1. Introduction
The skeletal muscles are some of the most important structures of the body; they make up more than 50% of the human body and are the ones that generate movement and help maintain its figure. These are composed of many fibers connected to various points of the bones through the tendons. To generate a movement, an electrical stimulus generates a contraction in the muscle fibers, temporarily increasing the force exerted on the tendons, and related bones.
Different approaches to simulating human tissue respond to different performance and functional requirements. For example, interactivity is required for real-time applications, such as virtual surgery simulators, but visual realism is more desirable in the entertainment industry. Moreover, biomechanical accuracy is most crucial in designing medical applications. A notable example is in Computer-Assisted Surgery (CAS), where a connection to biomechanics has helped by defining a theoretical and numerical framework that provides information about the mechanics of the tissues after a clinical treatment or surgical intervention [
1]. CAS has addressed a larger spectrum of clinical domains such as cardiology [
2], neurosurgery [
3], urology [
4], and abdominal surgery [
5]. For these applications, biomechanics faces a new challenge since the involved tissues are required to move and be deformed by stress generated by clinical actions. Moreover, soft tissues are difficult to model accurately since they typically exhibit complex, time dependent, non-linear, inhomogeneous and anisotropic behaviors. Such models are very computationally demanding and are therefore limited to pre-operative use, since the simulations often require many minutes or hours to compute. For clinical applications interactivity is critical, and reduced computational times are essential.
The Finite element Method (FEM) is one common numerical approach used to simulate skeletal muscles [
6,
7,
8,
9,
10,
11]. However, modeling of complicated 3D muscle geometries increases the complexity of mesh generation for Finite Element (FE) analysis. Poorly built meshes lead to mesh distortion and significant errors in the FE analysis. Standard FE approaches are still ineffective in handling extreme material distortions, as it could occur in muscle deformations. In 3D subject specific models, fiber direction is measured at each of the muscle pixel points, which need to be interpolated at the integration points in FE model, which introduces additional approximation errors in the FE analysis. Additionally, performance depends on the model’s mesh quality and complexity, and any change to the mesh during the analysis represents an extra computational cost, which is a significant drawback. Another complex step of FEM in electromechanics is the coupling between electrophysiology and mechanics. Meshless approaches are a possible solution to address these limitations.
Meshless methods [
12] have several advantages over the FEM, and as such have been used to simulate a wide range of biomechanical phenomena. The work of Doweidar et al. [
13] showed that meshless methods have apparent advantages over the FEM in biomechanical problems dealing with large strains, such as in the simulation of the human lateral collateral ligament and the human knee joint. Furthermore, Zhang et al. [
14] extended a meshless whose results confirmed the accuracy of meshless methods to deal with highly demanding nonlinear hyperelastic biomaterials. Soft tissue simulations have also benefited from the meshless formulation. Horton et al. [
15] presented a meshless method for computing the deformation of soft tissue. The model presented a speedup of 2 times the speed of a hexahedral-based FE simulation, and a speedup of 3 times when compared to a similar tetrahedral-based simulation.
In this work, a new particle-based meshless method to simulate skeletal muscle tissue was introduced. The muscle tissue was simulated using a highly viscous fluid which preserves its shape and volume, and it was controlled and deformed with the monodomain model and a biophysical cell model. Both the fluid and the electrophysiology simulations were solved using smoother particle hydrodynamics (SPH). We are focusing on creating simulations that can be used in interactive applications, such as CAS, where the tissue is visualized in a virtual world, and it is able to respond to interaction from the user in real-time. Specifically, the focus of this work is on the deformation and internal pressure of the muscle tissue by using a biophysical model. In order for the simulation of the muscle to be viable in such applications, the method was accelerated with the use of General Purpose Computing on Graphics Processing Units (GPGPU), achieving real time simulations at more than 60 frames per second (fps). To evaluate the accuracy of this approach, a comparison of the meshless method with a FEM implementation was made. Total pressure and displacement were compared, having an error of around 0.09, with up to a 10% difference in the models.
2. Related Work
2.1. Skeletal Muscle Simulations
Muscles drive body movement and anatomically characterize body shape, making them a central component of modeling animated human figures. Modeling the morphology of muscles requires that their deformations are accurately depicted. To this end, several approaches have been presented, including geometrically-based, and data-driven approaches. On the other hand, the simulation of physiological muscle functions aims to identify the biomechanical controls responsible for human motion. Estimating these muscle controls has been pursued through static and dynamic simulations. Please refer to the work by Lee et al. [
16] for a complete review of muscle simulations, and the several models that exist that are used to model them.
Researchers have developed increasingly sophisticated biomechanical models of individual body parts based on skeletal muscles, such as hands [
17,
18], head and neck [
19], legs [
20], facial animation [
21,
22,
23], and the upper body [
24]. However, even in the most detailed models, such as the ones presented by Zordan and Lee [
24,
25], muscles are grouped and treated as single rigid line objects, and their behaviour is computed following models such as the Hill muscle model [
26,
27]; which is a mechanically inspired simplification of the muscle’s behaviour based on mass-less spring systems. Even more recent applications and models, such as the ones that can be implemented in the OpenSim [
28,
29] software represent muscles as rigid lines, and focus on the analysis of movement, not on the complete tissue deformation and internal force, which limit their application in simulations such as CAS.
Specifically, for meshless skeletal muscle simulations, Basava [
30] presented the meshless Reproducing Kernel Collocation Method (RKCM) in context of nonlinear hyperelasticity. The method was able to provide both computational efficiency and controllable accuracy for large scale problems. Chen et al. [
31] introduced the meshfree Reproducing Kernel Particle Method (RKPM) for 3D image-based modeling of skeletal muscles. This approach uses pixel data obtained from medical images which are used as nodes for domain discretization in the meshless modeling. Valizadeh et al. [
32] implemented a 3D patient specific leg-muscle pixel-based model using isogeometric analysis (IGA) and the RKPM.
2.2. SPH for Biological Simulations
In recent years, SPH has become increasingly popular in computer graphics. It has been successfully used for the simulation of various fluid phenomena, including: rigid and elastic solids, deformable objects, spray, foam, tiny air bubbles, granular materials, and other complex scenes that consists of millions of points [
33,
34,
35]. Recently, SPH based methods have been used to solve non-hydrodynamic partial differential equations such as the wave equation, the diffusion equation, Maxwell’s equations and Poisson’s equation [
36], as well as to solve electronic structure calculations [
37].
The extension of SPH to simulate biological structures was relatively sparse, with a few examples of blood or biological fluids confined by meshes [
38,
39,
40], simulations of a virtual liver [
41], lips [
42], cartilage [
43], and generic biological tissues [
44].
Regarding the simulation of biological soft tissue with SPH, several works tackled the simulation while exposing some advantages of using the method: Gastélum et al. [
45] integrated the effect of internal and external forces, and demonstrated the advantage of using SPH for large tissue deformations; Palyanov et al. [
46] used a variation of SPH, Predictive-Corrective Incompressible SPH (PCISPH) to simulate different types of tissue, both solid and fluid, and introduced contractile fibers based on mass-spring systems; Rausch et al. [
44] used SPH to simulate tissue that experienced large deformations and damage, to the point of failure. Most of these works results were in agreement with analytical solutions, as well as with Finite Element Method (FEM) solutions.
These solutions do not consider biophysical models to control the activation and deformation of the tissues. Additionally, the tissues are modeled using mass-spring, elastic, or stress-based models that are not necessarily applicable to biological tissue simulations since they are non-volumetric methods where spring elements connecting point masses must be tuned for each desired scenario [
43]. This is an issue present in many other works that simulate skeletal muscles. Current computational models of skeletal muscle models typically focus on simplified phenomenological relationships mimicking the overall (mechanical) behavior of a single skeletal muscle.
3. Materials and Methods
In order to simulate skeletal muscles, specifically the muscle belly, the SPH method, in conjunction with Shape Matching (SM) [
47] and the monodomain model [
48], were proposed. The base structure of the tissue is composed of a particle based highly viscous fluid. The use of SM with a velocity correction scheme, similar to the one presented by Takahashi et al. [
49], allows the tissue to maintain its shape, and guarantee the conservation of volume, while avoiding the use of additional elastic or stress models. This will allow the fluid to behave as a deformable solid, showing deformations based on the activation of the tissue from an electric stimulation.
By using the monodomain model, an electric stimulation that innvervates the tissue is used to calculate an electric potential. Since the electric potential can be considered as pressure in hydraulic systems [
50,
51,
52], the resulting electric potential will be considered in the pressure force calculation of the fluid simulation, and the changes in pressure of the fluid will in turn deform the tissue. With this proposed method, the particles will move from a high pressure area to a low pressure area, and effects such as the bulging of the muscles will be present because of this movement. SPH will be used to solve both the fluid dynamics, and the monodomain model.
3.1. SPH Method Description
SPH is an computational method using particle systems that is used to simulate the mechanics of media such as solid mechanics or fluid dynamics. According to Muller [
53], a scalar quantity
A is interpolated at location
r by a weighted sum of contributions from all particles:
where
j iterates over all other particles,
is the mass of particle
j,
its position,
the density, and
the field quantity at
. The function
is called the smoothing kernel with core radius
h.
While the mass
is constant throughout the simulation, the density
varies and needs to be evaluated at every time step: through substitution of
into
in Equation (
1) we get, for the density at location
r:
When considering the SPH method, derivatives only affect the smoothing kernels. The gradient of
A is simply
while the Laplacian of
A evaluates to
3.2. SPH Applied to Fluid Simulations
In the Eulerian formulation, fluids are characterized by a velocity field
, a density field
and a pressure field
p. The progression of these fields over time is given by two equations. The first guarantees the conservation of mass
while the Navier-Stokes equation defines the conservation of momentum
where
is an external force and
the viscosity of the fluid.
Using particles instead of a grid based formulation, Equation (
6) can be expressed as
where
corresponds to the acceleration of particle
i and is integrated using the Leap-Frog scheme [
54]:
The time step size
that will be used must be adapted by the Courant-Freidrich-Levy condition [
55]:
where
d is the particle diameter, and
is the maximum particle velocity.
Substituting Equations (
3) and (
4) into the pressure and viscosity terms of the Navier-Stokes equation and solving according to Müller et al. [
53] yields:
where
p is the pressure,
the velocity, and
the viscosity coefficient. The pressure
of particle
i is computed via the modified gas state equation suggested by [
56]:
where
is the rest density, and
k is a stiffness constant that scales the pressure, and, thus, the pressure gradient and the respective pressure forces [
34].
SPH Smoothing Kernels for Fluid Simulations
Stability, accuracy, and speed of the SPH method rely upon the choice of smoothing kernels. For fluid simulations Müller et al. [
53] designed the following kernel
Unfortunately, if this kernel is used for the calculation of the pressure forces, particles build clusters under high pressure, and as they get close to each other, the repulsion force vanishes because the gradient of the kernel approaches zero. Desbrun [
56] solved this issue by using a spiky kernel with a non vanishing gradient near the center. That kernel generates necessary repulsion forces in pressure calculations and is defined as:
Viscosity is caused by friction and, thus, should only have a smoothing effect on the velocity field. However, if a standard kernel is used for viscosity, the resulting viscosity forces do not always have this property. For this reason, for the computation of viscosity forces, Müller et al. [
53] proposed a third kernel:
3.3. Bidomain Model
One of the most common approaches for modeling the electrical activity of biological tissues [
10], which do not describe the electrophysiology of a single cell, is to solve the bidomain model [
57]. It is the most complete description of electrical activity for biological tissue. It describes both the intracellular and extracellular potential fields, linking them through membrane behavior. The bidomain model, together with precise models of cell membrane kinetics, is generally accepted to provide a reasonable foundation for numerical simulations of electrophysiology [
58].
The basic bidomain equations [
48] are given by
where
and
are respectively the intracellular and extracellular conductivity tensors,
is the surface to volume ratio of the cardiac cells,
is the extracellular potential,
is the membrane capacitance per unit area,
is an electrical potential,
is the ionic current,
is the external stimulus applied to the intracellular space, and
is the external stimulus applied to the extracellular space. The ionic current
is calculated using an electrophysiological cell model.
