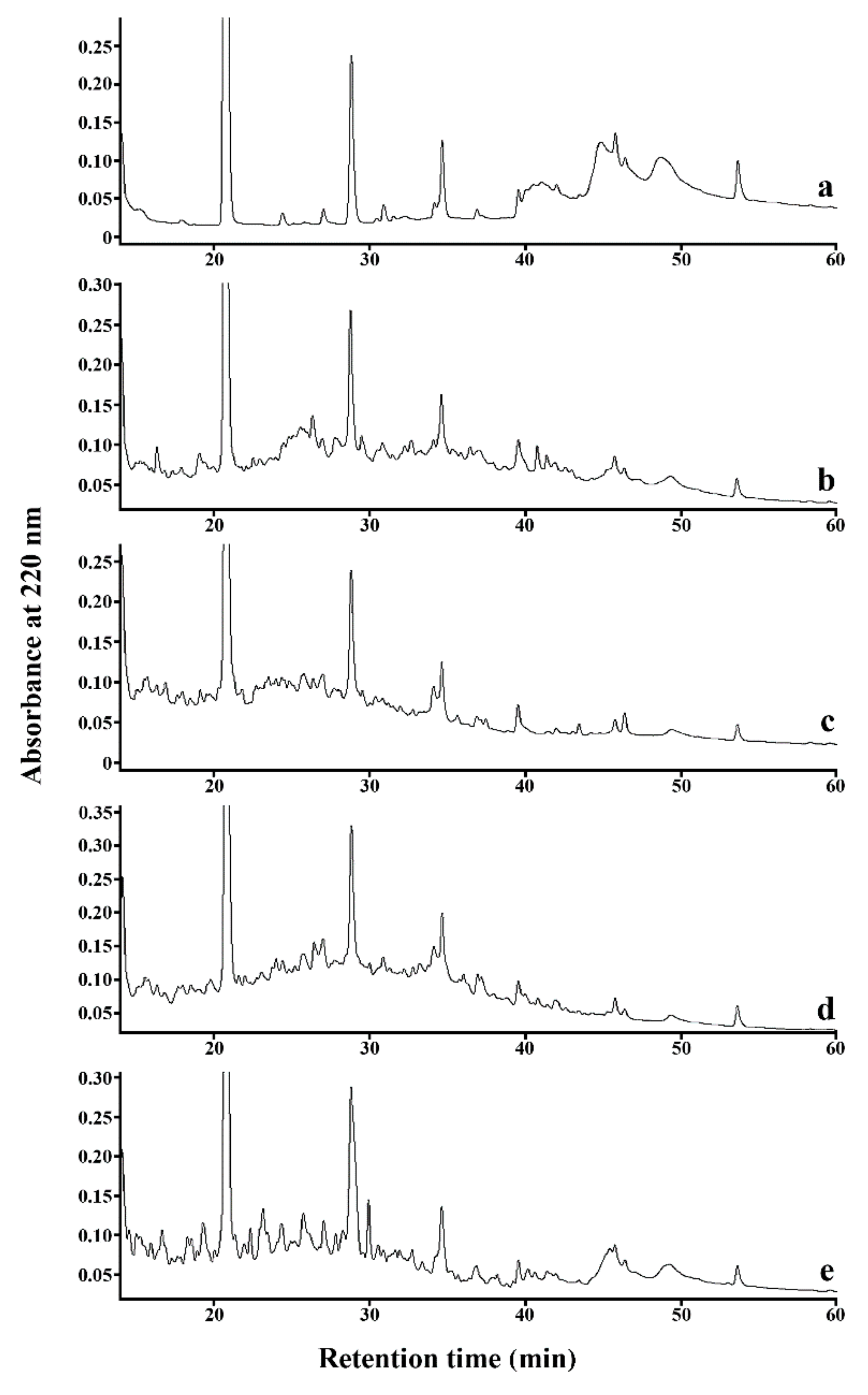
3.2. Monitoring the Process of SPC Hydrolysis
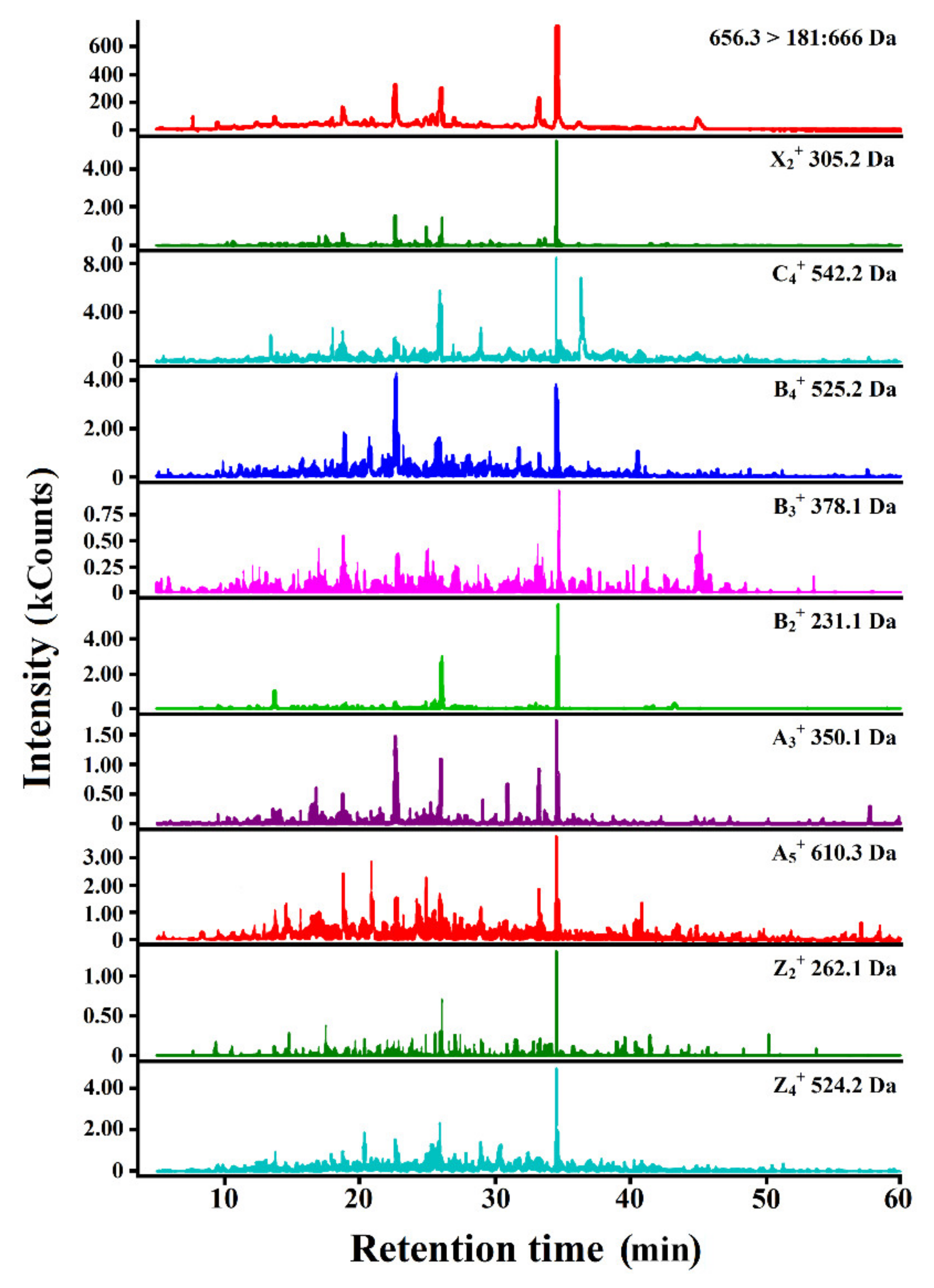
3.3. Identification of Peptides Likely to Be Bitter Derived from SPC Hydrolysates
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 0-SPC | Soybean protein concentrate before hydrolysis |
| A | Depending on the context: one-letter symbol alanine (amino acid) or A-type fragment (daughter) ion according to Roepstorff and Fohlman [25] |
| ACE | Angiotensin-converting enzyme (EC 3.4.15.1) |
| ACN | Acetonitrile |
| B | Depending on the context: bromelain (EC 3.4.22.32) or B-type fragment (daughter) ion according to Roepstorff and Fohlman [25] |
| BIOPEP-UWM | Depending on the context: database of bioactive peptide sequences [12] or database of sensory peptides and amino acids [20] |
| Bis-Tris | 2,2-bis(hydroxymethyl)-2,2′,2″-nitrilotriethanol |
| B-SPC | Soybean protein concentrate hydrolyzed by bromelain |
| C | Depending on the context: parameter to compare susceptibility of proteins to proteolytic enzymes on the basis of chromatograms (calculated using Equation (1)) or C-type fragment (daughter) ion according to Roepstorff and Fohlman [25] |
| DPP III | Dipeptidyl peptidase III (EC 3.4.14.4) |
| DPP IV | Dipeptidyl peptidase IV (EC 3.4.14.5) |
| F | Depending on the context: ficin (EC 3.4.22.3) or one-letter symbol of amino acid phenylalanine in peptide sequences |
| F-SPC | Soybean protein concentrate hydrolyzed by ficin |
| HMGR | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase) (EC 1.1.1.34) |
| HPLC | High performance liquid chromatography |
| MLR | Multivariate linear stepwise regression |
| MPC | Milk protein concentrate |
| P | Depending on the context: papain (EC 3.4.22.2) or one-letter symbol of amino acid proline in peptide sequences |
| P-SPC | Soybean protein concentrate hydrolyzed by papain |
| PK | Depending on the context: proteinase K (EC 3.4.21.64) or dipeptide prolyl-lysine annotated using one-letter code |
| PK-SPC | Soybean protein concentrate hydrolyzed by proteinase K |
| Q-value | Average free energy for the transfer of the amino acid chains from ethanol to water [65] |
| RP-HPLC | Reversed-phase high performance liquid chromatography |
| RP-HPLC-MS/MS | Reversed-phase high performance liquid chromatography online with tandem mass spectrometry |
| S | Depending on the context: symbol of relative area of peaks in defined data interval (in Equation (1)) or one-letter symbol of amino acid serine in peptide sequences |
| SPC | Soybean protein concentrate |
| SSRCalc | Sequence-Specific Retention Calculator |
| TFA | Trifluoroacetic acid |
| tR | Retention time (e.g., in Equation (2)) |
| UWM | University of Warmia and Mazury in Olsztyn |
| Y | Depending on the context: one-letter symbol of amino acid tyrosine in peptide sequences or Y-type fragment (daughter) ion according to Roepstorff and Fohlman [25] |
Appendix A
| Parent peptides released from profilin due to the simulated action of: | |||
|---|---|---|---|
| B1 | F | P | PK |
| VDDHLLCDIEG/VD, LL, EG IIG/II, IG QSTDFPQFK/FP PEEITA/EI MVIQG/VI VIRG/VI, RG LIIG/II, IG QCNMVVERPG/RP, RPG, VE LIDQG/LI | CDIEG/EG IIG/II, IG QSTDFPQFK/FP PEEITA/EI ERPG/RP, RPG | VDDH/VD LLCDIE/LL QSTDFPQFK/FP MVIQG/VI LIIG/II, IG, LI QCNMVVE/VE LIDQG/LI | EGNHL/EG QGEP/GE RGKKGP/RG, GP DQGY/GY |
| Parent peptides released from globulin 7S due to the simulated action of: | |||
| B | F | P | PK |