Monodomain Model
Considering that the bidomain model is composed of a complex Partial Differencial Equation (PDE) system, it is assumed that the intra and extracellular domains have equal anisotropy ratios to obtain a simplified model called the monodomain model [
48]. The monodomain model is entirely written in terms of the transmembrane potential, defined as the difference between the intra and extracellular potentials. It is given by
where
is a conductivity tensor given by
3.4. Tissue Simulation
The main idea for the viscoelastic tissue is similar to the work by Takahashi et al. [
49]: to simulate volume preserving viscoelastic fluids, the volume preservation and the viscoelasticity are dealt with indepentently. A velocity correction for viscoelastic effects based on SM is used, while volume preservation accomplished by enforcing the incompressibility of fluid using SPH.
Particle velocities are corrected to describe viscoelastic effects without changing particle positions. In order to obtain intermediate velocity
for particle
i as an input for SPH, the predicted velocity
is computed first with all forces
(external and gravity) excluding viscoelastic and pressure ones:
where
is the mass of each particle. Then, the particle’s velocities are corrected with the velocity correction vector
(which is obtained via a Shape Matching Scheme):
Finally, the unknown factor (X) SPH method, XSPH [
55] is used to reduce particle oscillations with
, and a velocity mixing parameter
:
For the viscous fluid simulation, the use of the smoothing kernels mentioned by Muller [
53] is proposed.
Shape Matching Scheme
In Shape Matching [
47], particle
i is pulled toward its goal position
to restore the original configuration of the particles, and individual goal positions are computed to match the original configuration of the particles defined by
with current particle distributions denoted as
after the particles are transferred. With the goal positions, a velocity correction vector
with a coefficient for the shape matching velocity correction
and
the lower limit of
:
3.5. Muscle Tissue Properties
Incorporating the skeletal muscle’s properties is fundamental for the correct functioning of the model. For the case of the proposed cell model used, the values need to correctly simulate the current propagation throughout the tissue. The values proposed for the monodomain model are based on the work of Röhrle et al. [
10]; for the cell model used, the parameters are based on Nickerson [
59]. These values can be seen in
Table 1.
For the density of the muscle belly, muscle architecture reports usually do not directly measure muscle density [
60]. Instead, several studies [
61,
62] use the value 1.0597 g/cm
, which was obtained from rabbit and canine muscle tissue [
63]. However, it was inaccurate for several reasons. First, a species effect could exist so that rabbit or canine muscle density differs from human muscle density. Second, the method and duration of fixation may cause shrinkage and thus dehydration, which alters muscle density. Ward et al. [
60] determined, through various experiments, that studies should instead use a value of 1.112 g/cm
for muscle density. As for the viscosity of the fluid, a coefficient of 15 Nm
s [
64] was selected.
Muscle fiber architecture is essential when considering muscle deformation. For this simulation, each particle has a direction vector that represents the fiber direction of the muscle fibers. The vectors are oriented following a line of action from the origin to the insertion of the muscle.
3.6. Monodomain Solved with SPH
The solution of the bidomain equations is usually done using standard FEM, or finite-difference (FD) methods. However, since the discretization of an object generates a mesh with many millions of nodes, the linear systems resulting from FD or FEM methods are very large. Several meshless methods have established the ability to provide a computational feasible model without the burden of mesh generation [
65]. For this work, SPH is proposed to numerically solve the monodomain model of electrophysiology.
The monodomain model can be written as:
where
represents the stimulus current that is applied to the tissue.
Applying the SPH formulation [
53] to the rewritten monodomain Equation (
24), and using XSPH to reduce particle oscillations with
yields:
The second derivative of the kernel, which is necessary to solve Equation (
25), is the following:
For time integration, a forward Euler method was used:
3.7. Activating and Deforming the Muscle
In order to control the activation of the muscles, the use of the Fitzhugh-Nagumo model [
66,
67] was proposed to control the propagation of electrical activity in the monodomain model. Once the muscle is activated, and a transmembrane potential is calculated, a way to deform the muscle is needed. Since the muscle bellies are going to be simulated using highly viscous fluids, and considering that transmembrane potentials can be considered as electrical pressure, the calculated transmembrane potentials were added to the pressure force term of the fluid simulation. Since the fluid moves from areas of high pressure to areas of low pressure, by increasing the pressure the tissue is forced to move in a given direction. After the particles’ velocity is calculated as part of the SPH fluid simulation step, the unit vector of the velocity and the direction vectors of the fibers are added, and then weighted by the magnitude of the velocity to calculate the direction of the contraction or expansion. To control the innervation of the muscle, a stimulus current
is applied to specific sections of the tissue: when the current is applied, the transmembrane potential increases, and thus the pressure is increased. When the stimulus current is removed, the transmembrane potential and the pressure begin to decrease, relaxing the muscle and allowing it to return to its original shape.
3.8. Gpu Considerations
Even though the proposed methods have been used to create real-time simulations of several deformable objects, in order to be able to simulate objects with much more detail, in the form of several thousands of points, or simulate many objects at the same time, the use of GPGPU is proposed.
Since the SPH method is going to be used to solve both the viscous fluid dynamics, as well as the monodomain model, special attention will be given to its implementation on a GPU. The main bottleneck for the parallelization of the method is the neighbor search step. For this work, the approach taken by several authors [
68,
69,
70] will be used.
3.9. Implementation
The proposed methods were applied to develop a simulation of the long head of the triceps brachii and the vastus lateralis. The focus of the simulations were the contraction and expansion of the muscle activated by the biophysical model, giving special attention to the displacement, pressure, and processing time. The muscle geometry for the simulations was obtained from the BodyParts3D database [
71], a dictionary-type database for anatomy in which anatomical concepts are represented by 3D structure data that specify corresponding segments of a three-dimensional whole-body model for an adult human male. The geometry was used as a “container” reference to initialize the particles that were used for the simulation.
To assess the accuracy of the proposed SPH-based model, another model that uses FEM was simulated for the same geometries, and results were compared for the displacement of the geometry, and the pressure generated by the tissue. The root mean squared error (RMSE) was used to compare the results of both methods. The error is defined by
where
is the value predicted by the SPH method, and
y is the value predicted by the FEM method. For the displacement, the resulting deformed mesh by both methods is compared. For the pressure, the total average pressure of the tissue by each method was calculated and then compared.
For the SPH method, 2 types of simulations were developed: one using only the CPU for processing, without any parallel processing; another using GPGPU. For the FEM simulation, only a CPU version was developed. The simulations were tested using the following setup, as shown in
Table 2:
3.10. SPH Simulation
The meshless simulations were developed using C++, CUDA for GPGPU processing, and rendered using OpenGL. For particle number, several values were proposed: 2231, 4944, 9888, and 18475. For the core radius h, the values of 0.02, 0.04, and 0.08 were selected. For the cell sizes, the values of 0.02, 0.04, 0.08, and 0.16 were selected.
The block and thread configuration for the GPU kernels launch was set to 128 threads, and 145 blocks. This was found to be the configuration that produced the best performance when considering execution time, occupancy, memory bandwidth, and compute resources utilization.
To optimize the neighbor search step of the algorithm, a grid of size was selected. The choice of a cell size double the size of the core radius was because of stability concerns: for the complex geometry of the muscle tested, if the cell size was the same as the radius, the particles would begin to vibrate and eventually collapse by breaking the geometry and moving erratically throughout the simulation space. While the added size for each cell increases the amount of particles that contribute to the approximation and thus help calculate a more accurate value of a property, the computational time was also increased.
Meshless simulations consisted on two phases: one where specific parts of the tissue were innervated with a stimulus current, and one where the stimulus current was removed. The purpose of the first phase was to test the effects of the stimulation current on the tissue, and try to simulate a contraction on the tissue. The second phase would allow the tissue to return to its original shape. Each phase ran for 250 time-steps.
3.10.1. Integration of SM, SPH, and Monodomain
With the transmembrane potential calculated using the monodomain model, the pressure force of the fluid was altered in order to simulate a contraction of the muscles. Since several steps of the model rely on SPH to calculate different properties, special care was taken to use the cycles of the SPH algorithm as best as possible. For this work, three SPH cycles were needed: to calculate the intermediate velocity; to calculate the pressure and density; and to calculate the pressure and viscosity forces, as well as the transmembrane potential.
The GPGPU version of the SPH algorithm did have additional constraints regarding how data was managed and processed. The SM section of the algorithm had no special considerations, and was “Embarrassingly Parallel”, that is, the same tasks were performed for each particle, and no special considerations or algorithms had to be considered besides thread synchronization.
A simple requirement to produce a better performing solution was to arrange data in a Structure of Arrays (SoA) manner to achieve memory coalescing. Each property of the simulation, from the position of the particles to their transmembrane current, were assigned to a one-dimensional array (the arrays were device arrays, that is, arrays whose data is only available on the graphics card itself), and the simulation data was stored sequentially.
3.10.2. Point-Based Tissue Simulation
The ends of the muscle geometry were fixed in space to emulate the origin and insertion of the muscle belly. For the first phase, the stimulation current was applied to the particles at the ends of the muscle geometry for 250 time-steps. The transmembrane potential of each particle gradually increased as the Ionic current was propagated throughout the tissue, and the particles of the muscle moved towards regions of lower pressure. For the second phase, the particles that had been innervated had the stimulation current removed and the tissue was allowed to return to its initial configuration.
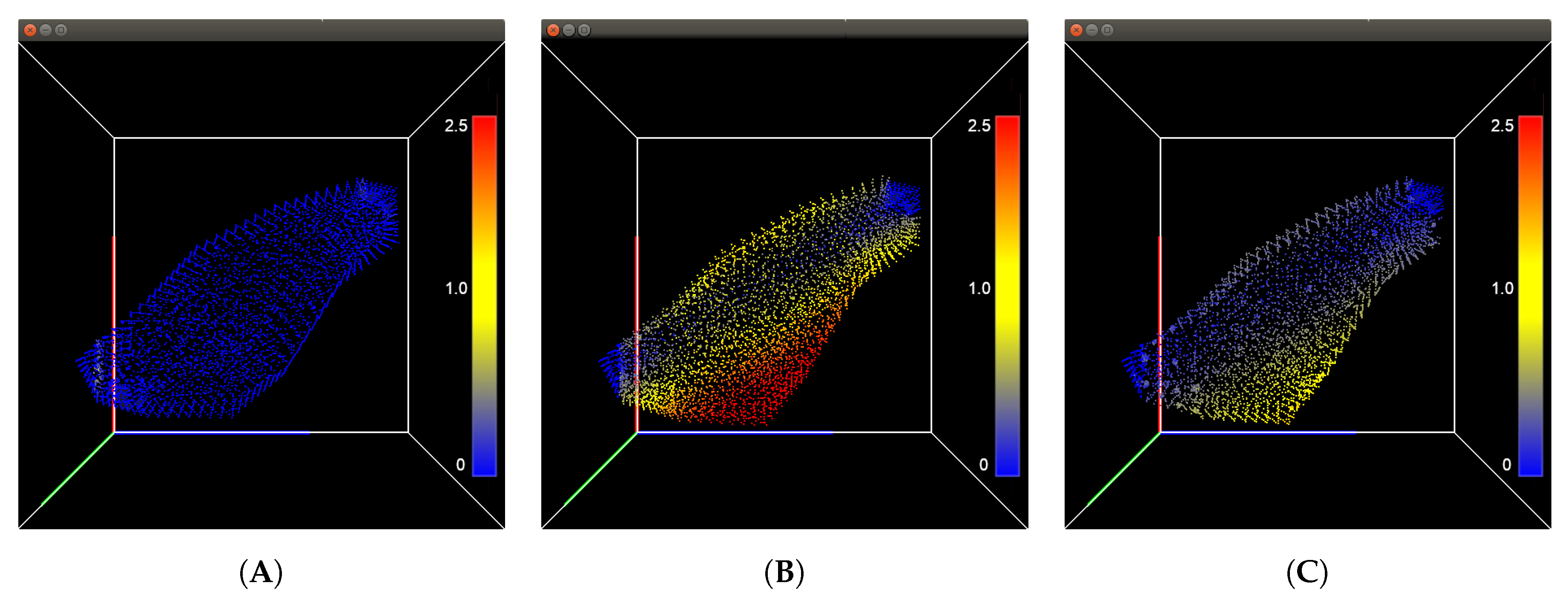
Figure 1 shows the displacement in millimeters of the triceps using the 18,475 particle resolution, while
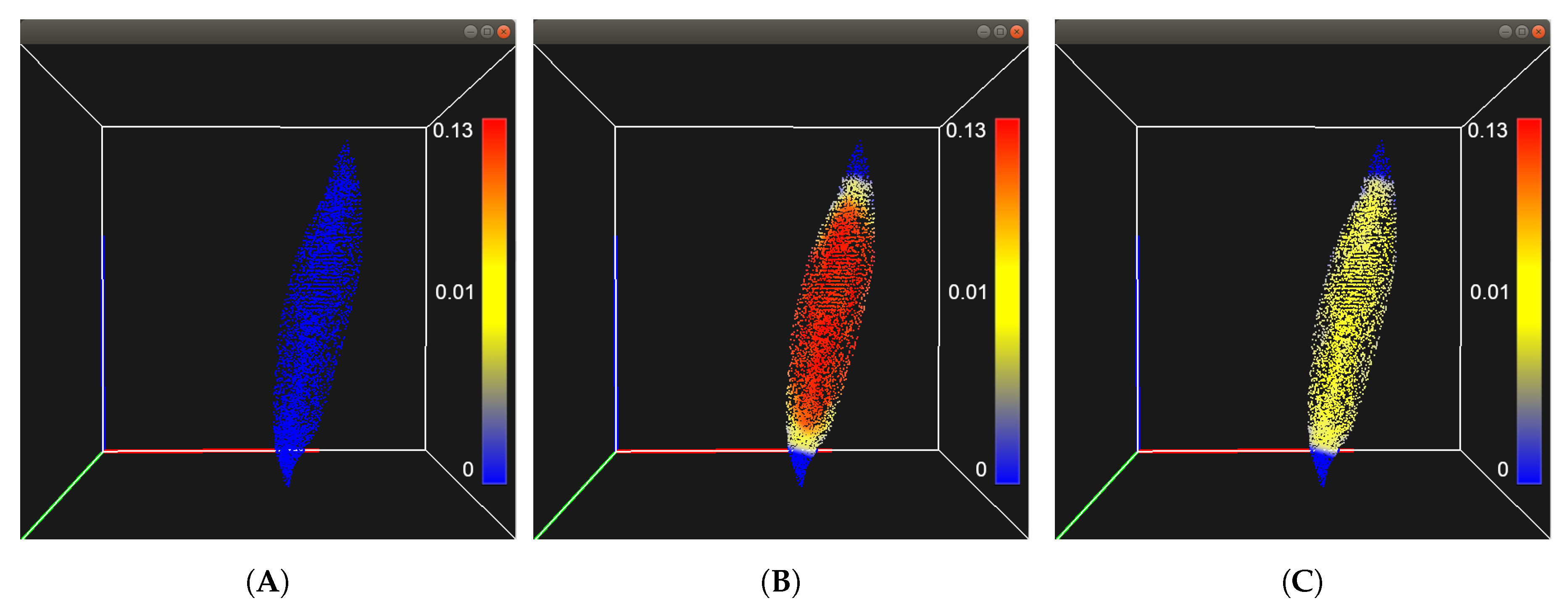
Figure 2 shows the displacement for the vastus lateralis with the same particle resolution.
3.11. FEM Simulation
To evaluate the performance of the proposed method, the FEM simulation was developed using the software framework FEBio [
72], and results were compared for the deformation and the pressure within the tissue. Particularly, the model was the one introduced by Blemker and Delp [
7]. Muscle is modeled as a fiber-reinforced composite with transversely-isotropic material symmetry. The model uses an uncoupled for of strain energy to simulate the nearly-incompressible behaviour of muscle tissue. This model was selected because it is capable of representing complex muscle geometry and architecture from MR images, it considers muscle fiber orientation and arrangement, and the predicted muscle shape was compared to MR images of the same movement, obtaining less than 5 mm of distance error for large muscles. The parameters used with the model can be seen in
Table 3.
One of the drawbacks of the FEM is the preprocessing steps needed before any computation can be performed. In this case, a mesh with finite elements had to be created, and muscle material properties, including fiber distribution, were integrated. Boundary conditions were similar to the meshless simulation: the ends of the tissue were fixed in space. A tetrahedral mesh was used, and six resolutions were selected: 1 thousand, 2 thousand, 5 thousand, 10 thousand, 20 thousand, and 40 thousand finite elements.
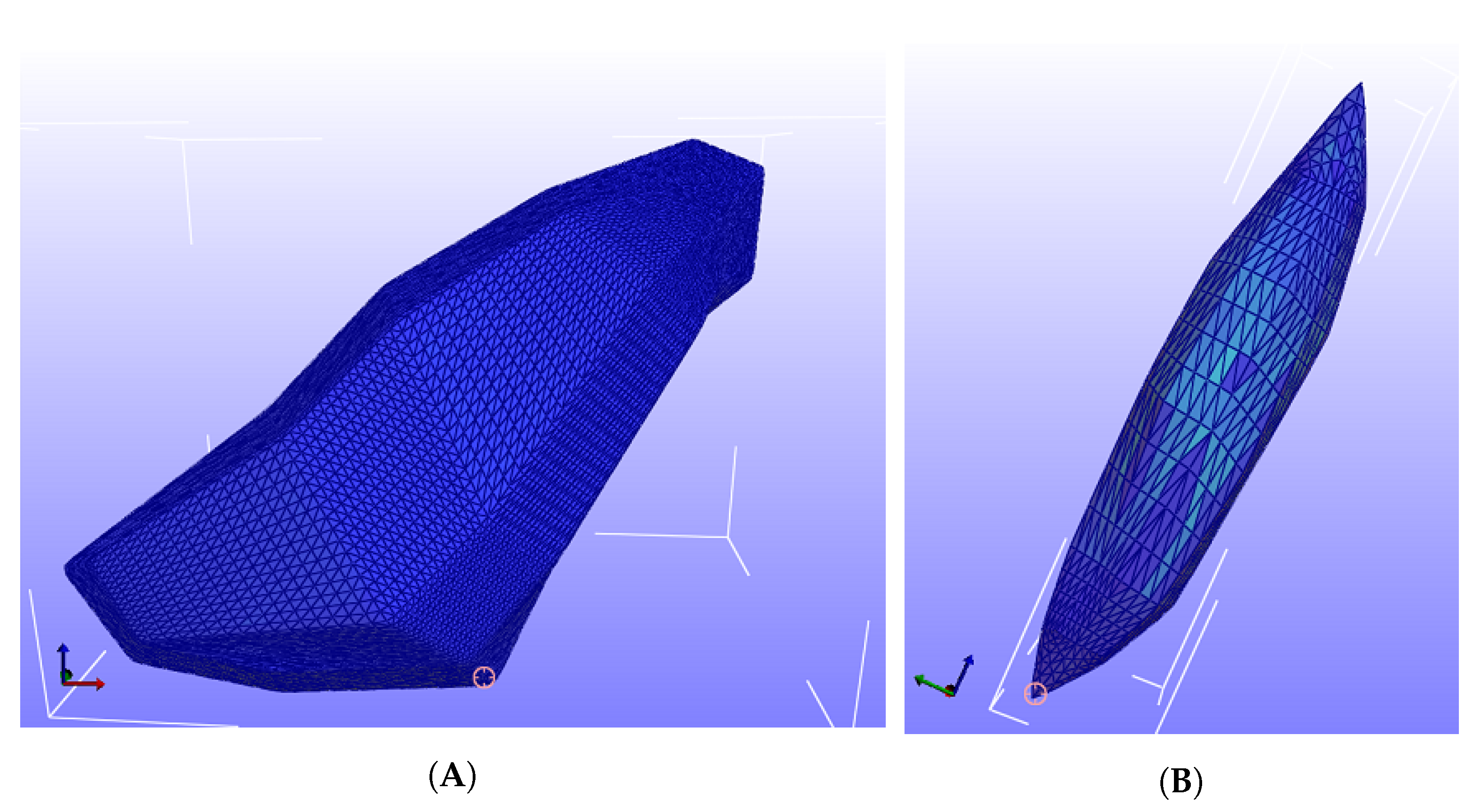
Figure 3 shows the mesh with around 40 thousand tetrahedral elements for the FEM simulation.
FEM simulations consist on the geometry being activated for 250 time steps, and then removing the activation and running for 250 time steps. A step size of 0.001 was selected to avoid convergence errors. As for boundary conditions, we selected 10% of the nodes around the origin and insertion of the muscles, respectively, and fixed their displacement in X, Y, and Z. A pressure load of 200 Pa was applied using a load curve, which ramps the pressure value from zero, to the rest of the nodes. Fiber direction was created similarly to the meshless solution: instead of each particle having a direction, each node had a direction vector which represented the fiber direction.
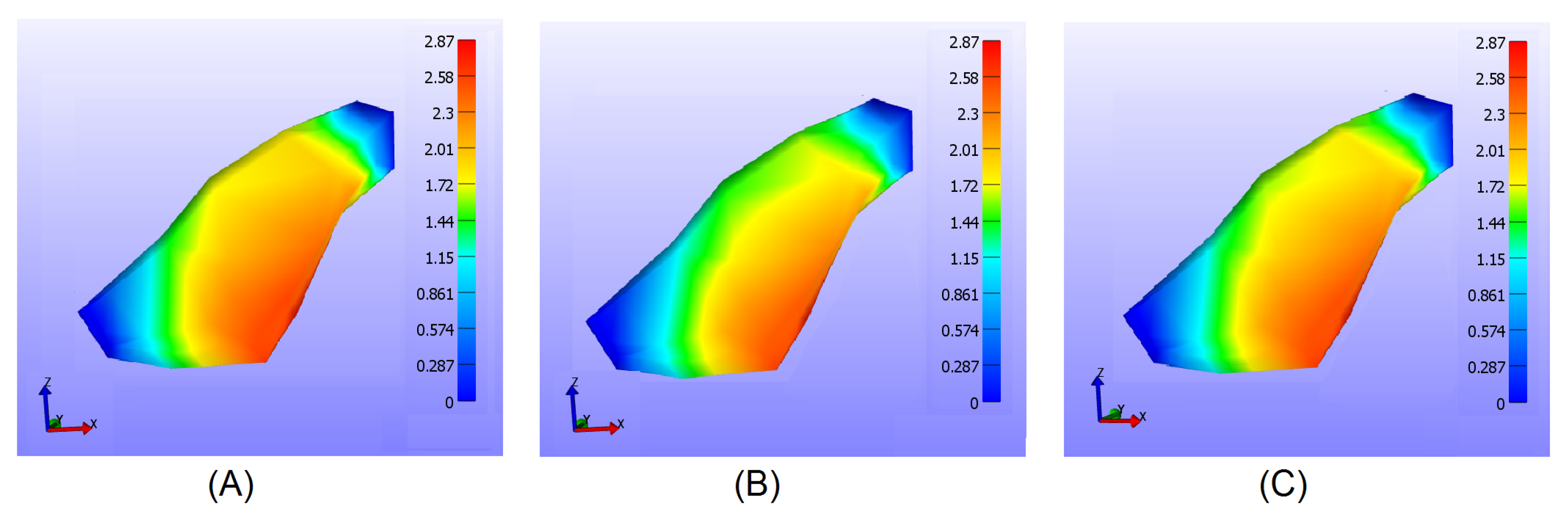
Figure 4 shows the deformation in millimeters of the triceps using the 40 thousand node resolution, while
Figure 5 shows the deformation of the vastus lateralis for the 40 thousand node resolution.
4. Results
Results are presented as follows: first, a quantitative comparison against the FEM solution for the displacement and the pressure is presented; then the stability of the SPH method is discussed; finally, the computation times for the different simulations are reported. The results are from the GPGPU simulation, even though the CPU simulation also yielded similar results but with a larger processing time per step. We also report the speedups obtained by using GPGPU. Speedup is a number that measures the relative performance of two systems calculating the same problem. In this case, it is the improvement of speed of execution when using a GPU vs using a CPU. The speedup is usually dimensionless. In this case, it means that the GPU version ran from 150 to 250 times faster than the CPU version.
4.1. Sensitivity Analysis
For the SPH method, a simulation for each of the proposed particle numbers, kernel, and cell sizes was developed. Similarly for the FEM, a simulation for each of the node resolutions was developed. Each of the simulations were paired against each other to calculate the RMSE.