| PINLVVLPVQNDG/VL, LV RTPLMQVPVLVDLNG/VL, VD, LV, DL PFCHSTQCSRA/PF SRPG/RP, RPG EDVLA/VL STQQLG/LG PLVTVPQFLFSCA/FL, LF, LV PSFLVQK/FL, LV LPRNTQG/PR SHFG/FG IIFG/FG, IF, II PNNMRQFQNQDIFHDLA/IF, DL NVRVNSIRINQHSVFPLNK/FP, VF ISSTIVG/IV TMISTSTPHMVLQQSVY/VY, VL FTQVFA/VF QQLPK/PK PSVDLVMDK/VD, LV, DL EDLMVQA/DL VTCLG/LG MQPRA/PR EITLG/LG, EI RQLEENLVVFDLA/VF, LE, LV, DL, EEN DLFNFA/LF, DL | PFCHSTQCSRA/PF SRPG/RP, RPG PQFL/FL PSFL/FL SHFG/FG IIFG/FG, IF, II QDIFHDL/IF, DL ISSTIV/IV MQPRA/PR EITL/EI | PINLVVLPVQNDG/VL, LV TPLMQVPVLVDLNG/VL, VD, LV, DL DVLA/VL STQQLG/LG PLVTVPQFLFSCA/FL, LF, LV PSFLVQK/FL, LV IIFG/FG, IF, II QFQNQDIFH/IF SVFPLNK/FP, VF ISSTIVG/IV MVLQQSVY/VY, VL FTQVFA/VF QQLPK/PK PSVDLVMDK/VD, LV, DL DLMVQA/DL VTCLG/LG MQPR/PR ITLG/LG NLVVFDLA/VF, LV, DL DLFNFA/LF, DL | QNDGSTGL/GL AASRP/RP GCHKNTCGL/GL TQQTGL/GL GEDV/GE QKGL/GL RNTQGV/GV GDAP/DA QGEY/GE, EY SGEDL/GE, DL RAEI/EI EENL/EEN HSHGV/GV KCADL/AD, DL |
| Parent peptides released from glycinin due to the simulated action of: | |||
| B | F | P | PK |
| LEPDHRVESEG/VE, LE, EG LIETWNSQHPELQCA/LI, EL PQMIIVVQG/IV, II, VV PQQQSSRRG/RR, RG IRHFNEG/EG DVLVIPLG/VL, VI, LG, LV DEPVVA/VV ISPLDTSNFNNQLDQNPRVFY/PR, VF, LD QHRQQEEEG/EG SVLSG/VL HFLA/FL QIVTVEG/VE, IV, EG LSVISPK/PK, VI WQEQEDEDEDEDEEY/EY PPRRPSHG/RR, PP, RP, PR HEDDEDEDEEEDQPRPDHPPQRPSRPEQQEPRG/PP, RG, RP, RP, RP, PR, PR VEENICTMK/VE, EEN RPSRA/RP LRQFG/FG VVLY/VV, VL NSVTMTRG/RG RVRVVNCQG/VV VFDG/VF ELRRG/RR, RG, EL QLLVVPQNPA/LL, LV, VV VVFK/VV, VF DVFRVIPSEVLSNSY/VF, VL, VI PLVNP/LV | ESEG/EG SQHPEL/EL PQMIIV/II, IV PQQQSSRRG/RR, RG QHRQQEEEG/EG ISPK/PK WQEQEDEDEDEDEEY/EY PPRRPSHG/RR, PP, RP, PR HEDDEDEDEEEDQPRPDHPPQRPSRPEQQEPRG/PP, RG, RP, RP, RP, PR, PR RPSRA/RP RQFG/FG TMTRG/RG | PQMIIVVQG/IV, II, VV DVLVIPLG/VL, VI, LG, LV PVVA/VV ISPLDTSNFNNQLDQNPR/PR, LD SVLSG/VL QIVTVE/VE, IV LSVISPK/PK, VI DQPR/PR PPQR/PP VVLY/VV, VL VVNCQG/VV VFDG/VF QLLVVPQNPA/LL, LV, VV VVFK/VF, VV DVFR/VF VIPSE/VI VLSNSY/VL PLVNP/LV | TSSKF/KF ESEGGL/EGG, GL, GGL, EG QCAGV/GV QQQSSRRGSRSQQQL/RR, RG NEGDV/EG QQQQQQKSHGGRKQGQHRQQEEEGGSV/GR, EGG, EG EGGL/EGG, EG, GGL, GL QEQEDEDEDEDEEY/EY GRTP/GR RGRGCQTRNGV/RG, RG, GR, GV SRADF/AD KAGRI/GR TRGKGRV/RG, GR DGEL/GE, EL RRGQL/RR, RG AEQGGEQGL/GE, GL QGNSGP/GP |
| Parent peptides released from β-conglycinin (β-chain) due to the simulated action of: | |||
| B | F | P | PK |
| VREDENNPFY/PF FRSSNSFQTLFENQNVRIRLLQRFNK/RF, LF, LL, RSPQLENLRDY/LE RIVQFQSK/IV PNTILLPHHA/LL, IL DFLLFVLSG/FV, FL, LF, VL, LL ILTLVNNDDRDSY/LV, IL LVNPHDHQNLK/LV DDFFLSSTQA/FF, FL FSHNILETSFHSEFEEINRVLFG/FG, LF, VL, LE, IL, EI, EF EEEEQRQQEG/EG VIVELSK/VI, VE, IV, EL EQIRQLSRRA/RR TISSEDEPFNLRSRNPIY/PF SNNFG/FG FFEITPEK/FF, EI NPQLRDLDIFLSSVDINEG/FL, IF, VD, LD, DL, EG LLLPHFNSK/LL, LL, LLL IVILVINEG/VI, VI, IV, LV, IL, EG NIELVG/LV, EL, VG QEEEPLEVQRY/LE ELSEDDVFVIPA/FV, VF, VI, EL PFVVNA/FV, PF, VV TSNLNFLA/FL ENNQRNFLA/FL DNVVRQIERQVQELA/EL, VV QDVERLLK/VE, LL FVDA/FV, VD, DA PFPSILG/FP, PF, PFP, LG, IL LY | QRFN/RF DDFFL/FF, FL ETSFHSEFEEIN/EI, EF EEEEQRQQEG/EG SRRA/RR TISSEDEPFN/PF FFEITPEK/FF, EI DIFL/FL, IF PFPSIL/FP, PF, PFP, IL | NNPFY/PF SSNSFQTLFE/LF LLQR/LL SPQLE/LE IVQFQSK/IV PNTILLPH/LL, IL DFLLFVLSG/FV, FL, LF, VL, LL ILTLVNNDDR/LV, IL LVNPH/LV DDFFLSSTQA/FF, FL NILE/LE, IL VLFG/VL, FG, LF VIVE/VI, VE, IV PFNLR/PF SNNFG/FG DLDIFLSSVDINE/FL, IF, VD, LD, DL LLLPH/LL, LL, LLL IVILVINE/VI, VI, IV, LV, IL DDVFVIPA/FV, VF, VI PFVVNA/FV, PF, VV TSNLNFLA/FL NFLA/FL DNVVR/VV QDVE/VE FVDA/FV, VD, DA PFPSILG/FP, PF, PFP, LG, IL | QSKP/KP HHADADF/AD, AD, DA SGRAI/GR GDAQRI/DA HSEF/EF GEEEEQRQQEGV/GV, GE, EG SRRAKSSSRKTI/RR NEGAL/EG NEGDANI/DA, EG RAEL/EL AGEKDNV/GE DAQP/DA QQKEEGSKGRKGP/GR, GP, EG |
| Parent peptides released from β-conglycinin (α-chain) due to the simulated action of: | |||
| B | F | P | PK |