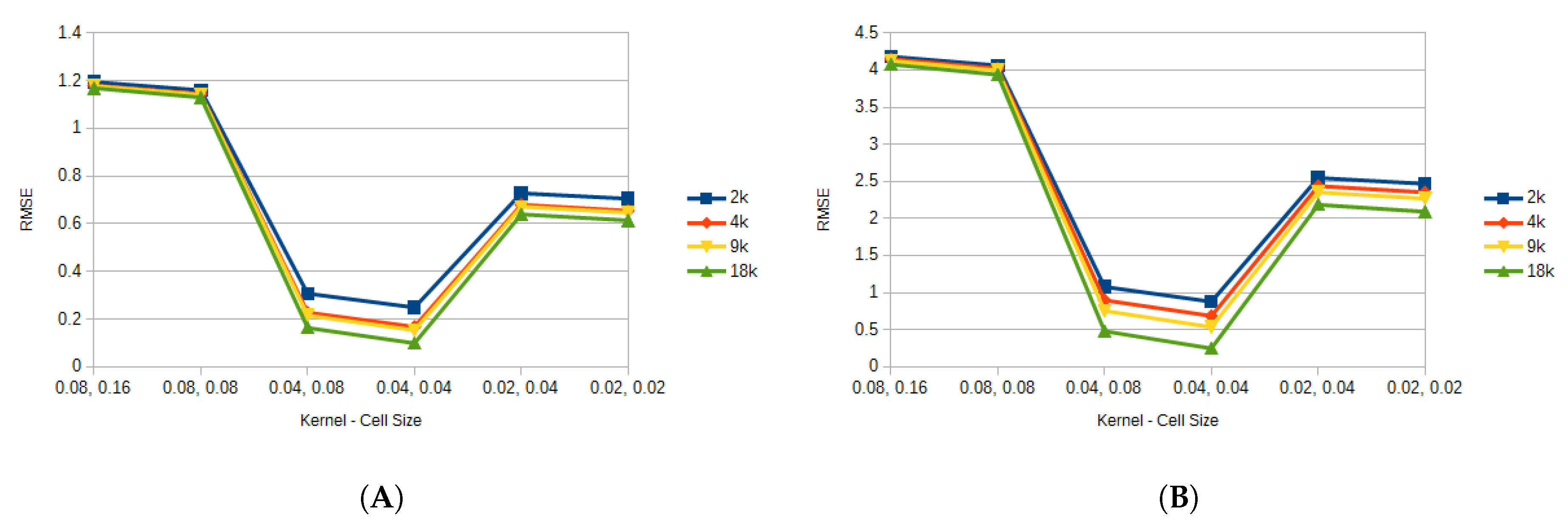
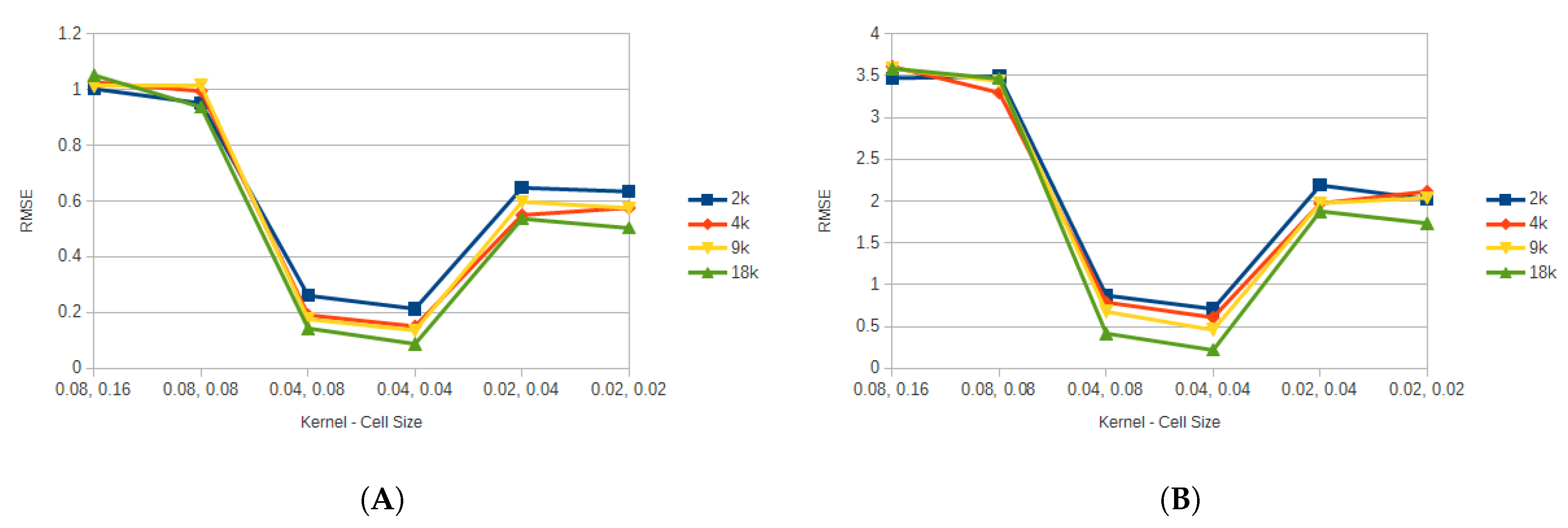
Figure 6 shows the average error for all the FEM resolutions against the different particle resolutions by kernel and cell size for the triceps, while
Figure 7 shows the average error for the vastus lateralis. It can be seen that the kernel and cell size selection is critical when considering the accuracy of the model. In this case, when those value were set to 0.08 and 0.16, and to 0.08 and 0.08, the model presented to greatest error. This could be attributed to the fact that too many particles were present at a given neighborhood, and particles that should not have contributed to updating a property were updating it. This also explains the results for the 0.02 and 0.04, and the 0.02 and 0.04 configurations, where too few particles were contributing to updating the properties. Since the 0.04 kernel and 0.04 cell size produced the least error with respect to the FEM, for the rest of the results this choice of parameters was used.
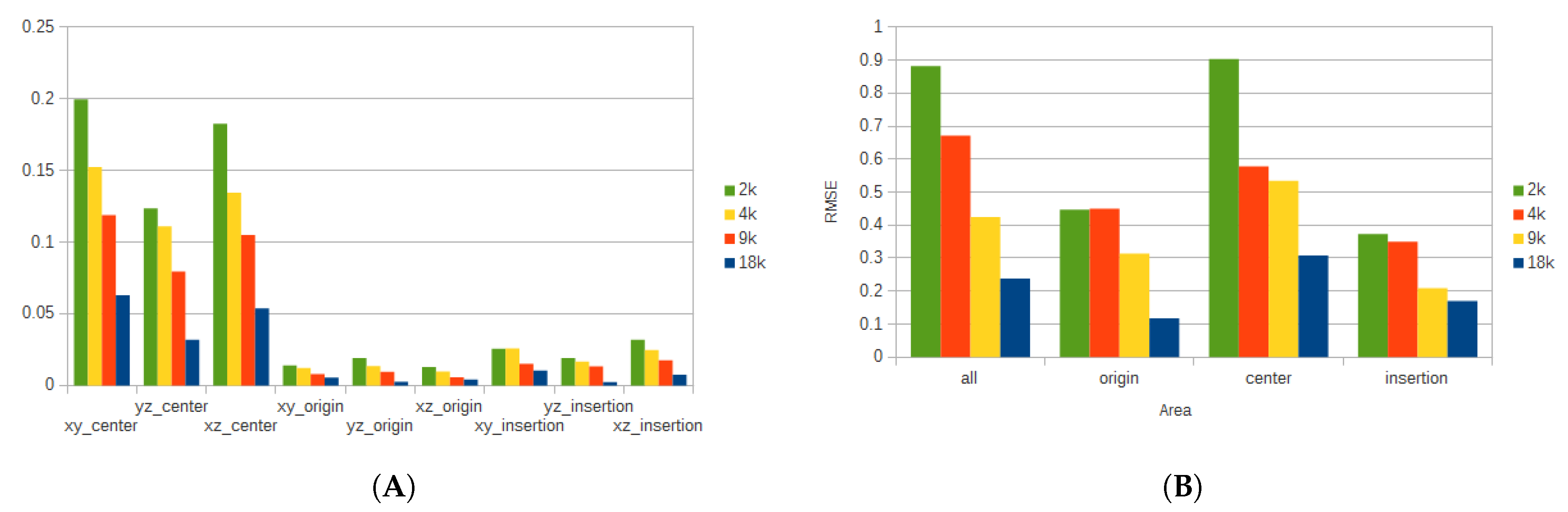
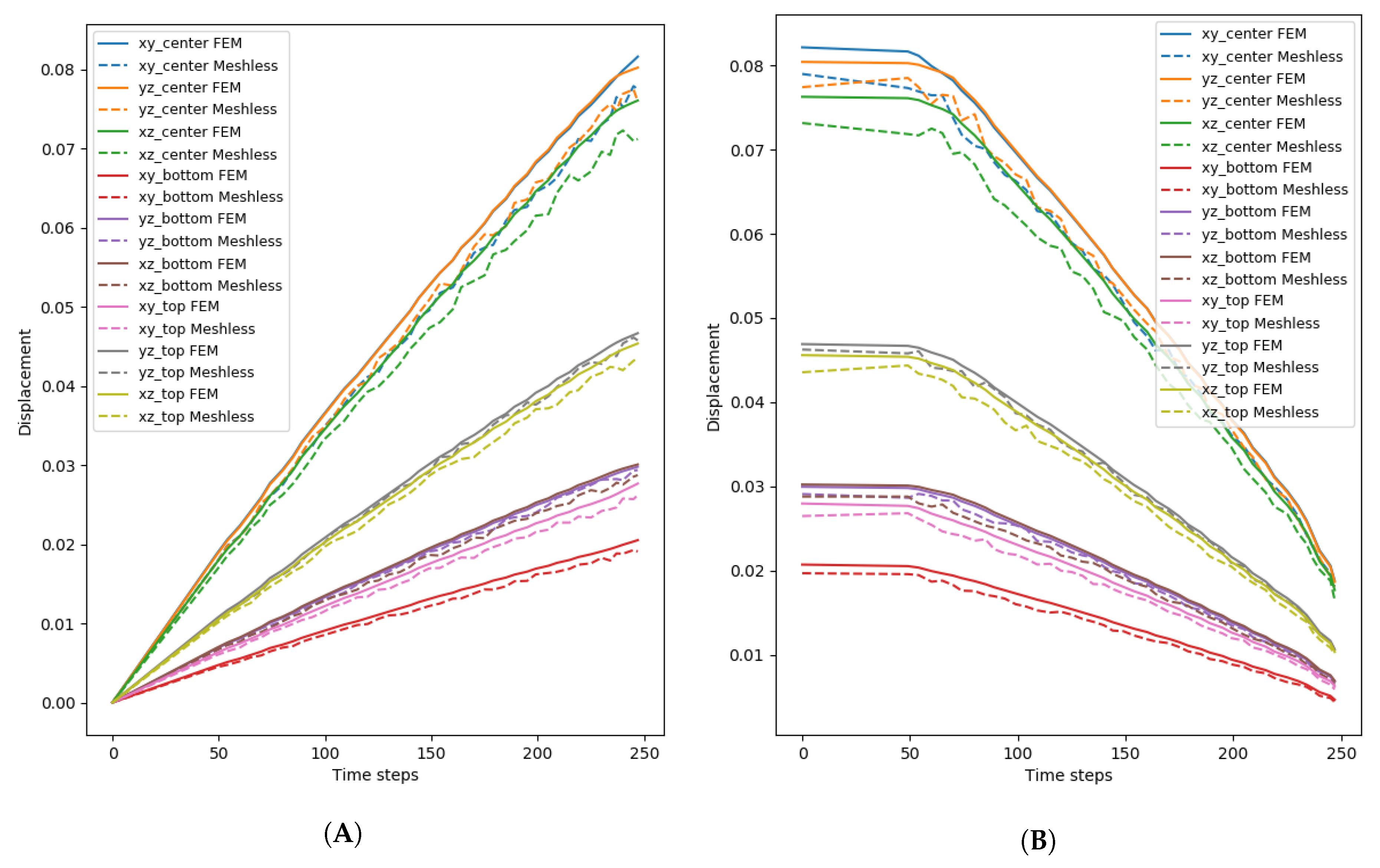
To get a better understanding of the deformation and the pressure of the tissue, additional considerations were taken. The muscle belly was divided into three regions: one around the origin of the muscle, another around the insertion, and the last comprised the rest of the muscle. From the origin and insertion, respectively, 20% of the tissue was selected to form each of the respective regions. The region at center of the muscle belly comprised the remaining 60%. Additionally, for the displacement, different coordinate planes were considered to analyze the deformation of the tissue along those planes.
Figure 8 shows the average error for each of the regions, and their respective planes for the deformation of the triceps, while
Figure 9 shows the average error for the vastus lateralis. In this case, when more particles are used for the simulation, the error is reduced considerably at the expense of additional processing time. For example, when comparing the error produced by the
coordinate plane on the center region for the 2k and 18k particle resolutions, the difference is more than 80%.
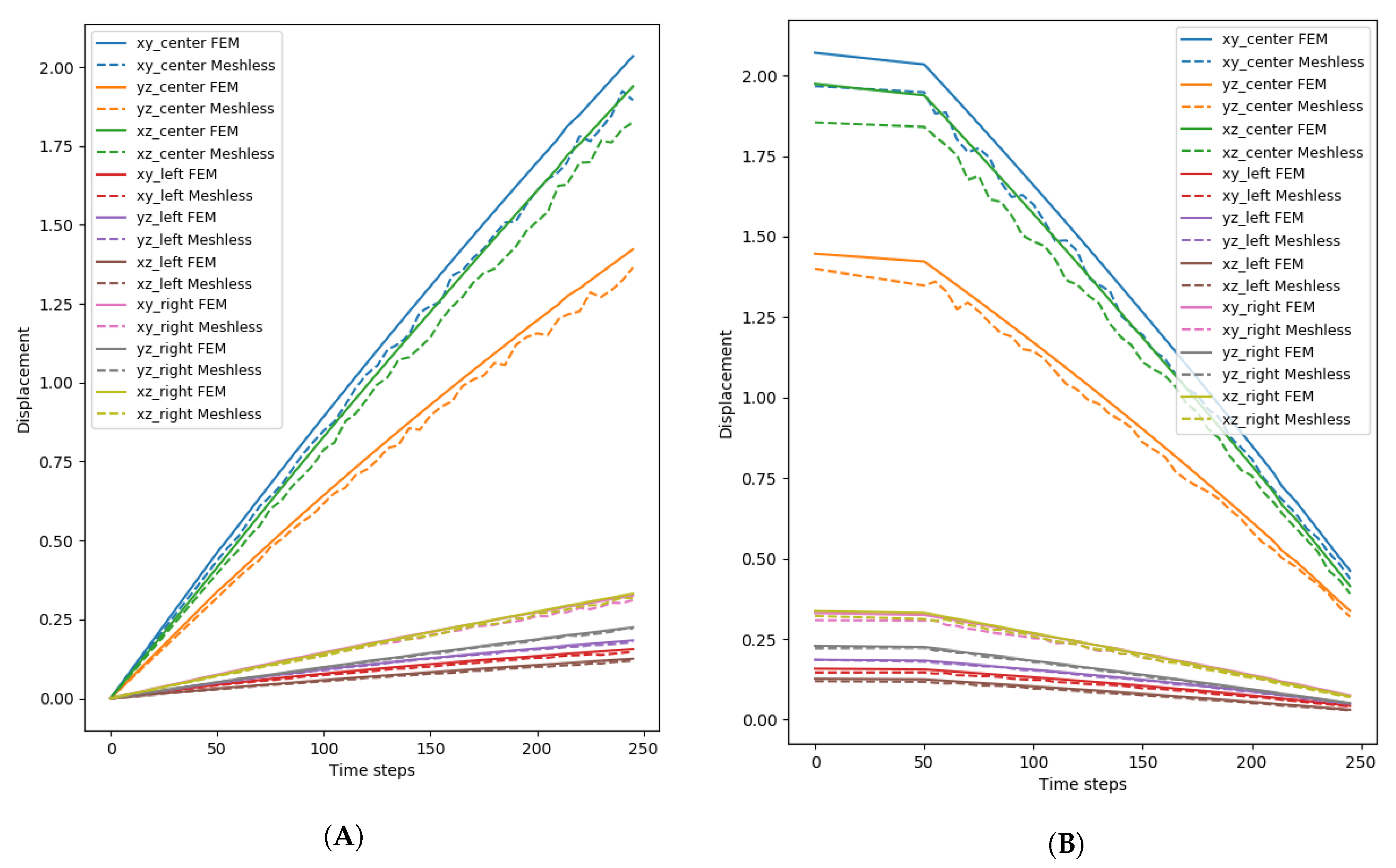
4.2. Displacement and Pressure Analysis
For the analysis of the displacement and the pressure, only one node configuration for FEM, and one particle configuration for the meshless method are presented. The FEM configuration was 40k nodes, while the meshless was 18k particles, with 0.04 cell size and kernel. These were chosen since more FEM nodes lead to better accuracy, and because with 18k nodes the error was the lower from all the tested configurations. It is worth noting that the results were similar for all the configurations, but with a larger error.
The mean displacement of the meshless and FEM simulations of the triceps can be seen in
Figure 10, while
Figure 11 shows the mean displacement for the vastus lateralis.
Figure 10A shows the displacement for the contraction part of the simulation for the triceps (
Figure 11A shows the same information for the vastus lateralis), while the displacement for the expansion part can be seen in
Figure 10B (
Figure 11B for the vastus lateralis). The area of the tissue that presented the most change was around the center, not only because it was the largest section, but also because the pressure exerted by the model made it so that the particles moved towards that direction. When both models were compared, a difference of around 10% was present throughout the simulations. Another point that became apparent was that the meshless simulation was not entirely stable when compared to the FEM simulation: the displacement of the model was not smooth and the data shows slight noise throughout. This was also apparent, if ever so slightly, in the rendered simulation: particles would oscillate while moving, creating visual artifacts. Contrary to the contraction of the tissue, where the displacement increased constantly, the expansion had a few moments, from time step 0 until around 50, where it experienced almost no change and then the displacement began to reduce constantly.
4.3. Stability and Deformation of the Model
The stability and deformations of the model depended mainly on the cell size, the core radius, and the number of particles. Deformation ranges were also similar. When the cell size and the core radius were set to 0.02 and 0.02, and to 0.02 and 0.04, the particles moved an average of 7% from their original positions. This behavior is due to an insufficient number of neighbor particles in each cell. Deformations became noticeable when the cell size and the core radius were set to 0.04 and 0.04, with a deformation of an average of 23%. This value ranged from around 20% to 26% depending on the number of particles; the simulation with 18475 particles yielded the 26% average deformation. This result is the closest to the 28% of optimal length during contraction that was reported by Murray et al. [
73]. When the cell size and core radius were set to 0.08 and 0.04, there was an average deformation of 37%. When the cell size and the core radius were set to 0.08, the particles deformed more than 70% from their original configuration, also presenting visual artifacts. Finally, the simulation became unstable in less than 100 time steps when the cell size was increased to 0.16 and the core radius was set to 0.08. The geometry deformed more than 70%, with several artifact forming before losing the shape completely. The FEM simulation, in contrast, did not present any instabilities, and also got closer to the 28% average deformation when more nodes were considered.
4.4. Computation Time
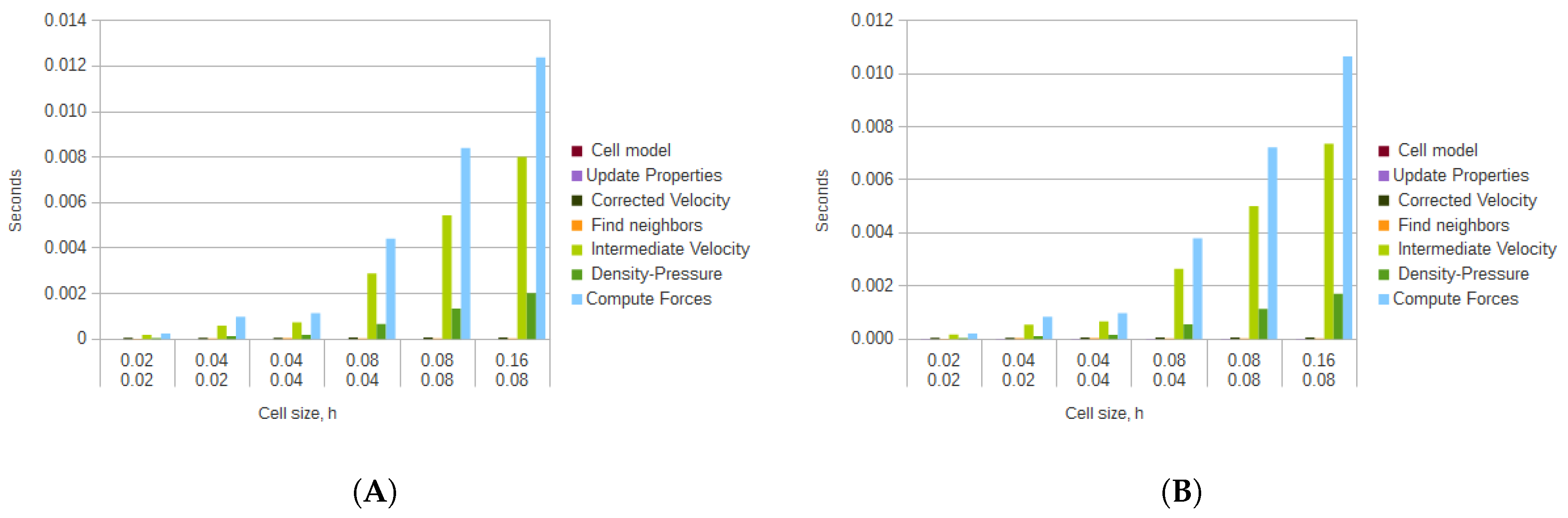
Using the GPU, the computation time was considerably sped-up when compared to the CPU version. The results for the average computation times for each of the algorithm steps, and for each of the particle sets are presented in
Figure 12.
The average times and speedups were reported for each of the main methods of the algorithm in order to showcase the differences in their performance. For the average times in
Figure 12, the functions that calculated the corrected velocity, the cell model, and updated the properties performed the best since simple calculations on each element of the data sets were executed; they did not have constructs such as
if-then blocks which lead to thread branching, and did not involve the more complex SPH method.
These simpler methods, however, did not gain much from being parallelized using GPGPU, as can be seen from the obtained speedups in
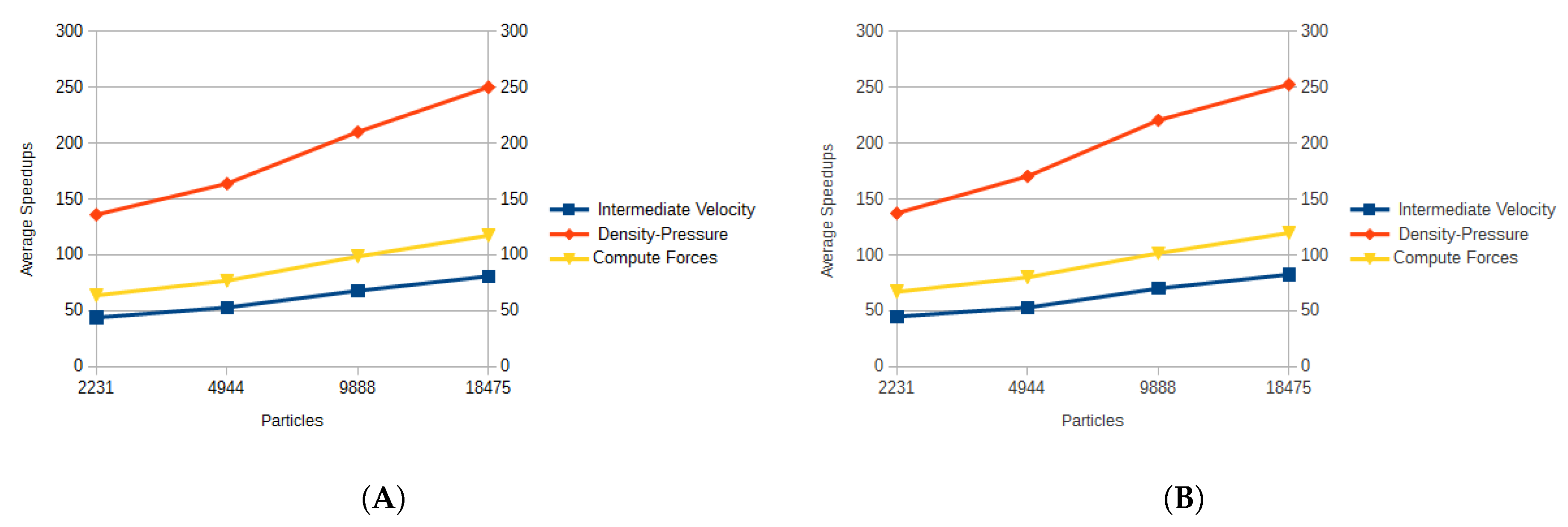
Figure 13. The function that gained the least speedup was the calculation of the corrected velocity, with an average speedup of 8. The calculation of the cell model had an average speedup of 15.473, while the updating of the properties achieved an average of 14.887. Even though the search for neighbors for the GPGPU version was more elaborate when compared to the CPU version, it also was not sped up by much, having an average speedup of 9.715.