| EECEEG/EG EIPRPRPRPQHPEREPQQPG/RP, RP, RP, PR, PR, PR, EI EEDEDEQPRPIPFPRPQPRQEEEHEQREEQEWPRK/RP, RP, PR, PR, PR, PR, FP, PF, PFP, PIP SEEEDEDEDEEQDERQFPFPRPPHQK/PP, RP, PR, FP, FP, FPF, PF, PFP EERNEEEDEDEEQQRESEESEDSELRRHK/RR, EL NPFLFG/FG, FL, LF, PF SNRFETLFK/RF, LF RIRVLQRFNQRSPQLQNLRDY/RF, VL RILEFNSK/LE, IL, EF PNTLLLPNHA/LL, LL, LLL LIVILNG/VI, IV, LI, IL ILSLVNNDDRDSY/LV, IL VVNPDNNENLRLITLA/LI, VV RFESFFLSSTEA/FF, RF, FL FSRNILEA/LE, IL FEEINK/EI VLFSREEG/LF, VL, EG EQRLQESVIVEISK/VI, VE, IV, EI PFNLRSRDPIY/PF FFEITPEK/FF, EI NPQLRDLDIFLSIVDMNEG/FL, IF, IV, VD, LD, DL, EG LLLPHFNSK/LL, LL, LLL IVILVINEG/VI, VI, IV, LV, IL, EG NIELVG/LV, EL, VG EQQQEQQQEEQPLEVRK/LE ELSEQDIFVIPA/FV, IF, VI, EL PVVVNA/VVV, VV, VV TSNLNFFA/FF ENNQRNFLA/FL SQDNVISQIPSQVQELA/VI, EL FVDA/FV, VD, DA PLSSILRA/IL | EECEEG/EG EIPRPRPRPQHPEREPQQPG/RP, RP, RP, PR, PR, PR, EI EEDEDEQPRPIPFPRPQPRQEEEHEQREEQEWPRK/RP, RP, PR, PR, PR, PR, FP, PF, PFP, PIP SEEEDEDEDEEQDERQFPFPRPPHQK/PP, RP, PR, FP, FP, FPF, PF, PFP EEEDEDEEQQRESEESEDSEL/EL RRHK/RR RFETL/RF QRFN/RF RFESFFL/FF, RF, FL FEEIN/EI FSREEG/EG EISK/EI FFEITPEK/FF, EI DIFL/FL, IF SEQDIFV/FV, IF SSIL/IL | PIPFPR/PR, FP, PF, PFP, PIP PQPR/PR QFPFPR/PR, FP, FP, FPF, PF, PFP NPFLFG/FG, FL, LF, PF TLFK/LF VLQR/VL PNTLLLPNH/LL, LL, LLL LIVILNG/VI, IV, LI, IL ILSLVNNDDR/LV, IL VVNPDNNE/VV LITLA/LI SFFLSSTE/FF, FL NILE/LE, IL VLFSR/LF, VL SVIVE/VI, VE, IV PFNLR/PF DLDIFLSIVDMNE/FL, IF, IV, VD, LD, DL LLLPH/LL, LL, LLL IVILVINE/VI, VI, IV, LV, IL QPLE/LE QDIFVIPA/FV, IF, VI PVVVNA/VVV, VV, VV TSNLNFFA/FF NFLA/FL SQDNVISQIPSQVQE/VI FVDA/FV, VD, DA PLSSILR/IL | EKEECEEGEI/GE, EG, EI GEKEEDEDEQP/GE RKEEKRGEKGSEEEDEDEDEEQDERQF/RG, GE HQKEERNEEEDEDEEQQRESEESEDSEL/EL RRHKNKNP/RR GSNRF/RF NSKP/KP NHADADY/AD, AD, DA QSGDAL/DA DTKF/KF SREEGQQQGEQRL/GE, EG SSEDKP/KP NEGAL/EG NEGDANI/DA, EG RAEL/EL DAQP/DA KKKEEGNKGRKGP/GR, GP, EG |
References
- Chatterjee, C.; Gleddie, S.; Xiao, C.W. Soybean bioactive peptides and their functional properties. Nutrients 2018, 10, 1211. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, G.; Baroni, L. Soy, soy foods and their role in vegetarian diets. Nutrients 2018, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-derived bioactive peptides in human health: Challenges and opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, X.; Kan, J. Isolation and identification of flavor peptides from douchi (traditional Chinese soybean food). Int. J. Food Prop. 2017, 20, 1982–1994. [Google Scholar] [CrossRef]
- Iwaniak, A.; Darewicz, M.; Mogut, D.; Minkiewicz, P. Elucidation of the role of in silico methodologies in approaches to studying bioactive peptides derived from foods. J. Funct. Foods 2019, 61, 103486. [Google Scholar] [CrossRef]
- Lyndqvist, M. Flavour Improvement of Water Solutions Comprising Bitter Amino Acids. Master’s Thesis, Swedish University of Agricultural Sciences, Department of Food Science, Uppsala, Sweden, 2010. Publication No. 277. [Google Scholar]
- Zamyatnin, A.A. Fragmentomics of natural peptide structures. Biochemistry (Moscow) 2009, 74, 1575–1585. [Google Scholar] [CrossRef]
- Liu, Z.P.; Wu, L.Y.; Wang, Y.; Zhang, X.S.; Chen, L. Bridging protein local structures and protein functions. Amino Acids 2008, 35, 627–650. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Hrynkiewicz, M.; Bucholska, J.; Darewicz, M. Hybrid approach in the analysis of bovine milk protein hydrolysates as a source of peptides containing di- and tripeptide bitterness indicators. Pol. J. Food Nutr. Sci. 2020, 70, 139–150. [Google Scholar] [CrossRef]
- Ashaolu, T.J. Application of soy protein hydrolysates in the emerging functional foods: A review. Int. J. Food Sci. Technol. 2019, 55, 421–428. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucl. Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Hrynkiewicz, M.; Bucholska, J.; Minkiewicz, P.; Darewicz, M. Understanding the nature of bitter-taste di- and tripeptides derived from food proteins based on chemometric analysis. J. Food Biochem. 2019, 43, e12500. [Google Scholar] [CrossRef] [PubMed]
- Otagiri, K.; Miyake, I.; Ishibashi, N.; Fukui, H.; Kanehisa, H.; Okai, H. Studies of bitter peptides from casein hydrolyzate. II. Syntheses of bitter peptide fragments and analogs of BPIa (Arg-Gly-Pro-Pro-Phe-Ile-Val) from casein hydrolysate. Bull. Chem. Soc. Jpn. 1983, 56, 1116–1119. [Google Scholar] [CrossRef]
- Peñta-Ramos, E.A.; Xiong, Y.L. Antioxidant activity of soy protein hydrolysates in a liposomal system. J. Food Sci. 2002, 67, 2952–2956. [Google Scholar] [CrossRef]
- Lee, J. Soy Protein Hydrolysate: Solubility, Thermal Stability, Bioactivity, and Sensory Acceptability in a Tea Beverage. Master’s Thesis, University of Minnesota, Minneapolis, MN, USA, 2011; pp. 1–137. [Google Scholar]
- Moruz, L.; Käll, L. Peptide retention time prediction. Mass Spectrom. Rev. 2017, 36, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Spicer, V.; Yamchuk, A.; Cortens, J.; Sousa, S.; Ens, W.; Standing, K.G.; Wilkins, J.A.; Krokhin, O.V. Sequence-specific retention calculator. A family of peptide retention time prediction algorithms in reversed-phase HPLC: Applicability to various chromatographic conditions and columns. Anal. Chem. 2007, 79, 8762–8768. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Vegarud, G.E.; Minkiewicz, P.; Iwaniak, A. Angiotensin I-converting enzyme (ACE) inhibitory activity and ACE inhibitory peptides of salmon (Salmo salar) protein hydrolysates obtained by human and porcine gastrointestinal enzymes. Int. J. Mol. Sci. 2014, 15, 14077–14101. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Sieniawski, K.; Starowicz, P. BIOPEP database of sensory peptides and amino acids. Food Res. Int. 2016, 85, 155–161. [Google Scholar] [CrossRef]
- Visser, S.; Slangen, C.J.; Rollema, H.S. Phenotyping of bovine milk poteins by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1991, 548, 361–370. [Google Scholar] [CrossRef]
- Bucholska, J.; Minkiewicz, P. The use of peptide markers of carp and herring allergens as an example of detection of sequenced and non-sequenced proteins. Food Technol. Biotechnol. 2016, 54, 266–274. [Google Scholar] [CrossRef]
- Monaci, L.; Losito, I.; Palmisano, F.; Visconti, A. Reliable detection of milk allergens in food using a high-resolution, stand-alone mass spectrometer. J. AOAC Int. 2011, 94, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Paizs, B.; Suhai, S. Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 2005, 24, 508–548. [Google Scholar] [CrossRef] [PubMed]
- Roepstorff, P.; Fohlman, J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 1984, 11, 601. [Google Scholar] [CrossRef] [PubMed]
- Kunda, P.B.; Benavente, F.; Catalá-Clariana, S.; Giménez, E.; Barbosa, J.; Sanz-Nebot, V. Identification of bioactive petides in a functional yogurt by liquid chromatography time-of-flight mass spectrometry assisted by retention time prediction. J. Chromatogr. A 2012, 1229, 121–128. [Google Scholar] [CrossRef]
- Murray, N.M.; O’Riordan, D.; Jacquier, J.-C.; O’Sullivan, M.; Holton, A.; Wyne, K.; Robinson, R.C.; Barile, D.; Nielsen, S.D.; Dallas, D.C. Peptidomic screening of bitter and nonbitter casein hydrolysate fractions for insulinogenic peptides. J. Dairy Sci. 2018, 101, 2826–2837. [Google Scholar] [CrossRef]
- Fan, J.; Saito, M.; Tatsumi, E.; Li, L. Preparation of angiotensin I-converting enzyme inhibiting peptides from soybean protein by enzymatic hydrolysis. Food Sci. Technol. Res. 2003, 9, 254–256. [Google Scholar] [CrossRef][Green Version]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Hrynkiewicz, M. Food protein-originating peptides as tastants—Physiological, technological, sensory, and bioinformatic approaches. Food Res. Int. 2016, 89, 27–38. [Google Scholar] [CrossRef]