The functions that took the longest to compute, even while using GPGPU, where the ones that implemented the SPH method. These functions, specially the one that computes the forces took, in some cases, more than two orders of magnitude more than the previously discussed simpler functions. However, since these functions are more elaborate, the contribution of using GPGPU was more noticeable. The intermediate velocity calculation was sped up by an average of 102.667, the computation of forces was speed up by an average of 151.551, while the calculation of the density and pressure by an average of 283.354. These results are dependent on the types of calculations and blocks that are involved in each function. The calculation of density and pressure had the most speedup since the executed operations were limited to additions and multiplications.
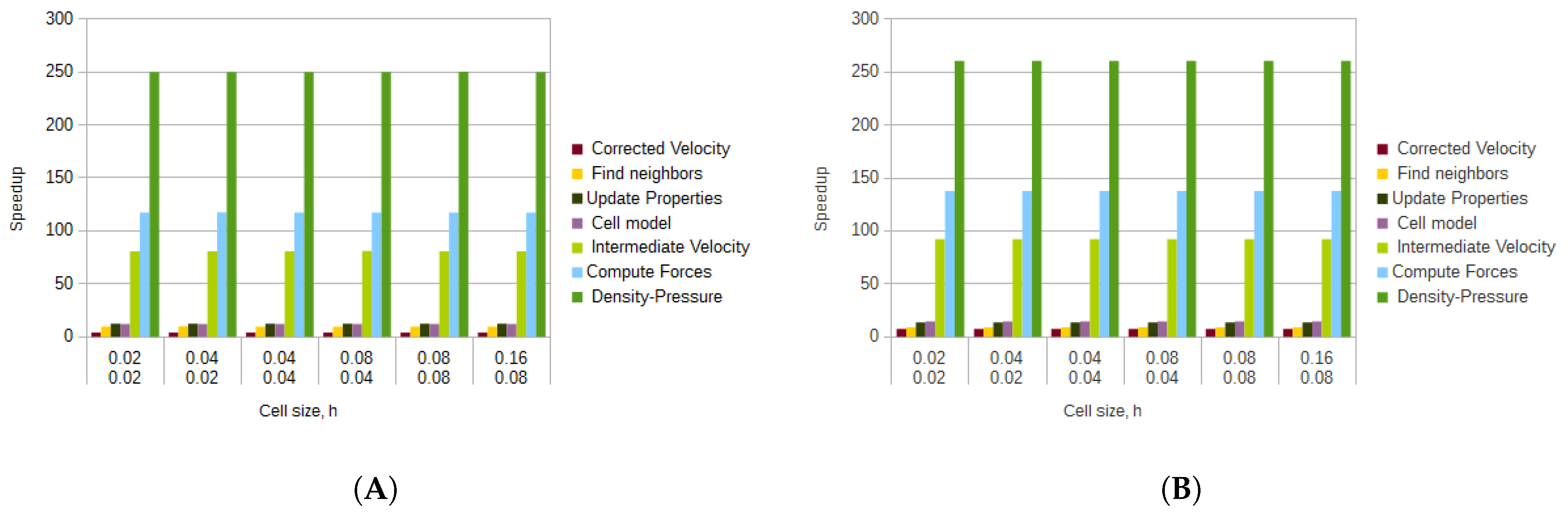
The average speedups for all cell and radius sizes of the kernels that implemented the SPH method can be seen in
Figure 14. As more particles were involved in the calculations, the speedup for each kernel was larger, indicating that the SPH method benefited more with the use of GPGPU when a larger number of particles was involved in the calculations.
Finally, it is worth noting that the computation times for the FEM were much higher than those for the SPH method. In particular for the 40 thousand node resolution, it took around 2 h to compute 500 time steps.
Figure 15 shows the average speedup when comparing the CPU version of the SPH method to the FEM simulation. The FEM times could be improved if another platform that solved the equations in parallel was used, such as OpenCMISS [
74]. Although there are several commercial FEM solvers, to our knowledge there are not many commercial FEM solvers that use GPUs. So far, the FEM model was only solved using CPU, and we plan to later run the model in GPU to more accurately compare the proposed model’s execution time.
5. Discussion
Even though SPH and Shape Matching had already been used to simulate viscoelastic fluids, to our knowledge, this was the first time a viscoelastic fluid was used in conjunction with a biophysical model, specifically, the monodomain model, to simulate biological tissue. Additionally, the use of SPH to provide an alternative mean to solve the monodomain model was implemented and used in the presented simulations. To our knowledge, this was also the first time the monodomain model was solved with SPH.
According to the results obtained, the following contributions of this research can be highlighted:
Biological tissue simulation. Most of the previous work related to the simulation of biological tissue used methods such as FEM. However, the definition and pre-processing of the mesh, in addition to the high computational cost, even with the use of GPGPU, made the method less than ideal for real time simulations. Here, an alternative mean to simulate biological tissue was presented. In this case, a skeletal muscle was simulated by using a skeletal muscle geometry and a specific activation model. By changing the geometry and the constitutive model, different biological tissues could be simulated. Since SPH was used to simulate the tissue, an integration with other particle-based models is also possible; for example, the inclusion of blood in the tissue.
Solution of the monodomain model with SPH. The monodomain model was usually solved with FEM or the FDM. The use of GPUs was also explored to speed the simulations up. Here, the model was solved with SPH, which is also paralellizable with GPUs, and the results were similar to the ones of a simulation with the FDM. These results show that biophysical models can be solved with the SPH method, and that the solved properties can be easily included in a more complex model.
Tissue deformation. The focus of this work was on the deformation of the muscle tissue by using a biophysical model. In order to apply the transmembrane potential to the tissue, and deform it, the potential was considered as a force of pressure that acted on the fluid. Since the fluid, and in this case, the tissue, flows from regions of high pressure to regions of low pressure, the added pressure made it so that the tissue contracted in a given direction when a stimulus current was applied, or it relaxed to its original shape when the current was removed. If the model were to be used in simulations, additional external forces could be applied to the tissue, and it would appropriately respond because it preserves volume, and the internal pressure force would change and deform the tissue. To our knowledge, this was the first time that such a model was used. The proposed method was able to achieve a contraction of around 23%, which is similar to the achieved contraction of a muscle. Additionally, the RMSE for the simulation was low when compared to a FEM simulation that has proven to be reliable to simulate skeletal muscles.
Achieved real time simulations. In order for the model to be viable in interactive simulations, it had to be able to be simulated in real time, at least 30 FPS. The CPU version, with around 9888 particles, was able to run in real time; at an average of 27.5 FPS for the mesh-based simulation. If 18,475 particles were simulated in CPU, an average of 3.7 FPS were obtained for the mesh-based simulation. In order to get a more detailed tissue, and to be able to simulate more than one, the use of GPGPU was proposed. The GPU version, with 18,475 particles, ran at 70.125 FPS for the mesh-based simulation. Even though the achieved speedups for FPS were not as high, speedups of more than 250 were achieved for specific parts of the method. Additionally, different techniques, such as reducing the neighbor search space, were used to reduce the computational complexity of the model.
In spite of the advantages of the method, there were several concerns that had to be worked around. The first was that some of the values, specifically the parameters for the elasticity, viscosity, and stiffness of the tissue, were obtained experimentally. Then, for the GPU version, the use of expensive arithmetic operations, such as divisions, hindered the performance of the simulation. Additionally, the algorithm had branching paths that were mostly idle, which also caused the performance to be lower.
Next, the model created visual artifacts in the simulation: when the cell size or the core radius were larger than 0.08, the particles would clump up at different points of the tissue, instead of returning to their original position. The clumps of particles was also the cause for the reconstructed mesh to have holes. By analyzing the particles’ properties in those areas, it could be seen that they were activated more than in other areas.
Finally, the accuracy and stability of the model could be considerably improved. To improve the accuracy of the simulation, the bidomain model could be used instead of the monodomain model. Different versions of SPH have been developed to address these issues [
75], and such modifications could also be considered to improve the model.
Future Work
Even though a novel muscle model was presented, there is still work to be performed before it can really be used to replace a muscle in a simulation. Some of the areas of opportunity include: using the bidomain model instead of the monodomain model for increased precision; fine tune the parameters to increase stability; redesign the GPU implementation to avoid thread divergence. Additionally, this work serves as the base for further muscle simulations; for example, using a GPU cluster to simulate, in real time, all the interacting tissues of the arm and forearm, including bone or ligaments, and even simulate a specific movement of the arm. Finally, force production also has to be considered so that the model can be compared to other established models. Currently, the model was focused on deformation and internal pressure, but force production is essential for muscle and joint movement, and will be considered in future iterations of the model.
Author Contributions
Conceptualization, O.N.-H. and M.A.-M.; methodology, O.N.-H. and M.A.-M.; software, O.N.-H.; validation, O.N.-H.; investigation, O.N.-H.; writing—original draft preparation, O.N.-H.; writing—review and editing, O.N.-H. and M.A.-M.; supervision, M.A.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by the 2015 Google Faculty Research Awards and Tides Foundation under grant number TFR15-00145, and by the Consejo Nacional de Ciencia y Tecnología (CONACYT) under grant number 342,814.