- Matsui, T.; Zhu, X.L.; Shiraishi, K.; Ueki, T.; Noda, Y.; Matsumoto, K. Antihypertensive effect of salt-free soy sauce, a new fermented seasoning, in spontaneously hypertensive rats. J. Food Sci. 2010, 75, H129–H134. [Google Scholar] [CrossRef]
- Wang, W.; Dia, V.P.; Vasconez, M.; de Mejia, E.G.; Nelson, R.L. Analysis of soybean protein-derived peptides and the effect of cultivar, environmental conditions, and processing on lunasin concentration in soybean and soy products. J. AOAC Int. 2008, 91, 936–994. [Google Scholar] [CrossRef]
- Zhong, F.; Liu, J.; Ma, J.; Shoemaker, C.F. Preparation of hypocholesterol peptides from soy protein and their hypocholesterolemic effect in mice. Food Res. Int. 2007, 40, 661–667. [Google Scholar] [CrossRef]
- De Mejia, E.G.; de Lumen, B.O. Soybean bioactive peptides: A new horizon in preventing chronic diseases. Sex. Reprod. Menop. 2006, 4, 91–95. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-originating ACE inhibitors, including antihypertensive peptides, as preventive food components in blood pressure reduction. Comp. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- Liu, R.; Cheng, J.; Wu, H. Discovery of food-derived dipeptidyl peptidase IV inhibitory peptides: A review. Int. J. Mol. Sci. 2019, 20, 463. [Google Scholar] [CrossRef] [PubMed]
- Khaket, T.P.; Redhu, D.; Dhanda, S.; Singh, J. In Silico evaluation of potential DPP-III inhibitor precursors from dietary proteins. Int. J. Food Prop. 2015, 18, 499–507. [Google Scholar] [CrossRef]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC Trends Analyt. Chem. 2018, 105, 7–17. [Google Scholar] [CrossRef]
- Udenigwe, C.C. Bioinformatic approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014, 36, 137–143. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, W.; Zhu, M.; Xiao, Z. In silico assessment of the potential of patatin as a precursor of bioactive peptides. J. Food Biochem. 2016, 40, 366–370. [Google Scholar] [CrossRef]
- Agyei, D.; Bambarandage, E.; Udenigwe, C. The role of bioinformatics in the discovery of bioactive peptides. In Encyclopedia of Food Chemistry, 2nd ed.; Melton, L., Shahidi, F., Varelis, P., Eds.; Elsevier Inc.: Cambridge, MA, USA, 2018; Volume 2, pp. 337–344. [Google Scholar] [CrossRef]
- Sutopo, C.C.Y.; Sutrisno, A.; Wang, L.-F.; Hsu, J.-L. Identification of a potent angiotensin-I converting enzyme inhibitory peptide from black cumin seed hydrolysate using orthogonal bioassay-guided fractionations coupled with in silico screening. Process Biochem. 2020, in press. [Google Scholar] [CrossRef]
- Alcaide-Hidalgo, J.M.; Romero, M.; Duarte, J.; López-Huertas, E. Antihypertensive effects of virgin olive oil (unfiltered) low molecular weight peptides with ACE inhibitory activity in spontaneously hypertensive rats. Nutrients 2020, 12, 271. [Google Scholar] [CrossRef]
- Kinariwala, D.; Panchal, G.; Sakure, A.; Hati, S. Exploring the potentiality of Lactobacillus cultures on the production of milk-derived bioactive peptides with antidiabetic activity. Int. J. Pept. Res. Ther. 2020. [Google Scholar] [CrossRef]
- Orts, A.; Revilla, E.; Rodriguez-Morgado, B.; Castaño, A.; Tejada, M.; Parrado, J.; Garciá-Quintanilla, A. Protease technology for obtaining a soy pulp extract enriched in bioactive compounds: Isoflavones and peptides. Heliyon 2018, 5, e01958. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vaquero, M.; Mora, L.; Hayes, M. In vitro and in silico approaches to generating and identifying angiotensin-converting enzyme I inhibitory peptides from green macroalga ulva lactuca. Mar. Drugs 2019, 17, 204. [Google Scholar] [CrossRef] [PubMed]
- Ashaolu, T.J. Health applications of soy protein hydrolysates. Int. J. Pept. Res. Ther. 2020. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska-Dziadkiewicz, J.; Pliszka, M. Carp proteins as a source of bioactive peptides—An in silico approach. Czech J. Food Sci. 2016, 34, 111–117. [Google Scholar] [CrossRef]
- Kęska, P.; Stadnik, J. Taste-active peptides and amino acids of pork meat as components of dry-cured meat products: An in-silico study. J. Sens. Stud. 2017, 32, e12301. [Google Scholar] [CrossRef]
- Sun, X.D. Enzymatic hydrolysis of soy proteins and the hydrolysates utilisation. Int. J. Food Sci. Technol. 2011, 46, 2447–2459. [Google Scholar] [CrossRef]
- Hrynkiewicz, M.; Iwaniak, A.; Bucholska, J.; Minkiewicz, P.; Darewicz, M. Structure-activity prediction of ACE inhibitory/bitter dipeptides—A chemometric approach based on stepwise regression. Molecules 2019, 24, 950. [Google Scholar] [CrossRef]
- Chanput, W.; Nakai, S.; Theerakulkait, C. Introduction of a new computer softwares for classification and prediction purposes of bioactive peptides: Case study in antioxidative peptides. Int. J. Food Prop. 2010, 13, 947–959. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E. Quantitative structure-activity relationship study of bitter di- and tri-peptides including relationship with angiotensin I-converting enzyme inhibitory activity. J. Pept. Sci. 2007, 13, 63–69. [Google Scholar] [CrossRef]
- Iwaniak, A.; Dziuba, J. BIOPEP-PBIL tool in the analysis of the structure of biologically active motifs derived from food proteins. Food Technol. Biotechnol. 2011, 49, 118–127. [Google Scholar]
- Iwaniak, A.; Dziuba, J. Animal and plant origin proteins as the precursors of peptides with ACE inhibitory activity. Proteins evaluation by means of in silico methods. Food Technol. Biotechnol. 2009, 47, 441–449. [Google Scholar]
- Arboleda, J.C.; Rojas, O.J.; Lucia, L.A. Acid-generated soy protein hydrolysates and their interfacial behavior on model surfaces. Biomacromolecules 2014, 15, 4336–4342. [Google Scholar] [CrossRef] [PubMed]
- Mallick, P.; Schirle, M.; Chen, S.S.; Flory, M.R.; Martin, D.; Ranish, J.; Raught, B.; Schmitt, R.; Werner, T.; Kustr, B.; et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 2007, 25, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Hrynkiewicz, M.; Bucholska, J.; Darewicz, M.; Minkiewicz, P. Structural characteristics of food protein-originating di- and tripeptides using principal component analysis. Eur. Food Res. Technol. 2018, 244, 1751–1758. [Google Scholar] [CrossRef]
- Panjaitan, F.C.A.; Gomez, H.L.R.; Chang, Y.-W. In silico analysis of bioactive peptides released from giant grouper (Epinephelus lanceolatus) roe proteins identified by proteomics approach. Molecules 2018, 23, 2910. [Google Scholar] [CrossRef]
- Vermeirssen, V.; van der Bent, A.; Van Camp, J.; van Amerongen, A.; Verstraete, W. A quantitative in silico analysis calculates angiotensin I converting enzyme (ACE) inhibitory activity in pea and whey protein digests. Biochimie 2004, 86, 231–239. [Google Scholar] [CrossRef]
- Pokora, M.; Zambrowicz, A.; Zabłocka, A.; Dąbrowska, A.; Szołtysik, M.; Babij, K.; Eckert, E.; Trziszka, T.; Chrzanowska, J. The use of serine protease from Yarrowia lipolytica yeasts in the production of biopeptides from denatured egg white proteins. Acta Biochim. Pol. 2017, 64, 245–253. [Google Scholar] [CrossRef]
- Khaldi, N. Bioinformatic approaches for identifying new therapeutic bioactive peptides in food. Funct. Food Health Dis. 2012, 2, 325–338. [Google Scholar] [CrossRef]