Acknowledgments
The authors would like to thank the Tecnológico de Monterrey IT & Computer Department for its support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Payan, Y. Soft Tissue Biomechanical Modeling for Computer Assisted Surgery; Springer: Berlin, Germany, 2012; Volume 11. [Google Scholar]
- Famaey, N.; Vander Sloten, J.; Kuhl, E. A three-constituent damage model for arterial clamping in computer-assisted surgery. Biomech. Model. Mechanobiol. 2013, 12, 123–136. [Google Scholar] [CrossRef]
- Ji, S.; Fan, X.; Hartov, A.; Roberts, D.W.; Paulsen, K.D. Estimation of intraoperative brain deformation. In Soft Tissue Biomechanical Modeling for Computer Assisted Surgery; Springer: Berlin, Germany, 2012; pp. 97–133. [Google Scholar]
- Salcudean, S.E.; Sahebjavaher, R.S.; Goksel, O.; Baghani, A.; Mahdavi, S.S.; Nir, G.; Sinkus, R.; Moradi, M. Biomechanical modeling of the prostate for procedure guidance and simulation. In Soft Tissue Biomechanical Modeling for Computer Assisted Surgery; Springer: Berlin, Germany, 2012; pp. 169–198. [Google Scholar]
- Maier-Hein, L.; Mountney, P.; Bartoli, A.; Elhawary, H.; Elson, D.; Groch, A.; Kolb, A.; Rodrigues, M.; Sorger, J.; Speidel, S.; et al. Optical techniques for 3D surface reconstruction in computer-assisted laparoscopic surgery. Med. Image Anal. 2013, 17, 974–996. [Google Scholar] [CrossRef]
- Yucesoy, C.A.; Koopman, B.H.; Huijing, P.A.; Grootenboer, H.J. Three-dimensional finite element modeling of skeletal muscle using a two-domain approach: Linked fiber-matrix mesh model. J. Biomech. 2002, 35, 1253–1262. [Google Scholar] [CrossRef]
- Blemker, S.S.; Delp, S.L. Three-dimensional representation of complex muscle architectures and geometries. Ann. Biomed. Eng. 2005, 33, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Courtecuisse, H.; Jung, H.; Allard, J.; Duriez, C.; Lee, D.Y.; Cotin, S. GPU-based real-time soft tissue deformation with cutting and haptic feedback. Prog. Biophys. Mol. Biol. 2010, 103, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Spyrou, L.A.; Aravas, N. Muscle and tendon tissues: Constitutive modeling and computational issues. J. Appl. Mech. 2011, 78, 041015. [Google Scholar] [CrossRef]
- Röhrle, O.; Davidson, J.B.; Pullan, A.J. A physiologically based, multi-scale model of skeletal muscle structure and function. Front. Physiol. 2012, 3, 1–14. [Google Scholar] [CrossRef]
- Spyrou, L.; Aravas, N. Muscle-driven finite element simulation of human foot movements. Comput. Methods Biomech. Biomed. Eng. 2012, 15, 925–934. [Google Scholar] [CrossRef]
- Liu, G.R.; Gu, Y.T. An Introduction to Meshfree Methods and Their Programming; Springer Science & Business Media: Berlin, Germany, 2005. [Google Scholar]
- Doweidar, M.; Calvo, B.; Alfaro, I.; Groenenboom, P.; Doblaré, M. A comparison of implicit and explicit natural element methods in large strains problems: Application to soft biological tissues modeling. Comput. Methods Appl. Mech. Eng. 2010, 199, 1691–1700. [Google Scholar] [CrossRef]
- Zhang, G.; Wittek, A.; Joldes, G.; Jin, X.; Miller, K. A three-dimensional nonlinear meshfree algorithm for simulating mechanical responses of soft tissue. Eng. Anal. Bound. Elem. 2014, 42, 60–66. [Google Scholar] [CrossRef]
- Horton, A.; Wittek, A.; Joldes, G.R.; Miller, K. A meshless Total Lagrangian explicit dynamics algorithm for surgical simulation. Int. J. Numer. Methods Biomed. Eng. 2010, 26, 977–998. [Google Scholar] [CrossRef]
- Lee, D.; Glueck, M.; Khan, A.; Fiume, E.; Jackson, K. A survey of modeling and simulation of skeletal muscle. ACM Trans. Graph. 2010, 28, 162. [Google Scholar]
- Tsang, W.; Singh, K.; Fiume, E. Helping hand: An anatomically accurate inverse dynamics solution for unconstrained hand motion. In Proceedings of the 2005 ACM SIGGRAPH/Eurographics symposium on Computer animation, Los Angeles, CA, USA, 29–31 July 2005; pp. 319–328. [Google Scholar]
- Albrecht, I.; Haber, J.; Seidel, H.P. Construction and animation of anatomically based human hand models. In Proceedings of the 2003 ACM SIGGRAPH/Eurographics Symposium on Computer Animation, San Diego, CA, USA, 26–27 July 2003; Eurographics Association: Aire-la-Ville, Switzerland, 2003; pp. 98–109. [Google Scholar]
- Lee, S.H.; Terzopoulos, D. Heads up!: Biomechanical modeling and neuromuscular control of the neck. In ACM Transactions on Graphics (TOG); ACM: New York, NY, USA, 2006; Volume 25, pp. 1188–1198. [Google Scholar]
- Dong, F.; Clapworthy, G.J.; Krokos, M.A.; Yao, J. An anatomy-based approach to human muscle modeling and deformation. IEEE Trans. Vis. Comput. Graph. 2002, 8, 154–170. [Google Scholar] [CrossRef]
- Waters, K. A Muscle model for animating three dimensional facial expressions. Comput. Graph. 1987, 21, 123–128. [Google Scholar] [CrossRef]
- Lee, Y.; Terzopoulos, D.; Waters, K. Realistic modeling for facial animation. In Proceedings of the 22nd Annual Conference on Computer Graphics And Interactive Techniques, Los Angeles, CA, USA, 6–11 August 1995; ACM: New York, NY, USA, 1995; pp. 55–62. [Google Scholar]
- Sifakis, E.; Neverov, I.; Fedkiw, R. Automatic Determination of Facial Muscle Activations from Sparse Motion Capture Marker Data. ACM Trans. Graph. 2005, 24, 417–425. [Google Scholar] [CrossRef]
- Lee, S.H.; Sifakis, E.; Terzopoulos, D. Comprehensive Biomechanical Modeling and Simulation of the Upper Body. ACM Trans. Graph. 2009, 28, 99:1–99:17. [Google Scholar] [CrossRef]
- Zordan, V.B.; Celly, B.; Chiu, B.; DiLorenzo, P.C. Breathe easy: Model and control of simulated respiration for animation. In Proceedings of the 2004 ACM SIGGRAPH/Eurographics Symposium on Computer Animation, Grenoble, France, 27–29 August 2004; Eurographics Association: Aire-la-Ville, Switzerland, 2004; pp. 29–37. [Google Scholar]
- Hill, A. The heat of shortening and the dynamic constants of muscle. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1938, 126, 136–195. [Google Scholar]
- Hill, A.V. First and Last Experiments in Muscle Mechanics; Cambridge University Press Cambridge: Cambridge, UK, 1970; Volume 32. [Google Scholar]
- Delp, S.L.; Anderson, F.C.; Arnold, A.S.; Loan, P.; Habib, A.; John, C.T.; Guendelman, E.; Thelen, D.G. OpenSim: Open-source software to create and analyze dynamic simulations of movement. IEEE Trans. Biomed. Eng. 2007, 54, 1940–1950. [Google Scholar] [CrossRef]
- Seth, A.; Hicks, J.L.; Uchida, T.K.; Habib, A.; Dembia, C.L.; Dunne, J.J.; Ong, C.F.; DeMers, M.S.; Rajagopal, A.; Millard, M.; et al. OpenSim: Simulating musculoskeletal dynamics and neuromuscular control to study human and animal movement. PLoS Comput. Biol. 2018, 14. [Google Scholar] [CrossRef]
- Basava, R.R. Meshfree Image-Based Reduced Order Modeling of Multiple Muscle Components with Connective Tissue and Fat. Ph.D. Thesis, University of California San Diego, San Diego, CA, USA, 2015. [Google Scholar]
- Chen, J.S.; Basava, R.R.; Zhang, Y.; Csapo, R.; Malis, V.; Sinha, U.; Hodgson, J.; Sinha, S. Pixel-based meshfree modelling of skeletal muscles. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2016, 4, 73–85. [Google Scholar] [CrossRef][Green Version]
- Valizadeh, N.; Bazilevs, Y.; Chen, J.; Rabczuk, T. A coupled IGA–Meshfree discretization of arbitrary order of accuracy and without global geometry parameterization. Comput. Methods Appl. Mech. Eng. 2015, 293, 20–37. [Google Scholar] [CrossRef]
- Liu, M.; Liu, G. Smoothed particle hydrodynamics (SPH): An overview and recent developments. Arch. Comput. Methods Eng. 2010, 17, 25–76. [Google Scholar] [CrossRef]
- Ihmsen, M.; Orthmann, J.; Solenthaler, B.; Kolb, A.; Teschner, M. SPH Fluids in Computer Graphics. In Eurographics 2014—State of the Art Reports; Lefebvre, S., Spagnuolo, M., Eds.; The Eurographics Association: Aire-la-Ville, Switzerland, 2014. [Google Scholar]
- Becker, M.; Ihmsen, M.; Teschner, M. Corotated SPH for Deformable Solids. In Eurographics Workshop on Natural Phenomena; Galin, E., Schneider, J., Eds.; The Eurographics Association: Aire-la-Ville, Switzerland, 2009; pp. 27–34. [Google Scholar]
- Zempo, Y.; Sugimoto, S. Development of the SSPH Method for Real-Space Electronic Structure Calculations. J. Phys. 2015, 640, 12–31. [Google Scholar] [CrossRef]
- Sugimoto, S.; Zempo, Y. Smoothed particle method for real-space electronic structure calculations. J. Phys. Conf. Ser. 2014, 510, 12–37. [Google Scholar] [CrossRef]
- Qin, J.; Pang, W.M.; Nguyen, B.P.; Ni, D.; Chui, C.K. Particle-based simulation of blood flow and vessel wall interactions in virtual surgery. In Proceedings of the 2010 Symposium on Information and Communication Technology, Hanoi, Vietnam, 27–28 August 2010; ACM: New York, NY, USA, 2010; pp. 128–133. [Google Scholar]
- Chui, Y.P.; Heng, P.A. A particle-based modeling framework for thrombo-emboli simulation. In Proceedings of the 11th ACM SIGGRAPH International Conference on Virtual-Reality Continuum and its Applications in Industry, Singapore, 2–4 December 2012; ACM: New York, NY, USA, 2012; pp. 213–222. [Google Scholar]
- Farazi, M.R.; Martin-Harris, B.; Harandi, N.M.; Fels, S.; Abugharbieh, R. A 3D dynamic biomechanical swallowing model for training and diagnosis of dysphagia. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn Bridge, NY, USA, 16–19 April 2015; pp. 1385–1388. [Google Scholar]
- Hieber, S.E.; Walther, J.H.; Koumoutsakos, P. Remeshed smoothed particle hydrodynamics simulation of the mechanical behavior of human organs. Technol. Health Care 2004, 12, 305–314. [Google Scholar] [CrossRef]
- Gastelum, A.; Krueger, M.; Marquez, J.; Gimel’farb, G.; Delmas, P. Automatic 3D lip shape segmentation and modelling. In Proceedings of the 2008 23rd International Conference Image and Vision Computing, Christchurch, New Zealand, 26–28 November 2008; pp. 1–6. [Google Scholar]
- Boyer, P.; Joslin, C. A Smoothed Particle Hydrodynamics Approach to Simulation of Articular Cartilage. Am. J. Biomed. Eng. 2014, 4, 41–52. [Google Scholar]
- Rausch, M.; Karniadakis, G.; Humphrey, J. Modelling soft tissue damage and failure using a combined particle/continuum approach. Biomech. Model. Mechanobiol. 2016, 16, 249–261. [Google Scholar] [CrossRef]
- Gastelum, A.; Mosso, J.L.; Delmas, P.; Marquez, J. A mesh-free mechanical model of the upper gastrointestinal system. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 555–558. [Google Scholar]
- Palyanov, A.; Khayrulin, S.; Larson, S.D. Application of smoothed particle hydrodynamics to modeling mechanisms of biological tissue. Adv. Eng. Softw. 2016, 98, 1–11. [Google Scholar] [CrossRef]
- Müller, M.; Heidelberger, B.; Teschner, M.; Gross, M. Meshless deformations based on shape matching. ACM Trans. Graph. (TOG) 2005, 24, 471–478. [Google Scholar] [CrossRef]
- Keener, J.; Sneyd, J. Mathematical Physiology, 2nd ed.; Springer: Berlin, Germany, 2009. [Google Scholar]
- Takahashi, T.; Fujishiro, I.; Nishita, T. A velocity correcting method for volume preserving viscoelastic fluids. In Proceedings of the Computer Graphics International, Sydney, Australia, 10–13 June 2014. [Google Scholar]
- Durfee, W.; Sun, Z.; Van de Ven, J. Fluid Power System Dynamics; Center for Compact and Efficient Fluid Power: Minneapolis, MN, USA, 2009. [Google Scholar]
- Abdi, M.; Karimi, A.; Navidbakhsh, M.; Rahmati, M.; Hassani, K.; Razmkon, A. Modeling the circle of willis using electrical analogy method under both normal and pathological circumstances. J. Biomed. Phys. Eng. 2013, 3, 45. [Google Scholar]
- Oh, K.W.; Lee, K.; Ahn, B.; Furlani, E.P. Design of pressure-driven microfluidic networks using electric circuit analogy. Lab Chip 2012, 12, 515–545. [Google Scholar] [CrossRef]
- Müller, M.; Charypar, D.; Gross, M. Particle-based fluid simulation for interactive applications. In Proceedings of the 2003 ACM SIGGRAPH/Eurographics symposium on Computer animation, San Diego, CA, USA, 26–27 July 2003; Eurographics Association: Aire-la-Ville, Switzerland, 2003; pp. 154–159. [Google Scholar]
- Pozrikidis, C. Numerical Computation in Science and Engineering; Oxford University Press: Oxford, UK, 1998; Volume 6. [Google Scholar]
- Monaghan, J.J. Smoothed particle hydrodynamics. Annu. Rev. Astron. Astrophys. 1992, 30, 543–574. [Google Scholar] [CrossRef]
- Desbrun, M.; Cani, M.P. Smoothed particles: A new paradigm for animating highly deformable bodies. In Proceedings of the Eurographics Workshop on Computer Animation And Simulation, Poitiers, France, 31 August–1 September 1996; Springer: Berlin, Germany, 1996; Volume 96, pp. 61–76. [Google Scholar]
- Tung, L. A bi-domain model for describing ischemic myocardial dc potentials. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 1978. [Google Scholar]
- Sundnes, J.; Nielsen, B.F.; Mardal, K.A.; Cai, X.; Lines, G.T.; Tveito, A. On the computational complexity of the bidomain and the monodomain models of electrophysiology. Ann. Biomed. Eng. 2006, 34, 1088–1097. [Google Scholar] [CrossRef]
- Nickerson, D.P. Cardiac electro-mechanics: From CellML to the whole heart. Ph.D. Thesis, ResearchSpace@ Auckland, Auckland, New Zealand, 2005. [Google Scholar]
- Ward, S.R.; Lieber, R.L. Density and hydration of fresh and fixed human skeletal muscle. J. Biomech. 2005, 38, 2317–2320. [Google Scholar] [CrossRef]
- Payne, R.C.; Crompton, R.H.; Isler, K.; Savage, R.; Vereecke, E.E.; Günther, M.M.; Thorpe, S.; D’Août, K. Morphological analysis of the hindlimb in apes and humans. I. Muscle architecture. J. Anat. 2006, 208, 709–724. [Google Scholar]
- Ward, S.R.; Eng, C.M.; Smallwood, L.H.; Lieber, R.L. Are current measurements of lower extremity muscle architecture accurate? Clin. Orthop. Relat. Res. 2009, 467, 1074–1082. [Google Scholar] [CrossRef]
- Méndez, J. Density and composition of mammalian muscle. Metabolism 1960, 9, 184–188. [Google Scholar]
- Uchiyama, T.; Saito, K. Stiffness and viscosity of the vastus lateralis muscle in cycling exercises at low constant power output. Adv. Biomed. Eng. 2018, 7, 124–130. [Google Scholar] [CrossRef]
- Corre, S.; Belmiloudi, A. Coupled Lattice Boltzmann Modeling of Bidomain Type Models in Cardiac Electrophysiology. In Mathematical and Computational Approaches in Advancing Modern Science and Engineering; Springer: Berlin, Germany, 2016; pp. 209–221. [Google Scholar]
- FitzHugh, R. Impulses and physiological states in theoretical models of nerve membrane. Biophys. J. 1961, 1, 445–466. [Google Scholar] [CrossRef]
- Nagumo, J.; Arimoto, S.; Yoshizawa, S. An active pulse transmission line simulating nerve axon. Proc. IRE 1962, 50, 2061–2070. [Google Scholar] [CrossRef]
- Hérault, A.; Bilotta, G.; Vicari, A.; Rustico, E.; Del Negro, C. Numerical simulation of lava flow using a GPU SPH model. Ann. Geophys. 2011, 54. [Google Scholar] [CrossRef]
- Green, S. Cuda particles. Nvidia Whitepaper 2008. Available online: http://developer.download.nvidia.com/compute/cuda/2_2/sdk/website/projects/particles/doc/particles.pdf (accessed on 3 March 2019).
- Gao, X.; Wang, Z.; Wan, H.; Long, X. Accelerate Smoothed Particle Hydrodynamics Using GPU. In Proceedings of the 2010 IEEE Youth Conference on Information, Computing and Telecommunications, Beijing, China, 28–30 November 2010; pp. 399–402. [Google Scholar]
- Mitsuhashi, N.; Fujieda, K.; Tamura, T.; Kawamoto, S.; Takagi, T.; Okubo, K. BodyParts3D: 3D structure database for anatomical concepts. Nucleic Acids Res. 2008, 37, D782–D785. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.A.; Ateshian, G.A.; Weiss, J.A. FEBio: History and advances. Annu. Rev. Biomed. Eng. 2017, 19, 279–299. [Google Scholar] [CrossRef]
- Murray, W.M.; Buchanan, T.S.; Delp, S.L. The isometric functional capacity of muscles that cross the elbow. J. Biomech. 2000, 33, 943–952. [Google Scholar] [CrossRef]
- Bradley, C.; Bowery, A.; Britten, R.; Budelmann, V.; Camara, O.; Christie, R.; Cookson, A.; Frangi, A.F.; Gamage, T.B.; Heidlauf, T.; et al. OpenCMISS: A multi-physics & multi-scale computational infrastructure for the VPH/Physiome project. Prog. Biophys. Mol. Biol. 2011, 107, 32–47. [Google Scholar]
- Shadloo, M.; Oger, G.; Le Touzé, D. Smoothed particle hydrodynamics method for fluid flows, towards industrial applications: Motivations, current state, and challenges. Comput. Fluids 2016, 136, 11–34. [Google Scholar] [CrossRef]
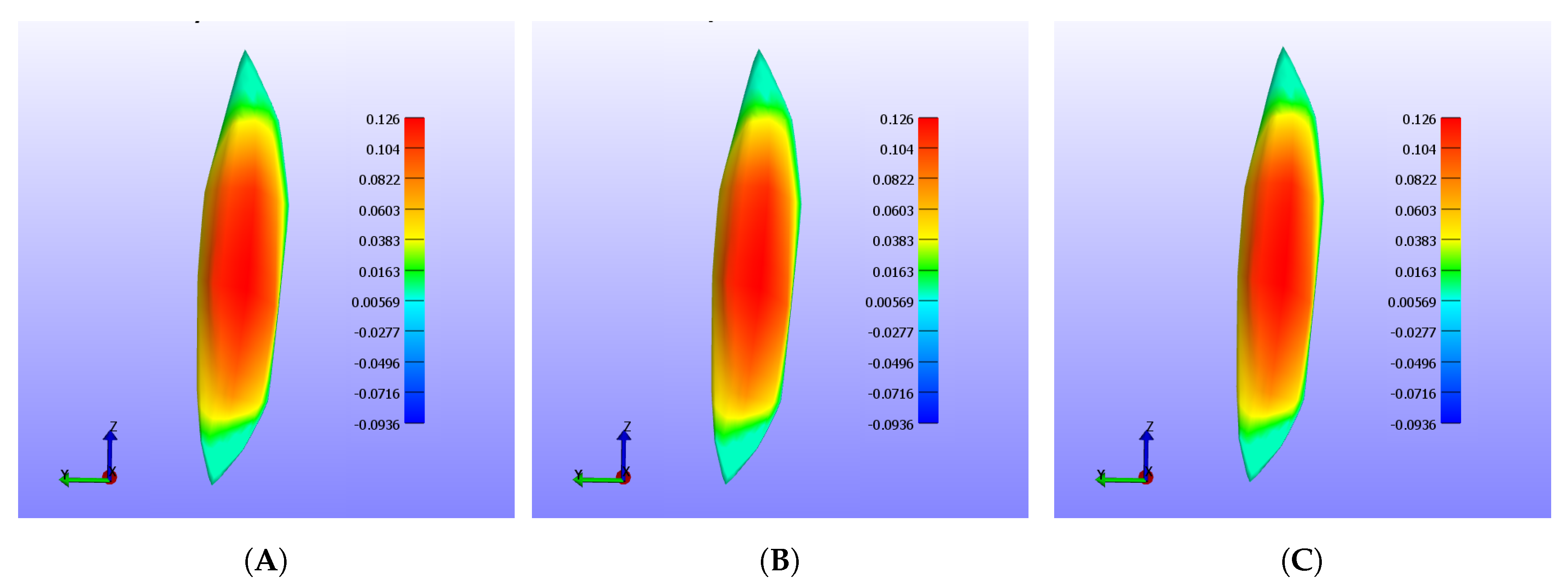
Figure 1.
Integrated point-based tissue model for the triceps. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated with a stimulus current, and (C) shows the muscle returning to its initial shape after the current was removed. The color of the particles represents its displacement with respect to its original position.
Figure 1.
Integrated point-based tissue model for the triceps. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated with a stimulus current, and (C) shows the muscle returning to its initial shape after the current was removed. The color of the particles represents its displacement with respect to its original position.
Figure 2.
Integrated point-based tissue model for the vastus lateralis. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated with a stimulus current, and (C) shows the muscle returning to its initial shape after the current was removed. The color of the particles represents its displacement with respect to its original position.
Figure 2.
Integrated point-based tissue model for the vastus lateralis. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated with a stimulus current, and (C) shows the muscle returning to its initial shape after the current was removed. The color of the particles represents its displacement with respect to its original position.
Figure 3.
Tetrahedral elements for the triceps and the vastus lateralis. (A) shows the tetrahedral elements for the triceps, while (B) shows the tetrahedral elements for the vastus lateralis.
Figure 3.
Tetrahedral elements for the triceps and the vastus lateralis. (A) shows the tetrahedral elements for the triceps, while (B) shows the tetrahedral elements for the vastus lateralis.
Figure 4.
FEM based tissue model for the triceps. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated, and (C) shows the muscle after the current is removed.
Figure 4.
FEM based tissue model for the triceps. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated, and (C) shows the muscle after the current is removed.
Figure 5.
FEM based tissue model for the vastus lateralis. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated, and (C) shows the muscle after the current is removed.
Figure 5.
FEM based tissue model for the vastus lateralis. (A) shows the initial state of the tissue, (B) shows the muscle after being innervated, and (C) shows the muscle after the current is removed.
Figure 6.
Average error for all the FEM resolutions for the triceps. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 6.
Average error for all the FEM resolutions for the triceps. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 7.
Average error for all the FEM resolutions for the vastus lateralis. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 7.
Average error for all the FEM resolutions for the vastus lateralis. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 8.
Average error for all the FEM resolutions for different regions for the triceps. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 8.
Average error for all the FEM resolutions for different regions for the triceps. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 9.
Average error for all the FEM resolutions for different regions for the vastus lateralis. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 9.
Average error for all the FEM resolutions for different regions for the vastus lateralis. (A) shows the average displacement error for all particle resolutions. (B) shows the average pressure error for all particle resolutions.
Figure 10.
Mean displacement of the triceps. (A) shows the mean displacement for the contraction step, (B) shows the mean displacement for the expansion step.
Figure 10.
Mean displacement of the triceps. (A) shows the mean displacement for the contraction step, (B) shows the mean displacement for the expansion step.
Figure 11.
Mean displacement of the vastus lateralis. (A) shows the mean displacement for the contraction step, (B) shows the mean displacement for the expansion step.
Figure 11.
Mean displacement of the vastus lateralis. (A) shows the mean displacement for the contraction step, (B) shows the mean displacement for the expansion step.
Figure 12.
Average times for the muscle tissues with 18,475 particles. (A) shows the average time for the triceps, (B) shows the average times for the vastus lateralis.
Figure 12.
Average times for the muscle tissues with 18,475 particles. (A) shows the average time for the triceps, (B) shows the average times for the vastus lateralis.
Figure 13.
Average speedups for the muscle geometry with 18475 particles. (A) shows the average speedups for the triceps, (B) shows the average speedups for the vastus lateralis.
Figure 13.
Average speedups for the muscle geometry with 18475 particles. (A) shows the average speedups for the triceps, (B) shows the average speedups for the vastus lateralis.
Figure 14.
Average speedups obtained for each kernel and each particle set. (A) shows the average speedups for the triceps, (B) shows the average speedups for the vastus lateralis.
Figure 14.
Average speedups obtained for each kernel and each particle set. (A) shows the average speedups for the triceps, (B) shows the average speedups for the vastus lateralis.
Figure 15.
Average speedups for smoothed particle hydrodynamics (SPH) kernels when compared to FEM using CPU.
Figure 15.
Average speedups for smoothed particle hydrodynamics (SPH) kernels when compared to FEM using CPU.
Table 1.
Values for the cell model, and the monodomain equations.
Table 1.
Values for the cell model, and the monodomain equations.
| Variable | Value | Description |
|---|
| 50 mm | Surface-to-volume ratio |
| 0.01 | Membrane capacitance |
| 0.893 mSmm | Internal fiber conductivity |
| 0.67 mSmm | External fiber conductivity |
| 8000 A/mm | External stimulus current |
| −85.0 mV | Resting Potential |
| 15.0 mV | Plateau Potential |
| −75.0 mV | Threshold Potential |
| 0.175 A mm | Excitation rate constant |
| 0.03 A mm | Excitation decay constant |
| b | 0.011 ms | Recovery rate constant |
| d | 0.55 ms | Recovery decay constant |
Table 2.
Specification for the computer where the experiments were conducted.
Table 2.
Specification for the computer where the experiments were conducted.
| Component | Specification |
|---|
| Processor | 12x Intel(R) Core(TM) i7-5820K CPU @ 3.30GHz |
| Memory | 16 GB |
| Operating System | Ubuntu 16.04.3 LTS |
| GPU | GeForce GTX TITANX 3072 CUDA Cores @1.08 GHz, 12GB Memory |
Table 3.
Properties used for the material model solved with the Finite Element Method (FEM).
Table 3.
Properties used for the material model solved with the Finite Element Method (FEM).
| Property | Value | Description |
|---|
| Density | 1112 kg/cm | Density of the tissue |
| 500 Pa | Fiber shear modulus |
| 500 Pa | Cross shear modulus |
| K | 1 × 10 Pa | Bulk modulus |
| 0.05 | Exponential stress coefficients |
| Lofl | 10.7 cm | Optimal fiber length |
| max | 3 × 10 Pa | Maximum isometric stress |
| 1 | Activation level |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).