- Rawlings, N.D. A large nad accurate collection of peptidase cleavages in the MEROPS database. Database (Oxford) 2009, 2009, bap015. [Google Scholar] [CrossRef]
- Worsztynowicz, P.; Białas, W.; Grajek, W. Integrated approach for obtaining bioactive peptides from whey proteins hydrolysed using a new proteolytic lactic acid bacteria. Food Chem. 2020, 312, 126035. [Google Scholar] [CrossRef]
- Li, X.; Xie, X.; Wang, J.; Xu, Y.; Yi, S.; Zhu, W. Identification, taste characteristics and molecular docking study of novel umami peptides derived from the aqueous extract of the clam Meretrix meretrix Linnaeus. Food Chem. 2020, 312, 126053. [Google Scholar] [CrossRef] [PubMed]
- Ney, K. Voraussage der Bitterkeit von Peptiden aus deren Aminosäurezu-sammensetzung. Prediction of bitterness of peptides from their amino acid composition. Z. Lebensm. Unters. Forsch. 1971, 147, 64–68. [Google Scholar] [CrossRef]
- Maehashi, K.; Huang, L. Bitter peptides and bitter taste receptors. Cell. Mol. Life Sci. 2009, 66, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, L.; Simard, R.E. Bitter flavour in dairy products. II. A review of bitter peptides from caseins: Their formation, isolation and identification, structure masking and inhibition. Láit 1992, 72, 335–382. [Google Scholar] [CrossRef]
- Wang, W.; Gonzalez de Mejia, E. A new frontier in soy bioactive peptides that may prevent age-related chronic diseases. Compr. Rev. Food Sci. Food Saf. 2005, 4, 63–78. [Google Scholar] [CrossRef]
- Cheung, L.K.Y.; Aluko, R.E.; Cliff, M.A.; Li-Chan, E.C.Y. Effects of exopeptidase treatment on antihypertensive activity and taste attributes of enzymatic whey protein hydrolysates. J. Funct. Foods 2015, 13, 262–275. [Google Scholar] [CrossRef]
- Gallego, M.; Mora, L.; Toldrá, F. The relevance of dipeptides and tripeptides in the bioactivity of dry-cured ham. Food Prod. Process. Nutr. 2019, 1, 2. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Cermeño, M.; Khalesi, M.; Kleekayai, T.; Amigo-Benavent, M. Application of in silico approaches for the generation of milk protein-derived bioactive peptides. J. Funct. Foods 2020, 64, 103636. [Google Scholar] [CrossRef]

| Parent Peptides Released from Profilin Due to the Simulated Action of: | |||
|---|---|---|---|
| B 1 | F | P | PK |
| Number of parent peptides/number of parent peptides containing bitter-tasting indicators | |||
| 9/7 | 5/2 | 7/5 | 4/1 |
| Total number of parent peptides/total number of parent peptides containing bitter-tasting indicators = 25/15 | |||
| Parent peptides released from globulin 7S due to the simulated action of: | |||
| B | F | P | PK |
| Number of parent peptides/number of parent peptides containing bitter-tasting indicators | |||
| 24/16 | 10/5 | 20/16 | 14/4 |
| Total number of parent peptides/total number of parent peptides containing bitter-tasting indicators = 68/41 | |||
| Parent peptides released from glycinin due to the simulated action of: | |||
| B | F | P | PK |
| Number of parent peptides/number of parent peptides containing bitter-tasting indicators | |||
| 28/17 | 12/4 | 18/11 | 17/8 |
| Total number of parent peptides/total number of parent peptides containing bitter-tasting indicators = 75/38 | |||
| Parent peptides released from β-conglycinin (β-chain) due to the simulated action of: | |||
| B | F | P | PK |
| Number of parent peptides/number of parent peptides containing bitter-tasting indicators | |||
| 30/18 | 9/4 | 26/17 | 13/4 |
| Total number of parent peptides/total number of parent peptides containing bitter-tasting indicators = 78/43 | |||
| Parent peptides released from β-conglycinin (α-chain) due to the simulated action of: | |||
| B | F | P | PK |
| Number of parent peptides/number of parent peptides containing bitter-tasting indicators | |||
| 32/23 | 16/8 | 27/17 | 17/5 |
| Total number of parent peptides/total number of parent peptides containing bitter-tasting indicators = 92/53 | |||
| Time Interval (min) | Relative Area of Chromatographic Peaks (%) 1 | ||||
|---|---|---|---|---|---|
| 0-SPC 2 | B-SPC | F-SPC | P-SPC | PK-SPC | |
| 14.00–39.99 | 50.1 | 83.3 | 96.7 | 93.6 | 85,2 |
| 40.00–60.00 | 49.9 | 16.7 | 3.3 | 6.4 | 14.8 |
| C 3 | 0.0 | 4.0 | 14.2 | 6.8 | 2.4 |
| Protein Source | Parent Peptide | m/z (Da) | tR predicted (min) | tR experimental (min) | Motif 1 |
|---|---|---|---|---|---|
| B-SPC 2 | |||||
| glycinin | LSVISPK | 743.466 | 25.430 | 22.580 | PK, VI |
| DVLVIPLG | 825.509 | 43.140 | 39.540 | VL, VI, LG, LV 3 | |
| β-conglycinin (β-chain) | LIVILNG | 741.487 | 38.280 | 39.390 | VI, IV, LI, IL |
| NPFLFG | 694.356 | 41.170 | 39.860 | FG, FL, LF, PF | |
| F-SPC | |||||
| 7S globulin | ISSTIV | 619.367 | 25.520 | 24.770 | IV |
| glycinin | PQMIIV | 700.407 | 34.500 | 40.620 | II, IV |
| β-conglycinin (β-chain) | PFPSIL | 673.392 | 27.530 | 27.050 | FP, PF, PFP, IL |
| DDFFL | 656.293 | 41.170 | 34.782 | FF, FL | |
| FFEITPEK | 1010.520 | 35.950 | 36.200 | FF, EI | |
| P-SPC | |||||
| β-conglycinin (α-chain) | LIVILNG | 741.487 | 38.280 | 39.390 | VI, IV, LI, IL |
| Parent Peptide | Motif 1 | Bioactivities |
|---|---|---|
| LSVISPK | PK; VI, SV, SP | DPP IV inhibitor |
| DVLVIPLG | VL; VI; LV; IP; PL | DPP IV inhibitor |
| LG; PL; IP; PLG | ACE inhibitor | |
| PLG | Opioid | |
| VL; LV | Stimulating glucose uptake | |
| LIVILNG | VI; LI; IL; LN; NG | DPP IV inhibitor |
| IL; LN; NG | ACE inhibitor | |
| IV; IL; LI | Stimulating glucose uptake | |
| NPFLFG | FL; PF; NP | DPP IV inhibitor |
| FL; PF | DPP III inhibitor | |
| FG; LF | ACE inhibitor | |
| ISSTIV | IV | Stimulating glucose uptake |
| ST | ACE inhibitor | |
| TI | DPP IV inhibitor | |
| PQMIIV | II; MI; PQ | DPP IV inhibitor |
| IV; II | Stimulating glucose uptake | |
| PQ | ACE inhibitor | |
| PFPSIL | FP; PFP; IL | ACE inhibitor |
| FP; PF; IL; PS; SI | DPP IV inhibitor | |
| IL | Stimulating glucose uptake | |
| PF | DPP III inhibitor | |
| DDFFL | FL | DPP IV inhibitor; DPP III inhibitor |
| FFL; DF | ACE inhibitor | |
| FFEITPEK | EI; TP; EK | DPP IV inhibitor; ACE inhibitor |
| PE | DPP III inhibitor |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwaniak, A.; Hrynkiewicz, M.; Minkiewicz, P.; Bucholska, J.; Darewicz, M. Soybean (Glycine max) Protein Hydrolysates as Sources of Peptide Bitter-Tasting Indicators: An Analysis Based on Hybrid and Fragmentomic Approaches. Appl. Sci. 2020, 10, 2514. https://doi.org/10.3390/app10072514
Iwaniak A, Hrynkiewicz M, Minkiewicz P, Bucholska J, Darewicz M. Soybean (Glycine max) Protein Hydrolysates as Sources of Peptide Bitter-Tasting Indicators: An Analysis Based on Hybrid and Fragmentomic Approaches. Applied Sciences. 2020; 10(7):2514. https://doi.org/10.3390/app10072514
Chicago/Turabian StyleIwaniak, Anna, Monika Hrynkiewicz, Piotr Minkiewicz, Justyna Bucholska, and Małgorzata Darewicz. 2020. "Soybean (Glycine max) Protein Hydrolysates as Sources of Peptide Bitter-Tasting Indicators: An Analysis Based on Hybrid and Fragmentomic Approaches" Applied Sciences 10, no. 7: 2514. https://doi.org/10.3390/app10072514
APA StyleIwaniak, A., Hrynkiewicz, M., Minkiewicz, P., Bucholska, J., & Darewicz, M. (2020). Soybean (Glycine max) Protein Hydrolysates as Sources of Peptide Bitter-Tasting Indicators: An Analysis Based on Hybrid and Fragmentomic Approaches. Applied Sciences, 10(7), 2514. https://doi.org/10.3390/app10072514